Dual Role of p73 in Cancer Microenvironment and DNA Damage Response
Abstract
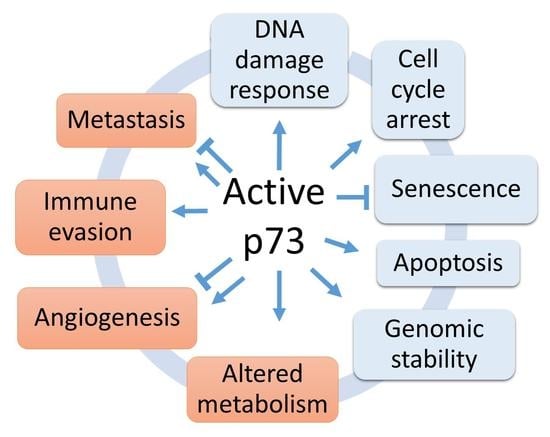
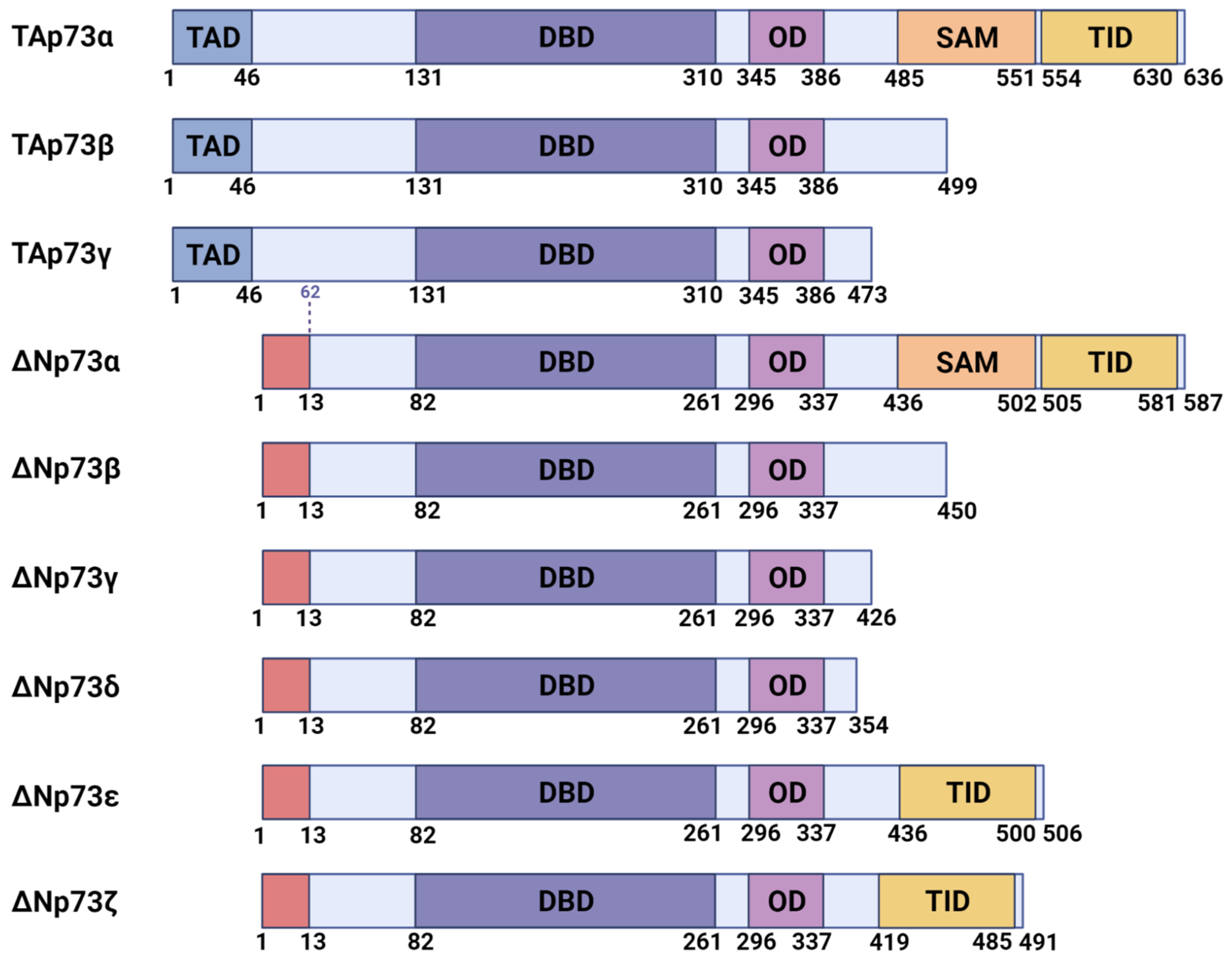
1. Introduction
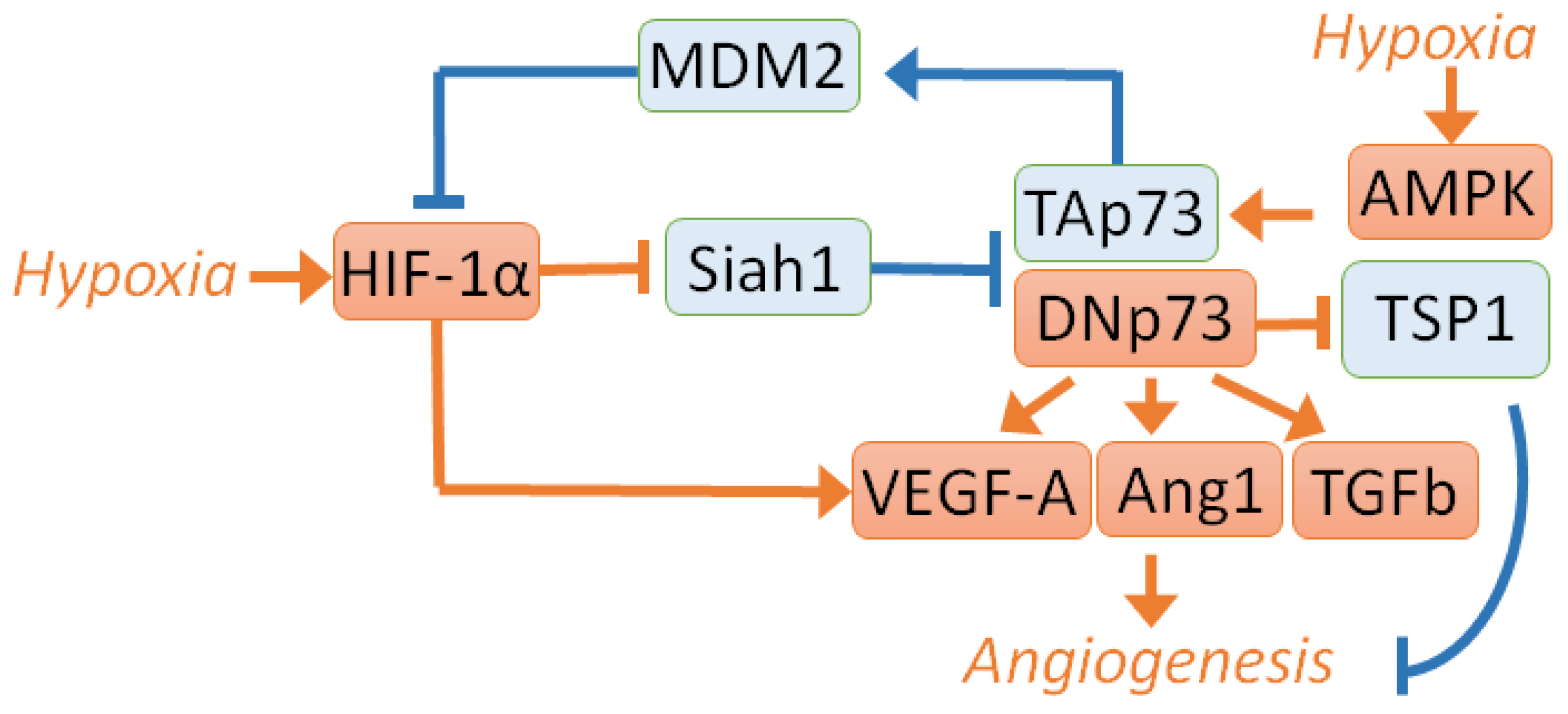
2. Angiogenesis Induction
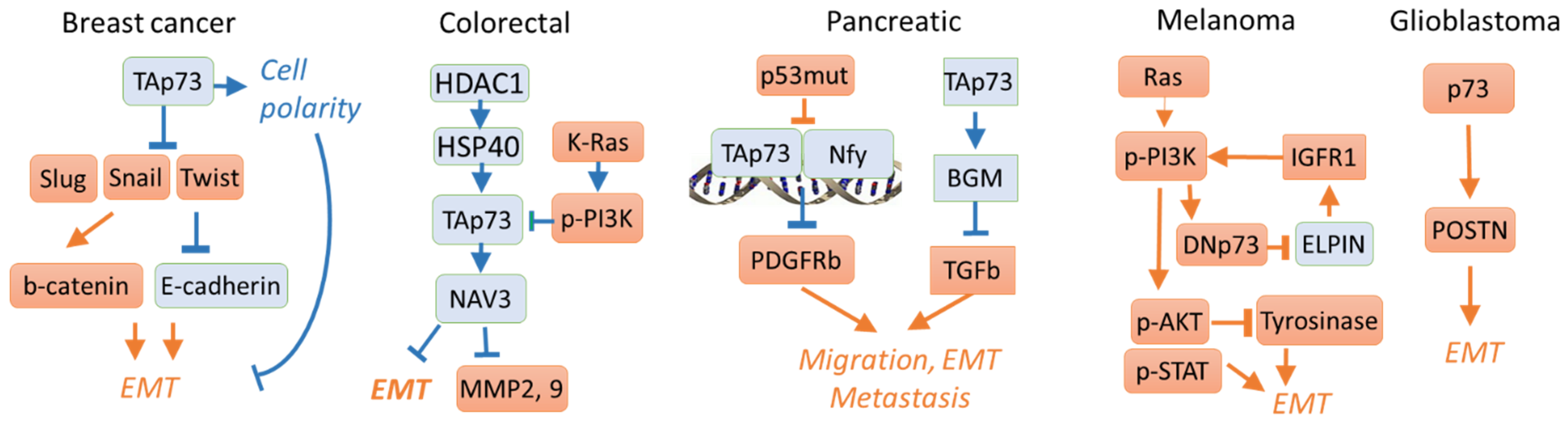
3. Activation of Invasion and Metastasis
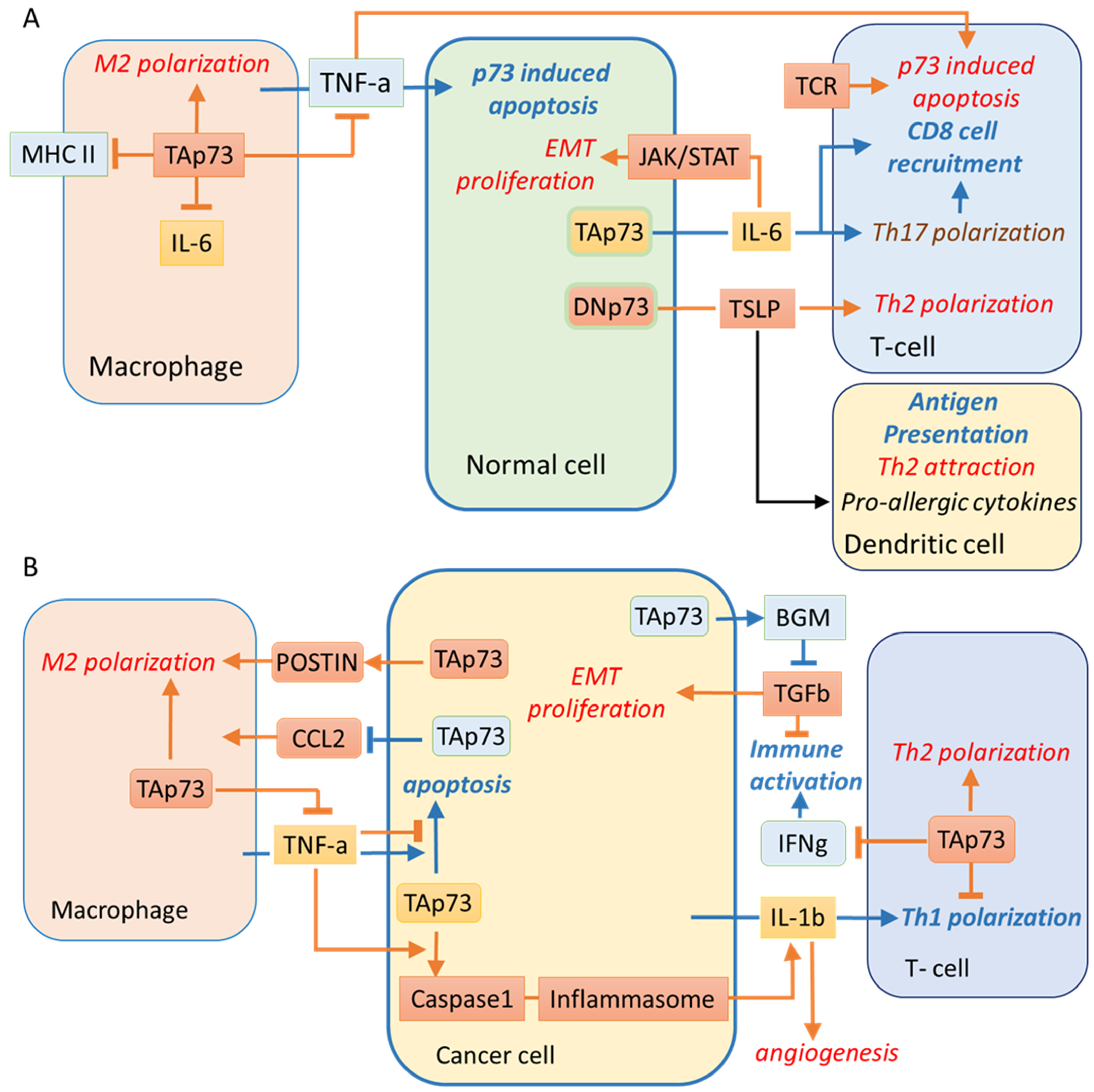
4. Tumour-Promoting Inflammation and Evading Immune Destruction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Levine, A.J.; Tomasini, R.; McKeon, F.D.; Mak, T.W.; Melino, G. The p53 family: Guardians of maternal reproduction. Nat. Rev. Mol. Cell Biol. 2011, 12, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Bernassola, F.; Federici, M.; Corazzari, M.; Terrinoni, A.; Hribal, M.L.; De Laurenzi, V.; Ranalli, M.; Massa, O.; Sesti, G.; McLean, W.H.I.; et al. Role of transglutaminase 2 in glucose tolerance: Knockout mice studies and a putative mutation in a MODY patient. FASEB J. 2002, 16, 1371–1378. [Google Scholar] [CrossRef]
- Bellomaria, A.; Barbato, G.; Melino, G.; Paci, M.; Melino, S. Recognition of p63 by the E3 ligase ITCH: Effect of an ectodermal dysplasia mutant. Cell Cycle 2010, 9, 3730–3739. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Cipollone, R.; Rivetti di Val Cervo, P.; Gonfloni, S.; Melino, G.; Knight, R. p63 in epithelial development. Cell Mol. Life Sci. 2008, 65, 3126–3133. [Google Scholar] [CrossRef]
- Candi, E.; Terrinoni, A.; Rufini, A.; Chikh, A.; Lena, A.M.; Suzuki, Y.; Sayan, B.S.; Knight, R.A.; Melino, G. p63 is upstream of IKK alpha in epidermal development. J. Cell Sci. 2006, 119, 4617–4622. [Google Scholar] [CrossRef]
- Shalom-Feuerstein, R.; Lena, A.M.; Zhou, H.; De La Forest Divonne, S.; Van Bokhoven, H.; Candi, E.; Melino, G.; Aberdam, D. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011, 18, 887–896. [Google Scholar] [CrossRef]
- Guilluy, C.; Rolli-Derkinderen, M.; Tharaux, P.-L.; Melino, G.; Pacaud, P.; Loirand, G. Transglutaminase-dependent RhoA activation and depletion by serotonin in vascular smooth muscle cells. J. Biol. Chem. 2007, 282, 2918–2928. [Google Scholar] [CrossRef]
- Jones, R.A.; Kotsakis, P.; Johnson, T.S.; Chau, D.Y.S.; Ali, S.; Melino, G.; Griffin, M. Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ. 2006, 13, 1442–1453. [Google Scholar] [CrossRef]
- Piredda, L.; Farrace, M.G.; Lo Bello, M.; Malorni, W.; Melino, G.; Petruzzelli, R.; Piacentini, M. Identification of “tissue” transglutaminase binding proteins in neural cells committed to apoptosis. FASEB J. 1999, 13, 355–364. [Google Scholar] [CrossRef]
- Popov, Y.; Sverdlov, D.Y.; Sharma, A.K.; Bhaskar, K.R.; Li, S.; Freitag, T.L.; Lee, J.; Dieterich, W.; Melino, G.; Schuppan, D. Tissue transglutaminase does not affect fibrotic matrix stability or regression of liver fibrosis in mice. Gastroenterology 2011, 140, 1642–1652. [Google Scholar] [CrossRef]
- Shweke, N.; Boulos, N.; Jouanneau, C.; Vandermeersch, S.; Melino, G.; Dussaule, J.-C.; Chatziantoniou, C.; Ronco, P.; Boffa, J.-J. Tissue transglutaminase contributes to interstitial renal fibrosis by favoring accumulation of fibrillar collagen through TGF-beta activation and cell infiltration. Am. J. Pathol. 2008, 173, 631–642. [Google Scholar] [CrossRef]
- Candi, E.; Oddi, S.; Terrinoni, A.; Paradisi, A.; Ranalli, M.; Finazzi-Agró, A.; Melino, G. Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J. Biol. Chem. 2001, 276, 35014–35023. [Google Scholar] [CrossRef]
- Cassidy, A.J.; van Steensel, M.A.M.; Steijlen, P.M.; van Geel, M.; van der Velden, J.; Morley, S.M.; Terrinoni, A.; Melino, G.; Candi, E.; McLean, W.H.I. A homozygous missense mutation in TGM5 abolishes epidermal transglutaminase 5 activity and causes acral peeling skin syndrome. Am. J. Hum. Genet. 2005, 77, 909–917. [Google Scholar] [CrossRef]
- Ikawa, S.; Nakagawara, A.; Ikawa, Y. p53 family genes: Structural comparison, expression and mutation. Cell Death Differ. 1999, 6, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Alonso, M.E.; Bello, M.J.; Lomas, J.; Gonzalez-Gomez, P.; Arjona, D.; De Campos, J.M.; Gutierrez, M.; Isla, A.; Vaquero, J.; Rey, J.A. Absence of mutation of the p73 gene in astrocytic neoplasms. Int. J. Oncol. 2001, 19, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Zaika, A.I.; Kovalev, S.; Marchenko, N.D.; Moll, U.M. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999, 59, 3257–3263. [Google Scholar] [PubMed]
- Yokozaki, H.; Shitara, Y.; Fujimoto, J.; Hiyama, T.; Yasui, W.; Tahara, E. Alterations of p73 preferentially occur in gastric adenocarcinomas with foveolar epithelial phenotype. Int. J. Cancer 1999, 83, 192–196. [Google Scholar] [CrossRef]
- Yasui, W.; Yokozaki, H.; Fujimoto, J.; Naka, K.; Kuniyasu, H.; Tahara, E. Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. J. Gastroenterol. 2000, 35 (Suppl. 2), 111–115. [Google Scholar]
- Ganini, C.; Amelio, I.; Bertolo, R.; Bove, P.; Buonomo, O.C.; Candi, E.; Cipriani, C.; Di Daniele, N.; Juhl, H.; Mauriello, A.; et al. Global mapping of cancers: The Cancer Genome Atlas and beyond. Mol. Oncol. 2021, 15, 2823–2840. [Google Scholar] [CrossRef]
- He, Y.; Fan, S.; Jiang, Y.; Xue, Z. Expression of ΔNp73 in human NSCLC and clinical implication. Zhongguo Fei Ai Za Zhi 2006, 9, 263–266. [Google Scholar]
- Dominguez, G.; Silva, J.M.; Silva, J.; Garcia, J.M.; Sanchez, A.; Navarro, A.; Gallego, I.; Provencio, M.; España, P.; Bonilla, F. Wild type p73 overexpression and high-grade malignancy in breast cancer. Breast Cancer Res. Treat. 2001, 66, 183–190. [Google Scholar] [CrossRef]
- Tomkova, K.; Belkhiri, A.; El-Rifai, W.; Zaika, A.I. p73 isoforms can induce T-cell factor-dependent transcription in gastrointestinal cells. Cancer Res. 2004, 64, 6390–6393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kamiya, M.; Nakazato, Y. The expression of p73, p21 and MDM2 proteins in gliomas. J. Neurooncol. 2002, 59, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Wager, M.; Guilhot, J.; Blanc, J.L.; Ferrand, S.; Milin, S.; Bataille, B.; Lapierre, F.; Denis, S.; Chantereau, T.; Larsen, C.J.; et al. Prognostic value of increase in transcript levels of Tp73 DeltaEx2-3 isoforms in low-grade glioma patients. Br. J. Cancer 2006, 95, 1062–1069. [Google Scholar] [CrossRef]
- Ugur, H.; Sayan, A.E.; Ozdamar, S.O.; Kanpolat, Y.; Ozturk, M. Expression of TAP73 and DeltaNP73 in malignant gliomas. Oncol. Rep. 2004, 11, 1337–1341. [Google Scholar]
- Inoue, K.; Fry, E.A. Alterations of p63 and p73 in human cancers. Subcell. Biochem. 2014, 85, 17–40. [Google Scholar]
- Engelmann, D.; Meier, C.; Alla, V.; Pützer, B.M. A balancing act: Orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 2015, 34, 4287–4299. [Google Scholar] [CrossRef]
- Jost, C.A.; Marin, M.C.; Kaelin, W.G. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 1997, 389, 191–194. [Google Scholar] [CrossRef]
- Kaghad, M.; Bonnet, H.; Yang, A.; Creancier, L.; Biscan, J.C.; Valent, A.; Minty, A.; Chalon, P.; Lelias, J.M.; Dumont, X.; et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90, 809–819. [Google Scholar] [CrossRef]
- Zeng, X.; Li, X.; Miller, A.; Yuan, Z.; Yuan, W.; Kwok, R.P.; Goodman, R.; Lu, H. The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell. Biol. 2000, 20, 1299–1310. [Google Scholar] [CrossRef]
