Wnt Inhibitory Factor 1 Binds to and Inhibits the Activity of Sonic Hedgehog
Abstract
:1. Introduction
2. Materials and Methods
2.1. Proteins, Cell Lines, Media and Reagents
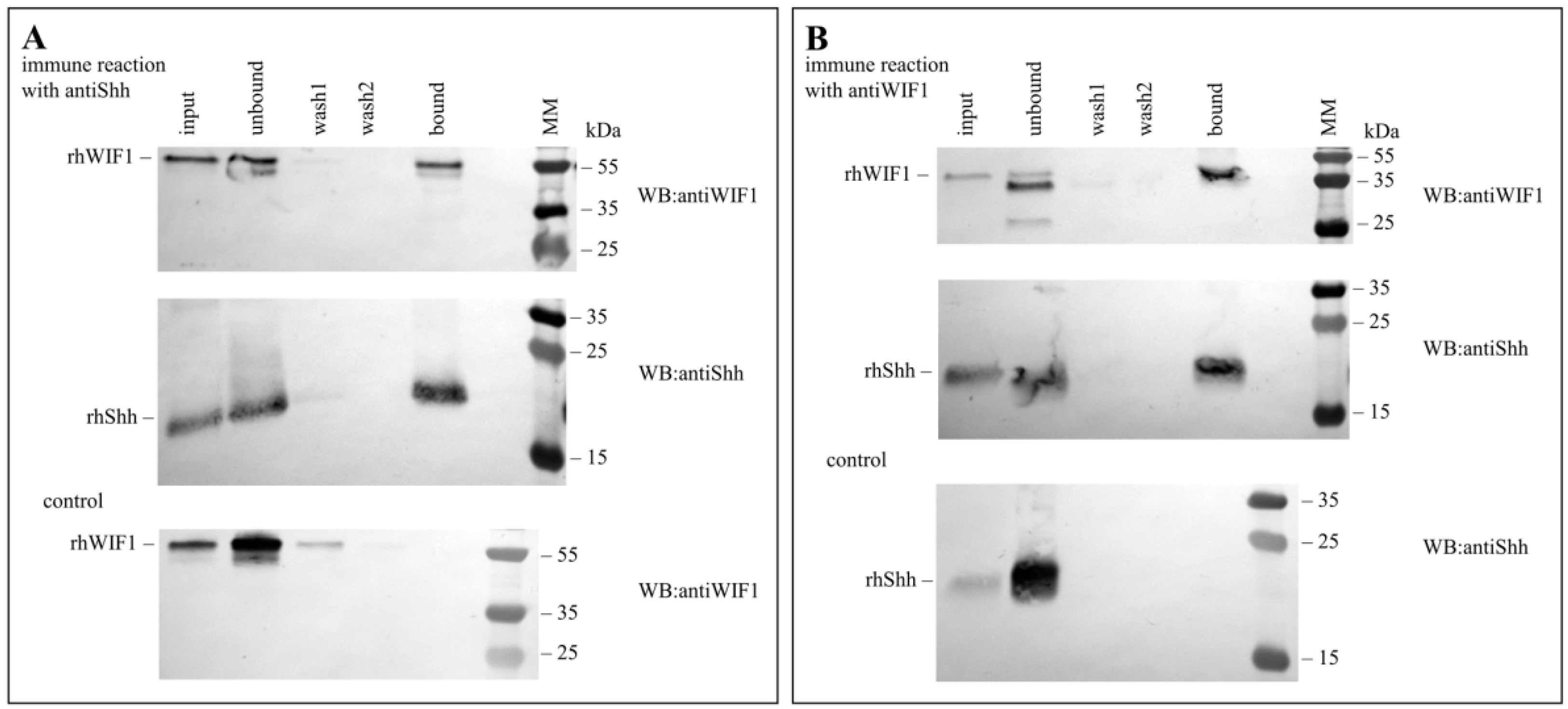
2.2. Pull-Down Assays
2.3. Surface Plasmon Resonance Analyses
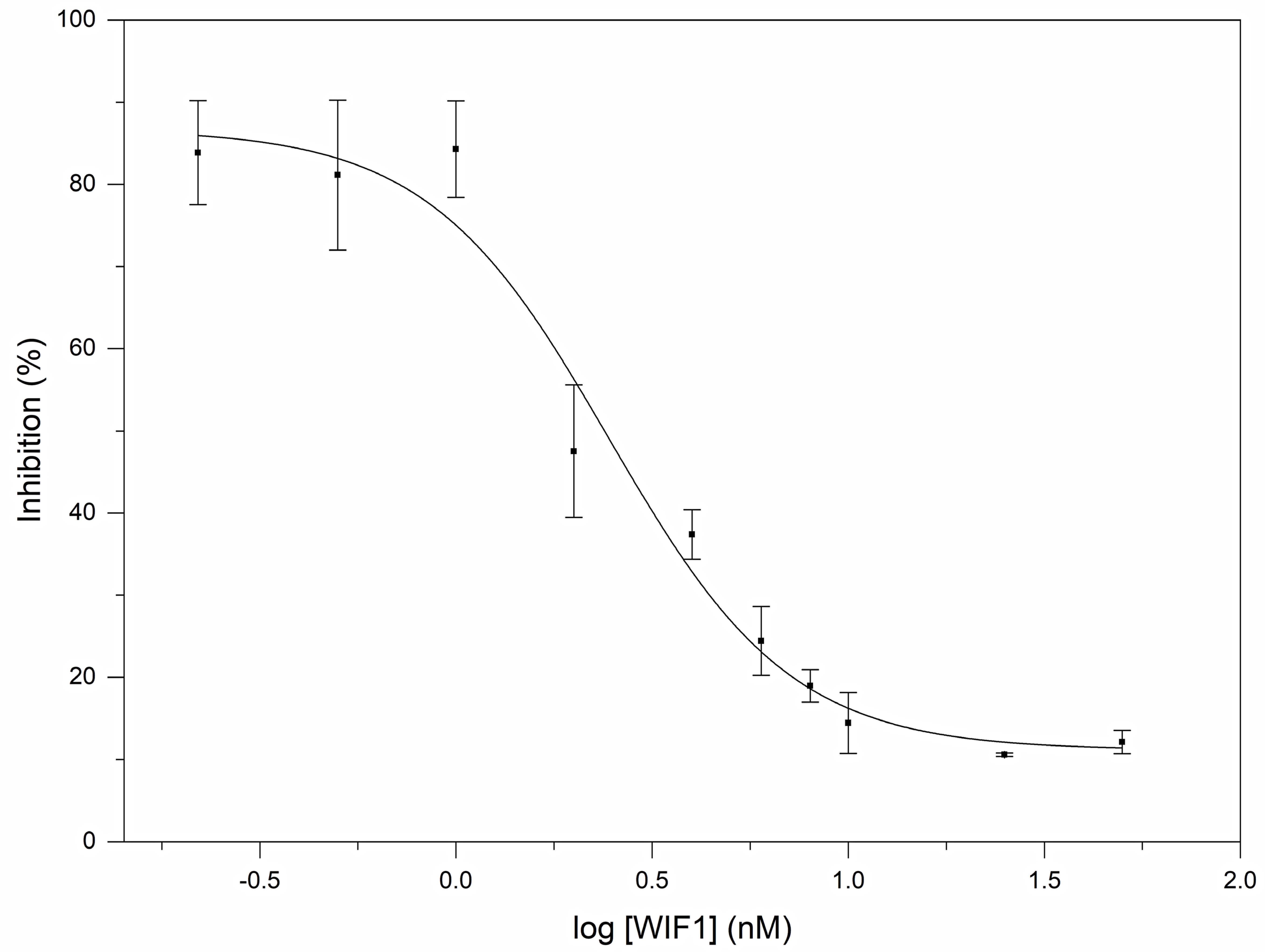
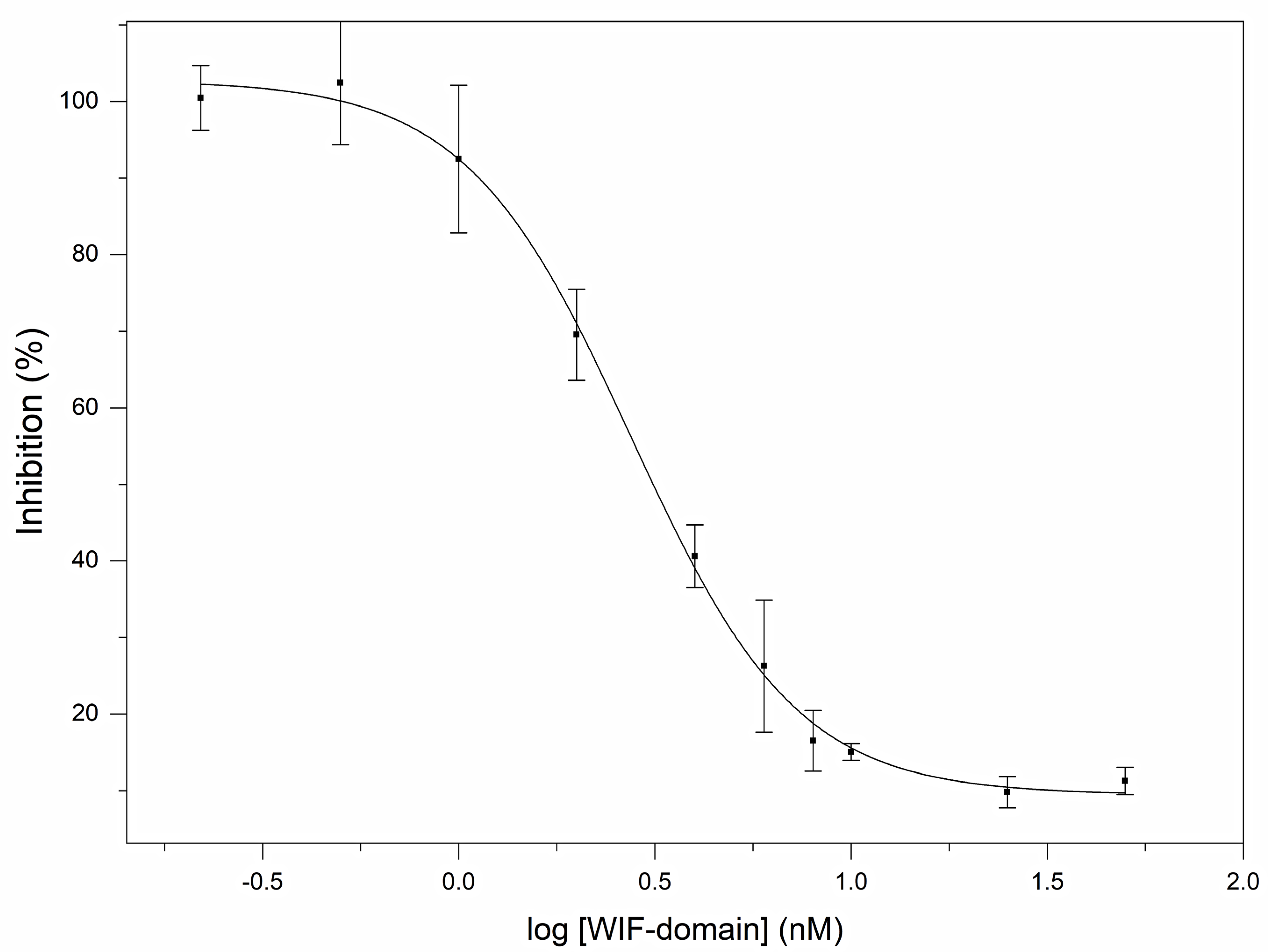
2.4. Reporter Assays
2.5. Protein Analyses
3. Results
3.1. Human Wnt Inhibitory Factor 1 Binds Human Shh with High Affinity
3.2. Human Wnt Inhibitory Factor 1 Is a Potent Antagonist of the Signaling Activity of Human Shh
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Van Amerongen, R.; Nusse, R. Towards an integrated view of Wnt signaling in development. Development 2009, 136, 3205–3214. [Google Scholar] [CrossRef] [Green Version]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog Signaling in Cancer: A Prospective Therapeutic Target for Eradicating Cancer Stem Cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Kalderon, D. Similarities between the Hedgehog and Wnt signaling pathways. Trends Cell Biol. 2002, 12, 523–531. [Google Scholar] [CrossRef]
- Nusse, R. Wnts and Hedgehogs: Lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development 2003, 130, 5297–52305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerekes, K.; Bányai, L.; Trexler, M.; Patthy, L. Structure, function and disease relevance of Wnt inhibitory factor 1, a secreted protein controlling the Wnt and hedgehog pathways. Growth Factors 2019, 37, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Brown, J.D.; Danenberg, E.; Duncan, A.W.; Weissman, I.L.; Reya, T.; Yates, J.R., III; Nusse, R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003, 423, 448–452. [Google Scholar] [CrossRef]