- Tomasini, R.; Tsuchihara, K.; Tsuda, C.; Lau, S.K.; Wilhelm, M.; Rufini, A.; Tsao, M.; Iovanna, J.L.; Jurisicova, A.; Melino, G.; et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc. Natl. Acad. Sci. USA 2009, 106, 797–802. [Google Scholar] [CrossRef]
- Melino, G.; Knight, R.A.; Nicotera, P. How many ways to die? How many different models of cell death? Cell Death Differ. 2005, 12 (Suppl. 2), 1457–1462. [Google Scholar] [CrossRef][Green Version]
- Nepravishta, R.; Sabelli, R.; Iorio, E.; Micheli, L.; Paci, M.; Melino, S. Oxidative species and S-glutathionyl conjugates in the apoptosis induction by allyl thiosulfate. FEBS J. 2012, 279, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Fazi, B.; Melino, S.; De Rubeis, S.; Bagni, C.; Paci, M.; Piacentini, M.; Di Sano, F. Acetylation of RTN-1C regulates the induction of ER stress by the inhibition of HDAC activity in neuroectodermal tumors. Oncogene 2009, 28, 3814–3824. [Google Scholar] [CrossRef]
- Sabelli, R.; Iorio, E.; De Martino, A.; Podo, F.; Ricci, A.; Viticchiè, G.; Rotilio, G.; Paci, M.; Melino, S. Rhodanese-thioredoxin system and allyl sulfur compounds. FEBS J. 2008, 275, 3884–3899. [Google Scholar] [CrossRef]
- Rufini, A.; Niklison-Chirou, M.V.; Inoue, S.; Tomasini, R.; Harris, I.S.; Marino, A.; Federici, M.; Dinsdale, D.; Knight, R.A.; Melino, G.; et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012, 26, 2009–2014. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A. p73, a sophisticated p53 family member in the cancer world. Cancer Sci. 2005, 96, 729–737. [Google Scholar] [CrossRef]
- Tebbi, A.; Guittet, O.; Cottet, M.-H.; Vesin, M.-F.; Lepoivre, M. TAp73 induction by nitric oxide: Regulation by checkpoint kinase 1 (CHK1) and protection against apoptosis. J. Biol. Chem. 2011, 286, 7873–7884. [Google Scholar] [CrossRef] [PubMed]
- Vossio, S.; Palescandolo, E.; Pediconi, N.; Moretti, F.; Balsano, C.; Levrero, M.; Costanzo, A. DN-p73 is activated after DNA damage in a p53-dependent manner to regulate p53-induced cell cycle arrest. Oncogene 2002, 21, 3796–3803. [Google Scholar] [CrossRef][Green Version]
- Liu, G.; Nozell, S.; Xiao, H.; Chen, X. DeltaNp73beta is active in transactivation and growth suppression. Mol. Cell. Biol. 2004, 24, 487–501. [Google Scholar] [CrossRef]
- Beeler, J.S.; Marshall, C.B.; Gonzalez-Ericsson, P.I.; Shaver, T.M.; Santos Guasch, G.L.; Lea, S.T.; Johnson, K.N.; Jin, H.; Venters, B.J.; Sanders, M.E.; et al. p73 regulates epidermal wound healing and induced keratinocyte programming. PLoS ONE 2019, 14, e0218458. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Carrera, D.; García-Puga, M.; Yáñez, L.; Romón, Í.; Pipaón, C. ∆Np73 is capable of inducing apoptosis by co-ordinately activating several BH3-only proteins. Biosci. Rep. 2015, 35, e00198. [Google Scholar] [CrossRef] [PubMed]
- Nyman, U.; Vlachos, P.; Cascante, A.; Hermanson, O.; Zhivotovsky, B.; Joseph, B. Protein kinase C-dependent phosphorylation regulates the cell cycle-inhibitory function of the p73 carboxy terminus transactivation domain. Mol. Cell. Biol. 2009, 29, 1814–1825. [Google Scholar] [CrossRef] [PubMed]
- Wildung, M.; Esser, T.U.; Grausam, K.B.; Wiedwald, C.; Volceanov-Hahn, L.; Riedel, D.; Beuermann, S.; Li, L.; Zylla, J.; Guenther, A.-K.; et al. Transcription factor TAp73 and microRNA-449 complement each other to support multiciliogenesis. Cell Death Differ. 2019, 26, 2740–2757. [Google Scholar] [CrossRef]
- Grob, T.J.; Novak, U.; Maisse, C.; Barcaroli, D.; Lüthi, A.U.; Pirnia, F.; Hügli, B.; Graber, H.U.; De Laurenzi, V.; Fey, M.F.; et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001, 8, 1213–1223. [Google Scholar] [CrossRef]
- Nakagawa, T.; Takahashi, M.; Ozaki, T.; Watanabe Ki, K.; Todo, S.; Mizuguchi, H.; Hayakawa, T.; Nakagawara, A. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol. Cell. Biol. 2002, 22, 2575–2585. [Google Scholar] [CrossRef]
- Kartasheva, N.N.; Contente, A.; Lenz-Stöppler, C.; Roth, J.; Dobbelstein, M. p53 induces the expression of its antagonist p73 Delta N, establishing an autoregulatory feedback loop. Oncogene 2002, 21, 4715–4727. [Google Scholar] [CrossRef]
- Stiewe, T.; Theseling, C.C.; Pützer, B.M. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: Implications for tumorigenesis. J. Biol. Chem. 2002, 277, 14177–14185. [Google Scholar] [CrossRef]
- Pozniak, C.D.; Radinovic, S.; Yang, A.; McKeon, F.; Kaplan, D.R.; Miller, F.D. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000, 289, 304–306. [Google Scholar] [CrossRef]
- Concin, N.; Becker, K.; Slade, N.; Erster, S.; Mueller-Holzner, E.; Ulmer, H.; Daxenbichler, G.; Zeimet, A.; Zeillinger, R.; Marth, C.; et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004, 64, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Takahashi, M.; Ozaki, T.; Watanabe, K.; Hayashi, S.; Hosoda, M.; Todo, S.; Nakagawara, A. Negative autoregulation of p73 and p53 by DeltaNp73 in regulating differentiation and survival of human neuroblastoma cells. Cancer Lett. 2003, 197, 105–109. [Google Scholar] [CrossRef]
- Oswald, C.; Stiewe, T. In good times and bad: p73 in cancer. Cell Cycle 2008, 7, 1726–1731. [Google Scholar] [CrossRef]
- Cam, M.; Charan, M.; Welker, A.M.; Dravid, P.; Studebaker, A.W.; Leonard, J.R.; Pierson, C.R.; Nakano, I.; Beattie, C.E.; Hwang, E.I.; et al. ΔNp73/ETS2 complex drives glioblastoma pathogenesis- targeting downstream mediators by rebastinib prolongs survival in preclinical models of glioblastoma. Neuro. Oncol. 2020, 22, 345–356. [Google Scholar] [CrossRef]
- Meier, C.; Hardtstock, P.; Joost, S.; Alla, V.; Pützer, B.M. p73 and IGF1R Regulate Emergence of Aggressive Cancer Stem-like Features via miR-885-5p Control. Cancer Res. 2016, 76, 197–205. [Google Scholar] [CrossRef]
- Soldevilla, B.; Díaz, R.; Silva, J.; Campos-Martín, Y.; Muñoz, C.; García, V.; García, J.M.; Peña, C.; Herrera, M.; Rodriguez, M.; et al. Prognostic impact of ΔTAp73 isoform levels and their target genes in colon cancer patients. Clin. Cancer Res. 2011, 17, 6029–6039. [Google Scholar] [CrossRef] [PubMed]
- Vikhreva, P.; Melino, G.; Amelio, I. p73 Alternative Splicing: Exploring a Biological Role for the C-Terminal Isoforms. J. Mol. Biol. 2018, 430, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chen, X. The C-terminal sterile alpha motif and the extreme C terminus regulate the transcriptional activity of the alpha isoform of p73. J. Biol. Chem. 2005, 280, 20111–20119. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.K.; Lee, H.J.; Jeong, M.-H.; Rhee, M.; Mo, J.-W.; Song, E.H.; Lim, J.-Y.; Choi, K.-H.; Jo, I.; Park, S.I.; et al. Role of activating transcription factor 3 on TAp73 stability and apoptosis in paclitaxel-treated cervical cancer cells. Mol. Cancer Res. 2008, 6, 1232–1249. [Google Scholar] [CrossRef]
- Vikhanskaya, F.; Toh, W.H.; Dulloo, I.; Wu, Q.; Boominathan, L.; Ng, H.H.; Vousden, K.H.; Sabapathy, K. p73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nat. Cell Biol. 2007, 9, 698–705. [Google Scholar] [CrossRef]
- Subramanian, D.; Bunjobpol, W.; Sabapathy, K. Interplay between TAp73 Protein and Selected Activator Protein-1 (AP-1) Family Members Promotes AP-1 Target Gene Activation and Cellular Growth. J. Biol. Chem. 2015, 290, 18636–18649. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Yang, X.; Duggal, P.; Allen, C.T.; Yan, B.; Cohen, J.; Nottingham, L.; Romano, R.-A.; Sinha, S.; King, K.E.; et al. TNF-α promotes c-REL/ΔNp63α interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res. 2011, 71, 6867–6877. [Google Scholar] [CrossRef]
- Koeppel, M.; van Heeringen, S.J.; Kramer, D.; Smeenk, L.; Janssen-Megens, E.; Hartmann, M.; Stunnenberg, H.G.; Lohrum, M. Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Res. 2011, 39, 6069–6085. [Google Scholar] [CrossRef]
- Si, H.; Lu, H.; Yang, X.; Mattox, A.; Jang, M.; Bian, Y.; Sano, E.; Viadiu, H.; Yan, B.; Yau, C.; et al. TNF-α modulates genome-wide redistribution of ΔNp63α/TAp73 and NF-κB cREL interactive binding on TP53 and AP-1 motifs to promote an oncogenic gene program in squamous cancer. Oncogene 2016, 35, 5781–5794. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, J.M.; Rogovaya, O.S.; Melino, G.; Barlev, N.A.; Kagansky, A. Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia. Cells 2020, 10, 25. [Google Scholar] [CrossRef]
- De Laurenzi, V.; Melino, G. Evolution of functions within the p53/p63/p73 family. Ann. N. Y. Acad. Sci. 2000, 926, 90–100. [Google Scholar] [CrossRef]
- Rozenberg, J.M.; Zvereva, S.; Dalina, A.; Blatov, I.; Zubarev, I.; Luppov, D.; Bessmertnyi, A.; Romanishin, A.; Alsoulaiman, L.; Kumeiko, V.; et al. The p53 family member p73 in the regulation of cell stress response. Biol. Direct 2021, 16, 23. [Google Scholar] [CrossRef]
- Urist, M.; Tanaka, T.; Poyurovsky, M.V.; Prives, C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004, 18, 3041–3054. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.; Raimundo, L.; Soares, J.; Loureiro, J.B.; Leão, M.; Ramos, H.; Monteiro, M.N.; Lemos, A.; Moreira, J.; Pinto, M.; et al. New inhibitor of the TAp73 interaction with MDM2 and mutant p53 with promising antitumor activity against neuroblastoma. Cancer Lett. 2019, 446, 90–102. [Google Scholar] [PubMed]
- Watson, I.R.; Blanch, A.; Lin, D.C.C.; Ohh, M.; Irwin, M.S. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J. Biol. Chem. 2006, 281, 34096–34103. [Google Scholar] [CrossRef]
- Nieto, A.; Hara, M.R.; Quereda, V.; Grant, W.; Saunders, V.; Xiao, K.; McDonald, P.H.; Duckett, D.R. βarrestin-1 regulates DNA repair by acting as an E3-ubiquitin ligase adaptor for 53BP1. Cell Death Differ. 2020, 27, 1200–1213. [Google Scholar] [CrossRef]
- Li, X.; Guo, M.; Cai, L.; Du, T.; Liu, Y.; Ding, H.-F.; Wang, H.; Zhang, J.; Chen, X.; Yan, C. Competitive ubiquitination activates the tumor suppressor p53. Cell Death Differ. 2020, 27, 1807–1818. [Google Scholar] [CrossRef]
- Rossi, M.; De Laurenzi, V.; Munarriz, E.; Green, D.R.; Liu, Y.-C.; Vousden, K.H.; Cesareni, G.; Melino, G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005, 24, 836–848. [Google Scholar] [CrossRef]
- Inoue, S.; Hao, Z.; Elia, A.J.; Cescon, D.; Zhou, L.; Silvester, J.; Snow, B.; Harris, I.S.; Sasaki, M.; Li, W.Y.; et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013, 27, 1101–1114. [Google Scholar] [CrossRef]
- Di Rita, A.; Peschiaroli, A.; Acunzo, P.D.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Rotblat, B.; Ansell, K.; Amelio, I.; Caraglia, M.; Misso, G.; Bernassola, F.; Cavasotto, C.N.; Knight, R.A.; Ciechanover, A.; et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014, 5, e1203. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, J.; Banerjee, S.; Ray, P.; Hossain, D.M.S.; Bhattacharyya, S.; Adhikary, A.; Chattopadhyay, S.; Das, T.; Sa, G. Gain of cellular adaptation due to prolonged p53 impairment leads to functional switchover from p53 to p73 during DNA damage in acute myeloid leukemia cells. J. Biol. Chem. 2010, 285, 33104–33112. [Google Scholar] [CrossRef] [PubMed]
- Long, J.S.; Schoonen, P.M.; Graczyk, D.; O’Prey, J.; Ryan, K.M. p73 engages A2B receptor signalling to prime cancer cells to chemotherapy-induced death. Oncogene 2015, 34, 5152–5162. [Google Scholar] [CrossRef]
- Kravchenko, J.E.; Ilyinskaya, G.V.; Komarov, P.G.; Agapova, L.S.; Kochetkov, D.V.; Strom, E.; Frolova, E.I.; Kovriga, I.; Gudkov, A.V.; Feinstein, E.; et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 6302–6307. [Google Scholar] [CrossRef]
- Ibrahim, N.; He, L.; Leong, C.-O.; Xing, D.; Karlan, B.Y.; Swisher, E.M.; Rueda, B.R.; Orsulic, S.; Ellisen, L.W. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer Res. 2010, 70, 7155–7165. [Google Scholar] [CrossRef]
- Gaiddon, C.; Lokshin, M.; Ahn, J.; Zhang, T.; Prives, C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 2001, 21, 1874–1887. [Google Scholar] [CrossRef]
- Strano, S.; Munarriz, E.; Rossi, M.; Cristofanelli, B.; Shaul, Y.; Castagnoli, L.; Levine, A.J.; Sacchi, A.; Cesareni, G.; Oren, M.; et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 2000, 275, 29503–29512. [Google Scholar] [CrossRef]
- Wolf, E.R.; McAtarsney, C.P.; Bredhold, K.E.; Kline, A.M.; Mayo, L.D. Mutant and wild-type p53 form complexes with p73 upon phosphorylation by the kinase JNK. Sci. Signal. 2018, 11, 524. [Google Scholar] [CrossRef]
- Gong, H.; Zhang, Y.; Jiang, K.; Ye, S.; Chen, S.; Zhang, Q.; Peng, J.; Chen, J. p73 coordinates with Δ133p53 to promote DNA double-strand break repair. Cell Death Differ. 2018, 25, 1063–1079. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.T.; Rufini, A.; Wetzel, M.K.; Tsuchihara, K.; Inoue, S.; Tomasini, R.; Itie-Youten, A.; Wakeham, A.; Arsenian-Henriksson, M.; Melino, G.; et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010, 24, 549–560. [Google Scholar] [CrossRef]
- Vikhanskaya, F.; D’Incalci, M.; Broggini, M. p73 competes with p53 and attenuates its response in a human ovarian cancer cell line. Nucleic Acids Res. 2000, 28, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.C.; Marques, M.M. Novel role of p73 as a regulator of developmental angiogenesis: Implication for cancer therapy. Mol. Cell. Oncol. 2016, 3, e1019973. [Google Scholar] [CrossRef][Green Version]
- Stantic, M.; Sakil, H.A.M.; Zirath, H.; Fang, T.; Sanz, G.; Fernandez-Woodbridge, A.; Marin, A.; Susanto, E.; Mak, T.W.; Arsenian Henriksson, M.; et al. TAp73 suppresses tumor angiogenesis through repression of proangiogenic cytokines and HIF-1α activity. Proc. Natl. Acad. Sci. USA 2015, 112, 220–225. [Google Scholar] [CrossRef]
- Uboveja, A.; Satija, Y.K.; Siraj, F.; Sharma, I.; Saluja, D. p73 NAV3 axis plays a critical role in suppression of colon cancer metastasis. Oncogenesis 2020, 9, 12. [Google Scholar] [CrossRef]
- Cui, X.-P.; Wang, C.-X.; Wang, Z.-Y.; Li, J.; Tan, Y.-W.; Gu, S.-T.; Qin, C.-K. LncRNA TP73-AS1 sponges miR-141-3p to promote the migration and invasion of pancreatic cancer cells through the up-regulation of BDH2. Biosci. Rep. 2019, 39, BSR20181937. [Google Scholar] [CrossRef]
- Bae, W.-K.; Hong, C.-S.; Park, M.-R.; Sun, E.-G.; Lee, J.-H.; Kang, K.; Ryu, K.-H.; Shim, H.-J.; Hwang, J.-E.; Cho, S.-H.; et al. TAp73 inhibits cell invasion and migration by directly activating KAI1 expression in colorectal carcinoma. Cancer Lett. 2018, 415, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Kuan, E.L.; Ziegler, S.F. A tumor-myeloid cell axis, mediated via the cytokines IL-1α and TSLP, promotes the progression of breast cancer. Nat. Immunol. 2018, 19, 366–374. [Google Scholar] [CrossRef]