- Doubravska, L.; Krausova, M.; Gradl, D.; Vojtechova, M.; Tumova, L.; Lukas, J.; Valenta, T.; Pospichalova, V.; Fafilek, B.; Plachy, J.; et al. Fatty acid modification of Wnt1 and Wnt3a at serine is prerequisite for lipidation at cysteine and is essential for Wnt signalling. Cell Signal. 2011, 23, 837–848. [Google Scholar] [CrossRef]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [Green Version]
- Buglino, J.A.; Resh, M.D. Palmitoylation of Hedgehog proteins. Vitam. Horm. 2012, 88, 229–252. [Google Scholar] [CrossRef] [Green Version]
- Porter, J.A.; Ekker, S.C.; Park, W.J.; von Kessler, D.P.; Young, K.E.; Chen, C.H.; Ma, Y.; Woods, A.S.; Cotter, R.J.; Koonin, E.V.; et al. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 1996, 86, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Porter, J.A.; Young, K.E.; Beachy, P.A. Cholesterol modification of hedgehog signaling proteins in animal development. Science 1996, 274, 255–259. [Google Scholar] [CrossRef]
- Williams, K.P.; Rayhorn, P.; Chi-Rosso, G.; Garber, E.A.; Strauch, K.L.; Horan, G.S.; Reilly, J.O.; Baker, D.P.; Taylor, F.R.; Koteliansky, V.; et al. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J. Cell Sci. 1999, 112, 4405–4414. [Google Scholar] [CrossRef]
- Kohtz, J.D.; Lee, H.Y.; Gaiano, N.; Segal, J.; Ng, E.; Larson, T.; Baker, D.P.; Garber, E.A.; Williams, K.P.; Fishell, G. N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 2001, 128, 2351–2363. [Google Scholar] [CrossRef]
- Petrova, E.; Rios-Esteves, J.; Ouerfelli, O.; Glickman, J.F.; Resh, M.D. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat. Chem. Biol. 2013, 9, 247–249. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Kypta, R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003, 116, 2627–2634. [Google Scholar] [CrossRef] [Green Version]
- Filipovich, A.; Gehrke, I.; Poll-Wolbeck, S.J.; Kreuzer, K.A. Physiological inhibitors of Wnt signaling. Eur. J. Haematol. 2011, 86, 453–465. [Google Scholar] [CrossRef]