- Chen, J.; Cao, X.; Li, B.; Zhao, Z.; Chen, S.; Lai, S.W.T.; Muend, S.A.; Nossa, G.K.; Wang, L.; Guo, W.; et al. Warburg effect is a cancer immune evasion mechanism against macrophage immunosurveillance. Front. Immunol. 2020, 11, 621757. [Google Scholar] [CrossRef]
- Chang, A.L.; Miska, J.; Wainwright, D.A.; Dey, M.; Rivetta, C.V.; Yu, D.; Kanojia, D.; Pituch, K.C.; Qiao, J.; Pytel, P.; et al. CCL2 Produced by the Glioma Microenvironment Is Essential for the Recruitment of Regulatory T Cells and Myeloid-Derived Suppressor Cells. Cancer Res. 2016, 76, 5671–5682. [Google Scholar] [CrossRef]
- Fortunato, O.; Belisario, D.C.; Compagno, M.; Giovinazzo, F.; Bracci, C.; Pastorino, U.; Horenstein, A.; Malavasi, F.; Ferracini, R.; Scala, S.; et al. CXCR4 inhibition counteracts immunosuppressive properties of metastatic NSCLC stem cells. Front. Immunol. 2020, 11, 02168. [Google Scholar] [CrossRef]
- Li, L.; Yu, R.; Cai, T.; Chen, Z.; Lan, M.; Zou, T.; Wang, B.; Wang, Q.; Zhao, Y.; Cai, Y. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int. Immunopharmacol. 2020, 88, 106939. [Google Scholar] [CrossRef] [PubMed]
- Wolfsberger, J.; Sakil, H.A.M.; Zhou, L.; van Bree, N.; Baldisseri, E.; de Souza Ferreira, S.; Zubillaga, V.; Stantic, M.; Fritz, N.; Hartman, J.; et al. TAp73 represses NF-κB-mediated recruitment of tumor-associated macrophages in breast cancer. Proc. Natl. Acad. Sci. USA 2021, 118, 10. [Google Scholar] [CrossRef]
- Vitali, A.; Botta, B.; Delle Monache, G.; Zappitelli, S.; Ricciardi, P.; Melino, S.; Petruzzelli, R.; Giardina, B. Purification and partial characterization of a peroxidase from plant cell cultures of Cassia didymobotrya and biotransformation studies. Biochem. J. 1998, 331, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Inoue, S.; Markert, E.K.; Levine, A.J.; Knight, R.A.; Mak, T.W.; Melino, G. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1α degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Dulloo, I.; Phang, B.H.; Othman, R.; Tan, S.Y.; Vijayaraghavan, A.; Goh, L.K.; Martin-Lopez, M.; Marques, M.M.; Li, C.W.; Wang, D.Y.; et al. Hypoxia-inducible TAp73 supports tumorigenesis by regulating the angiogenic transcriptome. Nat. Cell Biol. 2015, 17, 511–523. [Google Scholar] [CrossRef]
- Salimath, B.; Marmé, D.; Finkenzeller, G. Expression of the vascular endothelial growth factor gene is inhibited by p73. Oncogene 2000, 19, 3470–3476. [Google Scholar] [CrossRef][Green Version]
- Vikhanskaya, F.; Bani, M.R.; Borsotti, P.; Ghilardi, C.; Ceruti, R.; Ghisleni, G.; Marabese, M.; Giavazzi, R.; Broggini, M.; Taraboletti, G. p73 Overexpression increases VEGF and reduces thrombospondin-1 production: Implications for tumor angiogenesis. Oncogene 2001, 20, 7293–7300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, M.; Zhang, Z.; Ma, F.; Zhang, X.; Zhang, Z.; Tang, J.; Chen, P.; Zhou, C.; Wang, W. Association between TAp73, p53 and VASH1 expression in lung adenocarcinoma. Oncol. Lett. 2018, 15, 5175–5180. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Dulloo, I.; Sabapathy, K. Context-dependent AMPK activation distinctly regulates TAp73 stability and transcriptional activity. Signal Transduct. Target. Ther. 2018, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Dulloo, I.; Hooi, P.B.; Sabapathy, K. Hypoxia-induced DNp73 stabilization regulates Vegf-A expression and tumor angiogenesis similar to TAp73. Cell Cycle 2015, 14, 3533–3539. [Google Scholar] [CrossRef]
- Fernandez-Alonso, R.; Martin-Lopez, M.; Gonzalez-Cano, L.; Garcia, S.; Castrillo, F.; Diez-Prieto, I.; Fernandez-Corona, A.; Lorenzo-Marcos, M.E.; Li, X.; Claesson-Welsh, L.; et al. p73 is required for endothelial cell differentiation, migration and the formation of vascular networks regulating VEGF and TGFβ signaling. Cell Death Differ. 2015, 22, 1287–1299. [Google Scholar] [CrossRef] [PubMed]
- Mauretti, A.; Neri, A.; Kossover, O.; Seliktar, D.; Nardo, P.D.; Melino, S. Design of a Novel Composite H2 S-Releasing Hydrogel for Cardiac Tissue Repair. Macromol. Biosci. 2016, 16, 847–858. [Google Scholar] [CrossRef]
- Díaz, R.; Peña, C.; Silva, J.; Lorenzo, Y.; García, V.; García, J.M.; Sánchez, A.; Espinosa, P.; Yuste, R.; Bonilla, F.; et al. p73 Isoforms affect VEGF, VEGF165b and PEDF expression in human colorectal tumors: VEGF165b downregulation as a marker of poor prognosis. Int. J. Cancer 2008, 123, 1060–1067. [Google Scholar] [CrossRef]
- Guan, M.; Peng, H.-X.; Yu, B.; Lu, Y. p73 Overexpression and angiogenesis in human colorectal carcinoma. Jpn. J. Clin. Oncol. 2003, 33, 215–220. [Google Scholar] [CrossRef]
- Sabapathy, K. p73: A Positive or Negative Regulator of Angiogenesis, or Both? Mol. Cell. Biol. 2015, 36, 848–854. [Google Scholar] [CrossRef]
- Blomberg, O.S.; Spagnuolo, L.; de Visser, K.E. Immune regulation of metastasis: Mechanistic insights and therapeutic opportunities. Dis. Model. Mech. 2018, 11, dmm036236. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Hofstetter, G.; Berger, A.; Gehmacher, A.; Reimer, D.; Watrowski, R.; Tong, D.; Schuster, E.; Hefler, L.; Heim, K.; et al. Clinical relevance of dominant-negative p73 isoforms for responsiveness to chemotherapy and survival in ovarian cancer: Evidence for a crucial p53-p73 cross-talk in vivo. Clin. Cancer Res. 2005, 11, 8372–8383. [Google Scholar] [CrossRef]
- Bunch, B.; Krishnan, N.; Greenspan, R.D.; Ramakrishnan, S.; Attwood, K.; Yan, L.; Qi, Q.; Wang, D.; Morrison, C.; Omilian, A.; et al. TAp73 expression and P1 promoter methylation, a potential marker for chemoresponsiveness to cisplatin therapy and survival in muscle-invasive bladder cancer (MIBC). Cell Cycle 2019, 18, 2055–2066. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Campbell, K.R.; Greening, K.; Ho, G.C.; Hopkins, J.; Bui, M.; Douglas, J.M.; Sharlandjieva, V.; Munzur, A.D.; Lai, D.; et al. Single cell transcriptomes of normal endometrial derived organoids uncover novel cell type markers and cryptic differentiation of primary tumours. J. Pathol. 2020, 252, 201–214. [Google Scholar] [CrossRef]
- Hua, B.; Li, Y.; Yang, X.; Niu, X.; Zhao, Y.; Zhu, X. MicroRNA-361-3p promotes human breast cancer cell viability by inhibiting the E2F1/P73 signalling pathway. Biomed. Pharmacother. 2020, 125, 109994. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, W.; Jung, Y.S.; Chen, X. Mammary epithelial cell polarity is regulated differentially by p73 isoforms via epithelial-to-mesenchymal transition. J. Biol. Chem. 2012, 287, 17746–17753. [Google Scholar] [CrossRef] [PubMed]
- Persa, O.-D.; Niessen, C.M. Epithelial polarity limits EMT. Nat. Cell Biol. 2019, 21, 299–300. [Google Scholar] [CrossRef]
- Chatterjee, S.J.; Halaoui, R.; McCaffrey, L. Apical–basal polarity as a sensor for epithelial homeostasis: A matter of life and death. Curr. Pathobiol. Rep. 2016, 4, 99–106. [Google Scholar] [CrossRef]
- Zhang, Y.; Young, A.; Zhang, J.; Chen, X. P73 tumor suppressor and its targets, p21 and PUMA, are required for madin-darby canine kidney cell morphogenesis by maintaining an appropriate level of epithelial to mesenchymal transition. Oncotarget 2015, 6, 13994–14004. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, E.; Chen, X. TAp73 protein stability is controlled by histone deacetylase 1 via regulation of Hsp90 chaperone function. J. Biol. Chem. 2013, 288, 7727–7737. [Google Scholar] [CrossRef]
- Weissmueller, S.; Manchado, E.; Saborowski, M.; Morris, J.P.; Wagenblast, E.; Davis, C.A.; Moon, S.-H.; Pfister, N.T.; Tschaharganeh, D.F.; Kitzing, T.; et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 2014, 157, 382–394. [Google Scholar] [CrossRef]
- Thakur, A.K.; Nigri, J.; Lac, S.; Leca, J.; Bressy, C.; Berthezene, P.; Bartholin, L.; Chan, P.; Calvo, E.; Iovanna, J.L.; et al. TAp73 loss favors Smad-independent TGF-β signaling that drives EMT in pancreatic ductal adenocarcinoma. Cell Death Differ. 2016, 23, 1358–1370. [Google Scholar] [CrossRef]
- Groth, S.; Schulze, M.; Kalthoff, H.; Fändrich, F.; Ungefroren, H. Adhesion and Rac1-dependent regulation of biglycan gene expression by transforming growth factor-beta. Evidence for oxidative signaling through NADPH oxidase. J. Biol. Chem. 2005, 280, 33190–33199. [Google Scholar] [CrossRef]
- Soares, K.C.; Rucki, A.A.; Kim, V.; Foley, K.; Solt, S.; Wolfgang, C.L.; Jaffee, E.M.; Zheng, L. TGF-β blockade depletes T regulatory cells from metastatic pancreatic tumors in a vaccine dependent manner. Oncotarget 2015, 6, 43005–43015. [Google Scholar] [CrossRef]
- Sinha, N.; Meher, B.R.; Naik, P.P.; Panda, P.K.; Mukhapadhyay, S.; Maiti, T.K.; Bhutia, S.K. p73 induction by Abrus agglutinin facilitates Snail ubiquitination to inhibit epithelial to mesenchymal transition in oral cancer. Phytomedicine 2019, 55, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Beitzinger, M.; Hofmann, L.; Oswald, C.; Beinoraviciute-Kellner, R.; Sauer, M.; Griesmann, H.; Bretz, A.C.; Burek, C.; Rosenwald, A.; Stiewe, T. p73 poses a barrier to malignant transformation by limiting anchorage-independent growth. EMBO J. 2008, 27, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Steder, M.; Alla, V.; Meier, C.; Spitschak, A.; Pahnke, J.; Fürst, K.; Kowtharapu, B.S.; Engelmann, D.; Petigk, J.; Egberts, F.; et al. DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell 2013, 24, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Fürst, K.; Steder, M.; Logotheti, S.; Angerilli, A.; Spitschak, A.; Marquardt, S.; Schumacher, T.; Engelmann, D.; Herchenröder, O.; Rupp, R.A.W.; et al. DNp73-induced degradation of tyrosinase links depigmentation with EMT-driven melanoma progression. Cancer Lett. 2019, 442, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Stros, M.; Ozaki, T.; Bacikova, A.; Kageyama, H.; Nakagawara, A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 2002, 277, 7157–7164. [Google Scholar] [CrossRef]
- Uramoto, H.; Izumi, H.; Nagatani, G.; Ohmori, H.; Nagasue, N.; Ise, T.; Yoshida, T.; Yasumoto, K.; Kohno, K. Physical interaction of tumour suppressor p53/p73 with CCAAT-binding transcription factor 2 (CTF2) and differential regulation of human high-mobility group 1 (HMG1) gene expression. Biochem. J. 2003, 371, 301–310. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, L.; Guo, X.; Li, Z.; Liao, X.; Zhang, X.; Huang, L.; He, J. HMGB1-activated fibroblasts promote breast cancer cells metastasis via RAGE/aerobic glycolysis. Neoplasma 2020, 68, 71–78. [Google Scholar] [CrossRef]
- Okui, T.; Hiasa, M.; Ryumon, S.; Ono, K.; Kunisada, Y.; Ibaragi, S.; Sasaki, A.; Roodman, G.D.; White, F.A.; Yoneda, T. The HMGB1/RAGE axis induces bone pain associated with colonization of 4T1 mouse breast cancer in bone. J. Bone Oncol. 2021, 26, 100330. [Google Scholar] [CrossRef]
- Wu, X.-J.; Chen, Y.-Y.; Guo, W.-W.; Li, T.; Dong, H.-B.; Wang, W.; Xie, M.; Ma, G.-L.; Pei, D.-S. HMGB1 regulates SNAI1 during NSCLC metastasis, both directly, through transcriptional activation, and indirectly, in a RSF1-IT2-dependent manner. Mol. Oncol. 2020, 14, 1348–1364. [Google Scholar] [CrossRef]
- Xie, N.; Vikhreva, P.; Annicchiarico-Petruzzelli, M.; Amelio, I.; Barlev, N.; Knight, R.A.; Melino, G. Integrin-β4 is a novel transcriptional target of TAp73. Cell Cycle 2018, 17, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Landré, V.; Antonov, A.; Knight, R.; Melino, G. p73 promotes glioblastoma cell invasion by directly activating POSTN (periostin) expression. Oncotarget 2016, 7, 11785–11802. [Google Scholar] [CrossRef] [PubMed]
- Amelio, I.; Melino, G. The p53 family and the hypoxia-inducible factors (HIFs): Determinants of cancer progression. Trends Biochem. Sci. 2015, 40, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.S.; Barlev, N.A. Tumor microenvironment regulation by hypoxia-indicible factors (hifs), and p53 family proteins. Tsitologiia 2017, 59, 259–270. [Google Scholar] [PubMed]
- Anqi, C.; Takabatake, K.; Kawai, H.; Oo, M.W.; Yoshida, S.; Fujii, M.; Omori, H.; Sukegawa, S.; Nakano, K.; Tsujigiwa, H.; et al. Differentiation and roles of bone marrow-derived cells on the tumor microenvironment of oral squamous cell carcinoma. Oncol. Lett. 2019, 18, 6628–6638. [Google Scholar] [CrossRef]
- Zhang, Y.; Lazarus, J.; Steele, N.G.; Yan, W.; Lee, H.-J.; Nwosu, Z.C.; Halbrook, C.J.; Menjivar, R.E.; Kemp, S.B.; Sirihorachai, V.R.; et al. Regulatory T-cell Depletion Alters the Tumor Microenvironment and Accelerates Pancreatic Carcinogenesis. Cancer Discov. 2020, 10, 422–439. [Google Scholar] [CrossRef]
- Garufi, A.; Traversi, G.; Cirone, M.; D’Orazi, G. HIPK2 role in the tumor-host interaction: Impact on fibroblasts transdifferentiation CAF-like. IUBMB Life 2019, 71, 2055–2061. [Google Scholar] [CrossRef]
- Beyranvand Nejad, E.; Labrie, C.; Abdulrahman, Z.; van Elsas, M.J.; Rademaker, E.; Kleinovink, J.W.; van der Sluis, T.C.; van Duikeren, S.; Teunisse, A.F.A.S.; Jochemsen, A.G.; et al. Lack of myeloid cell infiltration as an acquired resistance strategy to immunotherapy. J. Immunother. Cancer 2020, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Naserian, S.; Abdelgawad, M.E.; Afshar Bakshloo, M.; Ha, G.; Arouche, N.; Cohen, J.L.; Salomon, B.L.; Uzan, G. The TNF/TNFR2 signaling pathway is a key regulatory factor in endothelial progenitor cell immunosuppressive effect. Cell Commun. Signal. 2020, 18, 94. [Google Scholar] [CrossRef] [PubMed]
- Pallucca, R.; Visconti, S.; Camoni, L.; Cesareni, G.; Melino, S.; Panni, S.; Torreri, P.; Aducci, P. Specificity of ε and non-ε isoforms of arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets. PLoS ONE 2014, 9, e90764. [Google Scholar] [CrossRef]
- Koshiba, S.; Ichimiya, S.; Nagashima, T.; Tonooka, A.; Kubo, T.; Kikuchi, T.; Himi, T.; Sato, N. Tonsillar crypt epithelium of palmoplantar pustulosis secretes interleukin-6 to support B-cell development via p63/p73 transcription factors. J. Pathol. 2008, 214, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Chonov, D.C.; Ignatova, M.M.K.; Ananiev, J.R.; Gulubova, M.V. IL-6 Activities in the Tumour Microenvironment. Part 1. Open Access Maced. J. Med. Sci. 2019, 7, 2391–2398. [Google Scholar] [CrossRef]
- Guo, H.; Yang, S.; Xu, L.; Li, D.; Tang, J.; Wang, S.; Wei, B.; Liu, Z. Association between the p73 gene G4C14-to-A4T14 single nucleotide polymorphism and risk of cervical cancer by high resolution melting and PCR with confronting two-pair primers in a Chinese population. Oncol. Lett. 2016, 12, 721–726. [Google Scholar] [CrossRef][Green Version]