- Cruciat, C.M.; Niehrs, C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 2013, 5, a015081. [Google Scholar] [CrossRef] [Green Version]
- Malinauskas, T.; Jones, E.Y. Extracellular modulators of Wnt signalling. Curr. Opin. Struct. Biol. 2014, 29, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, J.C.; Kodjabachian, L.; Rebbert, M.L.; Rattner, A.; Smallwood, P.M.; Samos, C.H.; Nusse, R.; Dawid, I.B.; Nathans, J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 1999, 398, 431–436. [Google Scholar] [CrossRef]
- Kansara, M.; Tsang, M.; Kodjabachian, L.; Sims, N.A.; Trivett, M.K.; Ehrich, M.; Dobrovic, A.; Slavin, J.; Choong, P.F.; Simmons, P.J.; et al. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J. Clin. Investig. 2009, 119, 837–851. [Google Scholar] [CrossRef] [Green Version]
- Glise, B.; Miller, C.A.; Crozatier, M.; Halbisen, M.A.; Wise, S.; Olson, D.J.; Vincent, A.; Blair, S.S. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev. Cell 2005, 8, 255–266. [Google Scholar] [CrossRef]
- Gorfinkiel, N.; Sierra, J.; Callejo, A.; Ibañez, C.; Guerrero, I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev. Cell. 2005, 8, 241–253. [Google Scholar] [CrossRef]
- Han, C.; Lin, X. Shifted from Wnt to Hedgehog signaling pathways. Mol. Cell 2005, 17, 321–322. [Google Scholar] [CrossRef]
- Sánchez-Hernández, D.; Sierra, J.; Ortigão-Farias, J.R.; Guerrero, I. The WIF domain of the human and Drosophila Wif-1 secreted factors confers specificity for Wnt or Hedgehog. Development 2012, 139, 3849–3858. [Google Scholar] [CrossRef] [Green Version]
- Avanesov, A.; Honeyager, S.M.; Malicki, J.; Blair, S.S. The role of glypicans in Wnt inhibitory factor-1 activity and the structural basis of Wif1’s effects on Wnt and Hedgehog signaling. PLoS Genet. 2012, 8, e1002503. [Google Scholar] [CrossRef] [Green Version]
- Bányai, L.; Kerekes, K.; Patthy, L. Characterization of a Wnt-binding site of the WIF-domain of Wnt inhibitory factor-1. FEBS Lett. 2012, 586, 3122–3126. [Google Scholar] [CrossRef] [Green Version]
- Noubissi, F.K.; Goswami, S.; Sanek, N.A.; Kawakami, K.; Minamoto, T.; Moser, A.; Grinblat, Y.; Spiegelman, V.S. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 2009, 69, 8572–8578. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lin, P.; Wang, Q.; Zheng, M.; Pang, L. Wnt3a-regulated TCF4/β-catenin complex directly activates the key Hedgehog signalling genes Smo and Gli1. Exp. Ther. Med. 2018, 16, 2101–2107. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.; Elmenofi, S.; Taschauer, A.; Anton, M.; Sami, H.; Ogris, M. Luminescent and fluorescent triple reporter plasmid constructs for Wnt, Hedgehog and Notch pathway. PLoS ONE 2019, 14, e0226570. [Google Scholar] [CrossRef] [PubMed]
- Dennler, S.; André, J.; Alexaki, I.; Li, A.; Magnaldo, T.; ten Dijke, P.; Wang, X.J.; Verrecchia, F.; Mauviel, A. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007, 67, 6981–6986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niewiadomski, P.; Niedziółka, S.M.; Markiewicz, Ł.; Uśpieński, T.; Baran, B.; Chojnowska, K. Gli Proteins: Regulation in Development and Cancer. Cells 2019, 8, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrobono, S.; Gagliardi, S.; Stecca, B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front. Genet. 2019, 10, 556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Reyniès, A.; Javelaud, D.; Elarouci, N.; Marsaud, V.; Gilbert, C.; Mauviel, A. Large-scale pan-cancer analysis reveals broad prognostic association between TGF-β ligands, not Hedgehog, and GLI1/2 expression in tumors. Sci. Rep. 2020, 10, 14491. [Google Scholar] [CrossRef] [PubMed]