- Zhou, X.; Hao, Q.; Zhang, Q.; Liao, J.M.; Ke, J.W.; Liao, P.; Cao, B.; Lu, H. Ribosomal proteins L11 and L5 activate TAp73 by overcoming MDM2 inhibition. Cell Death Differ. 2015, 22, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Kanneganti, T.-D. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 2016, 213, 617–629. [Google Scholar] [CrossRef]
- Kantono, M.; Guo, B. Inflammasomes and cancer: The dynamic role of the inflammasome in tumor development. Front. Immunol. 2017, 8, 1132. [Google Scholar] [CrossRef]
- Vikhreva, P.; Petrova, V.; Gokbulut, T.; Pestlikis, I.; Mancini, M.; Di Daniele, N.; Knight, R.A.; Melino, G.; Amelio, I. TAp73 upregulates IL-1β in cancer cells: Potential biomarker in lung and breast cancer? Biochem. Biophys. Res. Commun. 2017, 482, 498–505. [Google Scholar] [CrossRef]
- Kumagai, A.; Kubo, T.; Kawata, K.; Kamekura, R.; Yamashita, K.; Jitsukawa, S.; Nagaya, T.; Sumikawa, Y.; Himi, T.; Yamashita, T.; et al. Keratinocytes in atopic dermatitis express abundant ΔNp73 regulating thymic stromal lymphopoietin production via NF-κB. J. Dermatol. Sci. 2017, 88, 175–183. [Google Scholar] [CrossRef]
- Soumelis, V.; Reche, P.A.; Kanzler, H.; Yuan, W.; Edward, G.; Homey, B.; Gilliet, M.; Ho, S.; Antonenko, S.; Lauerma, A.; et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat. Immunol. 2002, 3, 673–680. [Google Scholar] [CrossRef]
- Ebner, S.; Nguyen, V.A.; Forstner, M.; Wang, Y.-H.; Wolfram, D.; Liu, Y.-J.; Romani, N. Thymic stromal lymphopoietin converts human epidermal Langerhans cells into antigen-presenting cells that induce proallergic T cells. J. Allergy Clin. Immunol. 2007, 119, 982–990. [Google Scholar] [CrossRef] [PubMed]
- King, K.E.; George, A.L.; Sakakibara, N.; Mahmood, K.; Moses, M.A.; Weinberg, W.C. Intersection of the p63 and NF-κB pathways in epithelial homeostasis and disease. Mol. Carcinog. 2019, 58, 1571–1580. [Google Scholar] [CrossRef]
- King, K.E.; Ponnamperuma, R.M.; Allen, C.; Lu, H.; Duggal, P.; Chen, Z.; Van Waes, C.; Weinberg, W.C. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008, 68, 5122–5131. [Google Scholar] [CrossRef] [PubMed]
- De Monte, L.; Reni, M.; Tassi, E.; Clavenna, D.; Papa, I.; Recalde, H.; Braga, M.; Di Carlo, V.; Doglioni, C.; Protti, M.P. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011, 208, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Corren, J.; Ziegler, S.F. TSLP: From allergy to cancer. Nat. Immunol. 2019, 20, 1603–1609. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Jeong, H.-J.; Kim, H.-M. Anti-allergic and anti-inflammatory effects of the Bcl-2 inhibitor ABT-737 on experimental allergic rhinitis models. Eur. J. Pharmacol. 2018, 833, 34–43. [Google Scholar] [CrossRef]
- Tomasini, R.; Secq, V.; Pouyet, L.; Thakur, A.K.; Wilhelm, M.; Nigri, J.; Vasseur, S.; Berthezene, P.; Calvo, E.; Melino, G.; et al. TAp73 is required for macrophage-mediated innate immunity and the resolution of inflammatory responses. Cell Death Differ. 2013, 20, 293–301. [Google Scholar] [CrossRef]
- Carswell, E.A.; Old, L.J.; Kassel, R.L.; Green, S.; Fiore, N.; Williamson, B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA 1975, 72, 3666–3670. [Google Scholar] [CrossRef]
- Salomon, B.L.; Leclerc, M.; Tosello, J.; Ronin, E.; Piaggio, E.; Cohen, J.L. Tumor necrosis factor α and regulatory T cells in oncoimmunology. Front. Immunol. 2018, 9, 444. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Zhao, G.; Li, H. Forward and Reverse Signaling Mediated by Transmembrane Tumor Necrosis Factor-Alpha and TNF Receptor 2: Potential Roles in an Immunosuppressive Tumor Microenvironment. Front. Immunol. 2017, 8, 1675. [Google Scholar] [CrossRef]
- Chau, B.N.; Chen, T.-T.; Wan, Y.Y.; DeGregori, J.; Wang, J.Y.J. Tumor necrosis factor alpha-induced apoptosis requires p73 and c-ABL activation downstream of RB degradation. Mol. Cell. Biol. 2004, 24, 4438–4447. [Google Scholar] [CrossRef][Green Version]
- Zhan, Q.; Korngold, R.; Lezcano, C.; McKeon, F.; Murphy, G.F. Graft-versus-host disease-related cytokine-driven apoptosis depends on p73 in cytokeratin 15-positive target cells. Biol. Blood Marrow Transplant. 2012, 18, 841–851. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rastogi, S.; Rizwani, W.; Joshi, B.; Kunigal, S.; Chellappan, S.P. TNF-α response of vascular endothelial and vascular smooth muscle cells involve differential utilization of ASK1 kinase and p73. Cell Death Differ. 2012, 19, 274–283. [Google Scholar] [CrossRef]
- Yang, A.; Walker, N.; Bronson, R.; Kaghad, M.; Oosterwegel, M.; Bonnin, J.; Vagner, C.; Bonnet, H.; Dikkes, P.; Sharpe, A.; et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000, 404, 99–103. [Google Scholar] [CrossRef]
- Strait, A.A.; Wang, X.-J. The role of transforming growth factor-beta in immune suppression and chronic inflammation of squamous cell carcinomas. Mol. Carcinog. 2020, 59, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.; Oronsky, B.; Carter, C.A.; Oronsky, A.; Knox, S.J.; Sher, D.; Reid, T.R. TGF-beta: A master immune regulator. Expert Opin. Ther. Targets 2020, 24, 427–438. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Srivenugopal, K.S.; Fan, S.; Jiang, Y. In vitro antitumor cytotoxic T lymphocyte response induced by dendritic cells transduced with DeltaNp73alpha recombinant adenovirus. Oncol. Rep. 2007, 18, 1085–1091. [Google Scholar]
- Eisel, D.; Das, K.; Dickes, E.; König, R.; Osen, W.; Eichmüller, S.B. Cognate Interaction With CD4+ T Cells Instructs Tumor-Associated Macrophages to Acquire M1-Like Phenotype. Front. Immunol. 2019, 10, 219. [Google Scholar] [CrossRef]
- Ren, M.; Kazemian, M.; Zheng, M.; He, J.; Li, P.; Oh, J.; Liao, W.; Li, J.; Rajaseelan, J.; Kelsall, B.L.; et al. Transcription factor p73 regulates Th1 differentiation. Nat. Commun. 2020, 11, 1475. [Google Scholar] [CrossRef]
- Senoo, M.; Manis, J.P.; Alt, F.W.; McKeon, F. p63 and p73 are not required for the development and p53-dependent apoptosis of T cells. Cancer Cell 2004, 6, 85–89. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Shao, C.; Huang, J.; Gan, J.; Huang, X.; Bucci, E.; Piacentini, M.; Ippolito, G.; Melino, G. COVID-19 infection: The perspectives on immune responses. Cell Death Differ. 2020, 27, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Lopez, E.; Ghia, E.M.; Antonucci, L.; Sharma, N.; Rassenti, L.Z.; Xu, J.; Sun, B.; Kipps, T.J.; Karin, M. NF-κB-p62-NRF2 survival signaling is associated with high ROR1 expression in chronic lymphocytic leukemia. Cell Death Differ. 2020, 27, 2206–2216. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Kawaguchi, A.; Kanayama, T.; Hayano, A.; Yamanaka, R. Correlation between lower balance of Th2 helper T-cells and expression of PD-L1/PD-1 axis genes enables prognostic prediction in patients with glioblastoma. Oncotarget 2018, 9, 19065–19078. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, H.; Xu, J.; Lu, Y.; Ji, X.; Yao, Y.; Chao, H.; Zhang, J.; Zhang, X.; Yao, S.; et al. Different T-cell subsets in glioblastoma multiforme and targeted immunotherapy. Cancer Lett. 2021, 496, 134–143. [Google Scholar] [CrossRef]
- Lee, H.L.; Jang, J.W.; Lee, S.W.; Yoo, S.H.; Kwon, J.H.; Nam, S.W.; Bae, S.H.; Choi, J.Y.; Han, N.I.; Yoon, S.K. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci. Rep. 2019, 9, 3260. [Google Scholar] [CrossRef]
- Lauerova, L.; Dusek, L.; Simickova, M.; Kocák, I.; Vagundová, M.; Zaloudík, J.; Kovarík, J. Malignant melanoma associates with Th1/Th2 imbalance that coincides with disease progression and immunotherapy response. Neoplasma 2002, 49, 159–166. [Google Scholar] [PubMed]
- Tokumaru, Y.; Le, L.; Oshi, M.; Katsuta, E.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Association of Th2 high tumors with aggressive features of breast cancer. JCO 2020, 38, e12584. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, J.; Ge, S.; Chen, C.; Li, S.; Wu, X.; Feng, X.; Wang, Y.; Cai, D. Saikosaponin A inhibits breast cancer by regulating th1/th2 balance. Front. Pharmacol. 2019, 10, 624. [Google Scholar] [CrossRef]
- Coppari, E.; Yamada, T.; Bizzarri, A.R.; Beattie, C.W.; Cannistraro, S. A nanotechnological, molecular-modeling, and immunological approach to study the interaction of the anti-tumorigenic peptide p28 with the p53 family of proteins. Int. J. Nanomed. 2014, 9, 1799–1813. [Google Scholar] [PubMed]
- Biton, J.; Mansuet-Lupo, A.; Pécuchet, N.; Alifano, M.; Ouakrim, H.; Arrondeau, J.; Boudou-Rouquette, P.; Goldwasser, F.; Leroy, K.; Goc, J.; et al. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin. Cancer Res. 2018, 24, 5710–5723. [Google Scholar] [CrossRef] [PubMed]
- Rashed, H.E.; Abdelrahman, A.E.; Abdelgawad, M.; Balata, S.; Shabrawy, M.E. Prognostic Significance of Programmed Cell Death Ligand 1 (PD-L1), CD8+ Tumor-Infiltrating Lymphocytes and p53 in Non-Small Cell Lung Cancer: An Immunohistochemical Study. Turk Patoloji Derg. 2017, 1, 211–222. [Google Scholar] [CrossRef][Green Version]
- Rao, N.; Qiu, J.; Wu, J.; Zeng, H.; Su, F.; Qiu, K.; Wu, J.; Yao, H. Significance of Tumor-Infiltrating Lymphocytes and the Expression of Topoisomerase IIα in the Prediction of the Clinical Outcome of Patients with Triple-Negative Breast Cancer after Taxane-Anthracycline-Based Neoadjuvant Chemotherapy. Chemotherapy 2017, 62, 246–255. [Google Scholar] [CrossRef]
- Busuttil, V.; Droin, N.; McCormick, L.; Bernassola, F.; Candi, E.; Melino, G.; Green, D.R. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl. Acad. Sci. USA 2010, 107, 18061–18066. [Google Scholar] [CrossRef]
- Qiao, H.-B.; Li, J.; Lv, L.-J.; Nie, B.-J.; Lu, P.; Xue, F.; Zhang, Z.-M. The effects of interleukin 2 and rAd-p53 as a treatment for glioblastoma. Mol. Med. Rep. 2018, 17, 4853–4859. [Google Scholar] [CrossRef] [PubMed]
- Uz, U.; Eskiizmir, G. Association Between Interleukin-6 and Head and Neck Squamous Cell Carcinoma: A Systematic Review. Clin Exp Otorhinolaryngol 2021, 14, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Lee, O.-Y.; Park, Y.; Seo, M.W.; Lee, D.-S. IL-1β induces IL-6 production and increases invasiveness and estrogen-independent growth in a TG2-dependent manner in human breast cancer cells. BMC Cancer 2016, 16, 724. [Google Scholar] [CrossRef] [PubMed]
- Fiering, S.N.; Ho, G.W. Speed kills: Advancement in th17 cell adoptive cell therapy for solid tumors. Cancer Res. 2020, 80, 3795–3796. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, X.; Liu, Q.; Guan, Z.; Luo, J.; Cao, G.; Cai, R.; Li, Z.; Xu, Y.; Wu, Z.; et al. Roles of mTORC1 and mTORC2 in controlling γδ T1 and γδ T17 differentiation and function. Cell Death Differ. 2020, 27, 2248–2262. [Google Scholar] [CrossRef]
- Nunez, N.P.; Oh, W.-J.; Rozenberg, J.; Perella, C.; Anver, M.; Barrett, J.C.; Perkins, S.N.; Berrigan, D.; Moitra, J.; Varticovski, L.; et al. Accelerated tumor formation in a fatless mouse with type 2 diabetes and inflammation. Cancer Res. 2006, 66, 5469–5476. [Google Scholar] [CrossRef]
- Bent, R.; Moll, L.; Grabbe, S.; Bros, M. Interleukin-1 Beta-A Friend or Foe in Malignancies? Int. J. Mol. Sci. 2018, 19, 2155. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Sudhakar, C.; Swarup, G. Tumor necrosis factor-alpha-induced caspase-1 gene expression. Role of p73. FEBS J. 2007, 274, 4396–4407. [Google Scholar] [CrossRef]
- Roberts, J.Z.; Holohan, C.; Sessler, T.; Fox, J.; Crawford, N.; Riley, J.S.; Khawaja, H.; Majkut, J.; Evergren, E.; Humphreys, L.M.; et al. The SCFSkp2 ubiquitin ligase complex modulates TRAIL-R2-induced apoptosis by regulating FLIP(L). Cell Death Differ. 2020, 27, 2726–2741. [Google Scholar] [CrossRef]
- Damgaard, R.B.; Jolin, H.E.; Allison, M.E.D.; Davies, S.E.; Titheradge, H.L.; McKenzie, A.N.J.; Komander, D. OTULIN protects the liver against cell death, inflammation, fibrosis, and cancer. Cell Death Differ. 2020, 27, 1457–1474. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Zhou, S.; He, X.; Cao, X.; Wu, C.; Hu, H.; Qin, J.; Wei, G.; Wang, H.; et al. Membrane-bound TNF mediates microtubule-targeting chemotherapeutics-induced cancer cytolysis via juxtacrine inter-cancer-cell death signaling. Cell Death Differ. 2020, 27, 1569–1587. [Google Scholar] [CrossRef]
- Li, L.; Li, L.; Li, W.; Chen, T.; Zou, B.; Zhao, L.; Wang, H.; Wang, X.; Xu, L.; Liu, X.; et al. TAp73-induced phosphofructokinase-1 transcription promotes the Warburg effect and enhances cell proliferation. Nat. Commun. 2018, 9, 4683. [Google Scholar] [CrossRef]
- Lu, J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019, 38, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Lissy, N.A.; Davis, P.K.; Irwin, M.; Kaelin, W.G.; Dowdy, S.F. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 2000, 407, 642–645. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Lonetto, G.; Koifman, G.; Silberman, A.; Attery, A.; Solomon, H.; Levin-Zaidman, S.; Goldfinger, N.; Porat, Z.; Erez, A.; Rotter, V. Mutant p53-dependent mitochondrial metabolic alterations in a mesenchymal stem cell-based model of progressive malignancy. Cell Death Differ. 2019, 26, 1566–1581. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, Y.; Xiao, J.; Shang, J.; Tan, Q.; Ping, F.; Huang, W.; Wu, F.; Zhang, H.; Zhang, X. Inhibitor of apoptosis-stimulating protein of p53 inhibits ferroptosis and alleviates intestinal ischemia/reperfusion-induced acute lung injury. Cell Death Differ. 2020, 27, 2635–2650. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Pan, X.; Abali, G.K.; Little, J.B.; Yuan, Z.-M. Functional interplay between p53 and Δ133p53 in adaptive stress response. Cell Death Differ. 2020, 27, 1618–1632. [Google Scholar] [CrossRef]
- Radine, C.; Peters, D.; Reese, A.; Neuwahl, J.; Budach, W.; Jänicke, R.U.; Sohn, D. The RNA-binding protein RBM47 is a novel regulator of cell fate decisions by transcriptionally controlling the p53-p21-axis. Cell Death Differ. 2020, 27, 1274–1285. [Google Scholar] [CrossRef]
- Billon, N.; Terrinoni, A.; Jolicoeur, C.; McCarthy, A.; Richardson, W.D.; Melino, G.; Raff, M. Roles for p53 and p73 during oligodendrocyte development. Development 2004, 131, 1211–1220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gonzalez-Cano, L.; Fuertes-Alvarez, S.; Robledinos-Anton, N.; Bizy, A.; Villena-Cortes, A.; Fariñas, I.; Marques, M.M.; Marin, M.C. p73 is required for ependymal cell maturation and neurogenic SVZ cytoarchitecture. Dev. Neurobiol. 2016, 76, 730–747. [Google Scholar] [CrossRef]