- Liepinsh, E.; Bányai, L.; Patthy, L.; Otting, G. NMR structure of the WIF domain of the human Wnt-inhibitory factor-1. J. Mol. Biol. 2006, 357, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Janda, C.Y.; Waghray, D.; Levin, A.M.; Thomas, C.; Garcia, K.C. Structural basis of Wnt recognition by Frizzled. Science 2012, 337, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Schmiege, P.; Coutavas, E.; Wang, J.; Li, X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 2018, 560, 128–132. [Google Scholar] [CrossRef]
- Rudolf, A.F.; Kinnebrew, M.; Kowatsch, C.; Ansell, T.B.; El Omari, K.; Bishop, B.; Pardon, E.; Schwab, R.A.; Malinauskas, T.; Qian, M.; et al. The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat. Chem. Biol. 2019, 15, 975–982. [Google Scholar] [CrossRef]
- Ashe, H.L.; Levine, M. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature 1999, 398, 427–431. [Google Scholar] [CrossRef]
- O’Connor, M.B.; Umulis, D.; Othmer, H.G.; Blair, S.S. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 2006, 133, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Wierbowski, B.M.; Petrov, K.; Aravena, L.; Gu, G.; Xu, Y.; Salic, A. Hedgehog Pathway Activation Requires Coreceptor-Catalyzed, Lipid-Dependent Relay of the Sonic Hedgehog Ligand. Dev. Cell 2020, 23, 450–467. [Google Scholar] [CrossRef]
- Mo, R.; Kim, J.H.; Zhang, J.; Chiang, C.; Hui, C.C.; Kim, P.C. Anorectal malformations caused by defects in sonic hedgehog signaling. Am. J. Pathol. 2001, 159, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Ng, R.C.; Matsumaru, D.; Ho, A.S.; Garcia-Barceló, M.M.; Yuan, Z.W.; Smith, D.; Kodjabachian, L.; Tam, P.K.; Yamada, G.; Lui, V.C. Dysregulation of Wnt inhibitory factor 1 (Wif1) expression resulted in aberrant Wnt-β-catenin signaling and cell death of the cloaca endoderm, and anorectal malformations. Cell Death Differ. 2014, 21, 978–989. [Google Scholar] [CrossRef] [Green Version]
- Epstein, E.H. Basal cell carcinomas: Attack of the hedgehog. Nat. Rev. Cancer 2008, 8, 743–754. [Google Scholar] [CrossRef]