- Melino, G.; De Laurenzi, V.; Vousden, K.H. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2002, 2, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Agostini, M.; Melino, G.; Bernassola, F. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: Regulators and effectors. Hum. Mutat. 2014, 35, 702–714. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Uramoto, H.; Sugio, K.; Oyama, T.; Nakata, S.; Ono, K.; Morita, M.; Funa, K.; Yasumoto, K. Expression of deltaNp73 predicts poor prognosis in lung cancer. Clin. Cancer Res. 2004, 10, 6905–6911. [Google Scholar] [CrossRef]
- Wang, B.; Liu, X.; Liu, H.; Guo, J.; Zhang, T.; Zhou, N.; Ma, Y.; Yu, H.; Chen, L.; Ren, Z.; et al. Differential expressions of MDM2 and TAP73 in cancer and cancer-adjacent tissues in patients with non-small-cell lung carcinoma. Pulmonology 2018, 24, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Uramoto, H.; Sugio, K.; Oyama, T.; Nakata, S.; Ono, K.; Nozoe, T.; Yasumoto, K. Expression of the p53 family in lung cancer. Anticancer Res. 2006, 26, 1785–1790. [Google Scholar]
- Wakatsuki, M.; Ohno, T.; Iwakawa, M.; Ishikawa, H.; Noda, S.; Ohta, T.; Kato, S.; Tsujii, H.; Imai, T.; Nakano, T. p73 protein expression correlates with radiation-induced apoptosis in the lack of p53 response to radiation therapy for cervical cancer. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Mega Tiber, P.; Baloglu, L.; Ozden, S.; Ozgen, Z.; Ozyurt, H.; Eren, M.; Orun, O. The association of apoptotic protein expressions sensitive to apoptosis gene, p73 and p53 with the prognosis of cervical carcinoma. Onco. Targets. Ther. 2014, 7, 2161–2168. [Google Scholar]
- Ye, H.; Guo, X. TP73 is a credible biomarker for predicting clinical progression and prognosis in cervical cancer patients. Biosci. Rep. 2019, 39, BSR20190095. [Google Scholar] [CrossRef] [PubMed]
- Casciano, I.; Mazzocco, K.; Boni, L.; Pagnan, G.; Banelli, B.; Allemanni, G.; Ponzoni, M.; Tonini, G.P.; Romani, M. Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ. 2002, 9, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Zitterbart, K.; Zavrelova, I.; Kadlecova, J.; Spesna, R.; Kratochvilova, A.; Pavelka, Z.; Sterba, J. p73 expression in medulloblastoma: TAp73/DeltaNp73 transcript detection and possible association of p73alpha/DeltaNp73 immunoreactivity with survival. Acta Neuropathol. 2007, 114, 641–650. [Google Scholar] [CrossRef]
- Nemajerova, A.; Petrenko, O.; Trümper, L.; Palacios, G.; Moll, U.M. Loss of p73 promotes dissemination of Myc-induced B cell lymphomas in mice. J. Clin. Investig. 2010, 120, 2070–2080. [Google Scholar] [CrossRef] [PubMed]
- Gomez, L.C.; Sottile, M.L.; Guerrero-Gimenez, M.E.; Zoppino, F.C.M.; Redondo, A.L.; Gago, F.E.; Orozco, J.I.; Tello, O.M.; Roqué, M.; Nadin, S.B.; et al. TP73 DNA methylation and upregulation of ΔNp73 are associated with an adverse prognosis in breast cancer. J. Clin. Pathol. 2018, 71, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, G.; Peña, C.; Silva, J.; García, J.M.; García, V.; Rodríguez, R.; Cantos, B.; Citores, M.J.; España, P.; Bonilla, F. The presence of an intronic deletion in p73 and high levels of ZEB1 alter the TAp73/DeltaTAp73 ratio in colorectal carcinomas. J. Pathol. 2006, 210, 390–397. [Google Scholar] [CrossRef]
- Vilgelm, A.E.; Hong, S.M.; Washington, M.K.; Wei, J.; Chen, H.; El-Rifai, W.; Zaika, A. Characterization of ΔNp73 expression and regulation in gastric and esophageal tumors. Oncogene 2010, 29, 5861–5868. [Google Scholar] [CrossRef]
- Hofstetter, G.; Berger, A.; Chamson, M.; Müller-Holzner, E.; Reimer, D.; Ulmer, H.; Uramoto, H.; Marth, C.; Zeimet, A.G.; Zeillinger, R.; et al. Clinical relevance of TAp73 and ΔNp73 protein expression in ovarian cancer: A series of 83 cases and review of the literature. Int J Gynecol Pathol 2011, 30, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, D.; Killary, A.M.; Sen, S.; Amos, C.I.; Evans, D.B.; Abbruzzese, J.L.; Frazier, M.L. Polymorphisms of p16, p27, p73, and MDM2 modulate response and survival of pancreatic cancer patients treated with preoperative chemoradiation. Ann. Surg. Oncol. 2009, 16, 431–439. [Google Scholar] [CrossRef]
- Ito, Y.; Takeda, T.; Wakasa, K.; Tsujimoto, M.; Sakon, M.; Matsuura, N. Expression of p73 and p63 proteins in pancreatic adenocarcinoma: p73 overexpression is inversely correlated with biological aggressiveness. Int. J. Mol. Med. 2001, 8, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, X.; Zhou, X. [Expression and prognosis significance of p73 and PCNA in laryngeal squamous cell carcinoma]. Lin Chuang Er Bi Yan Hou Ke Za Zhi 2005, 19, 1121–1124. [Google Scholar] [PubMed]
- Choi, H.-R.; Batsakis, J.G.; Zhan, F.; Sturgis, E.; Luna, M.A.; El-Naggar, A.K. Differential expression of p53 gene family members p63 and p73 in head and neck squamous tumorigenesis. Hum. Pathol. 2002, 33, 158–164. [Google Scholar] [CrossRef] [PubMed]
| Cancer | Invasion and Metastasis | Angiogenesis | Inflammation | Role of p73 Isoforms in Cancer | Relative tp73 Expression in Cancer Tissue |
|---|---|---|---|---|---|
| Non-small cell lung carcinoma (NSCLC) | [134] | [150] | [210] | [211] | |
| Lung adenocarcinomas | [103] | [193] | [99,212] | 2, 95 | |
| Hepatocellular carcinoma (HCC) | |||||
| Cervix carcinoma | [126] | [213,214,215] | 15, 2 | ||
| Melanoma | [126,128] | ||||
| Osteosarcoma | [99,100,101] | 5, 4 | |||
| Glioblastoma | [126,135] | [135] | [216] | 5, 2 | |
| Medulloblastoma | [217] | --- | |||
| B-cell lymphoma | [88] | [218] | 33, 6 | ||
| Breast cancer | [116] | [88,100] | [150] | [219] | 1, 2 |
| Colorectal cancer | [89] | [88,100,108,109] | [163] | [55,220] | 9, 6 |
| Esophageal adenocarcinoma | [125] | [221] | 1, 1 | ||
| Thyroid cancer | |||||
| Ovarian cancer | [102] | [222] | 4, 1 | ||
| Pancreatic cancer | [121] | [156] | [223,224] | 3, 15 | |
| Neuroblastoma | [135] | ||||
| Squamous carcinoma | [61,63] | [225,226] | 1, 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozenberg, J.M.; Zvereva, S.; Dalina, A.; Blatov, I.; Zubarev, I.; Luppov, D.; Bessmertnyi, A.; Romanishin, A.; Alsoulaiman, L.; Kumeiko, V.; et al. Dual Role of p73 in Cancer Microenvironment and DNA Damage Response. Cells 2021, 10, 3516. https://doi.org/10.3390/cells10123516
Rozenberg JM, Zvereva S, Dalina A, Blatov I, Zubarev I, Luppov D, Bessmertnyi A, Romanishin A, Alsoulaiman L, Kumeiko V, et al. Dual Role of p73 in Cancer Microenvironment and DNA Damage Response. Cells. 2021; 10(12):3516. https://doi.org/10.3390/cells10123516
Chicago/Turabian StyleRozenberg, Julian M., Svetlana Zvereva, Alexandra Dalina, Igor Blatov, Ilya Zubarev, Daniil Luppov, Alexander Bessmertnyi, Alexander Romanishin, Lamak Alsoulaiman, Vadim Kumeiko, and et al. 2021. "Dual Role of p73 in Cancer Microenvironment and DNA Damage Response" Cells 10, no. 12: 3516. https://doi.org/10.3390/cells10123516
APA StyleRozenberg, J. M., Zvereva, S., Dalina, A., Blatov, I., Zubarev, I., Luppov, D., Bessmertnyi, A., Romanishin, A., Alsoulaiman, L., Kumeiko, V., Kagansky, A., Melino, G., & Barlev, N. A. (2021). Dual Role of p73 in Cancer Microenvironment and DNA Damage Response. Cells, 10(12), 3516. https://doi.org/10.3390/cells10123516