- Grachtchouk, V.; Grachtchouk, M.; Lowe, L.; Johnson, T.; Wei, L.; Wang, A.; de Sauvage, F.; Dlugosz, A.A. The magnitude of hedgehog signaling activity defines skin tumor phenotype. EMBO J. 2003, 22, 2741–2751. [Google Scholar] [CrossRef] [Green Version]
- Schlüter, H.; Stark, H.J.; Sinha, D.; Boukamp, P.; Kaur, P. WIF1 is expressed by stem cells of the human interfollicular epidermis and acts to suppress keratinocyte proliferation. J. Investig. Dermatol. 2013, 133, 1669–1673. [Google Scholar] [CrossRef] [Green Version]
- Becker, M.; Bauer, J.; Pyczek, J.; König, S.; Müllen, A.; Rabe, H.; Schön, M.P.; Uhmann, A.; Hahn, H. WIF1 Suppresses the Generation of Suprabasal Cells in Acanthotic Skin and Growth of Basal Cell Carcinomas upon Forced Overexpression. J. Investig. Dermatol. 2020, 140, 1556–1565. [Google Scholar] [CrossRef]
- So, P.L.; Langston, A.W.; Daniallinia, N.; Hebert, J.L.; Fujimoto, M.A.; Khaimskiy, Y.; Aszterbaum, M.; Epstein, E.H., Jr. Long-term establishment, characterization and manipulation of cell lines from mouse basal cell carcinoma tumors. Exp. Dermatol. 2006, 15, 742–750. [Google Scholar] [CrossRef]
- Alfaro, A.C.; Roberts, B.; Kwong, L.; Bijlsma, M.F.; Roelink, H. Ptch2 mediates the Shh response in Ptch1−/− cells. Development 2014, 141, 3331–3339. [Google Scholar] [CrossRef] [Green Version]
- Veenstra, V.L.; Dingjan, I.; Waasdorp, C.; Damhofer, H.; van der Wal, A.C.; van Laarhoven, H.W.; Medema, J.P.; Bijlsma, M.F. Patched-2 functions to limit Patched-1 deficient skin cancer growth. Cell Oncol. 2018, 41, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Lee, J.; Seppala, M.; Cobourne, M.T.; Kim, S.H. Ptch2/Gas1 and Ptch1/Boc differentially regulate Hedgehog signalling in murine primordial germ cell migration. Nat. Commun. 2020, 11, 1994, Erratum in 2020, 11, 2275. [Google Scholar] [CrossRef] [Green Version]
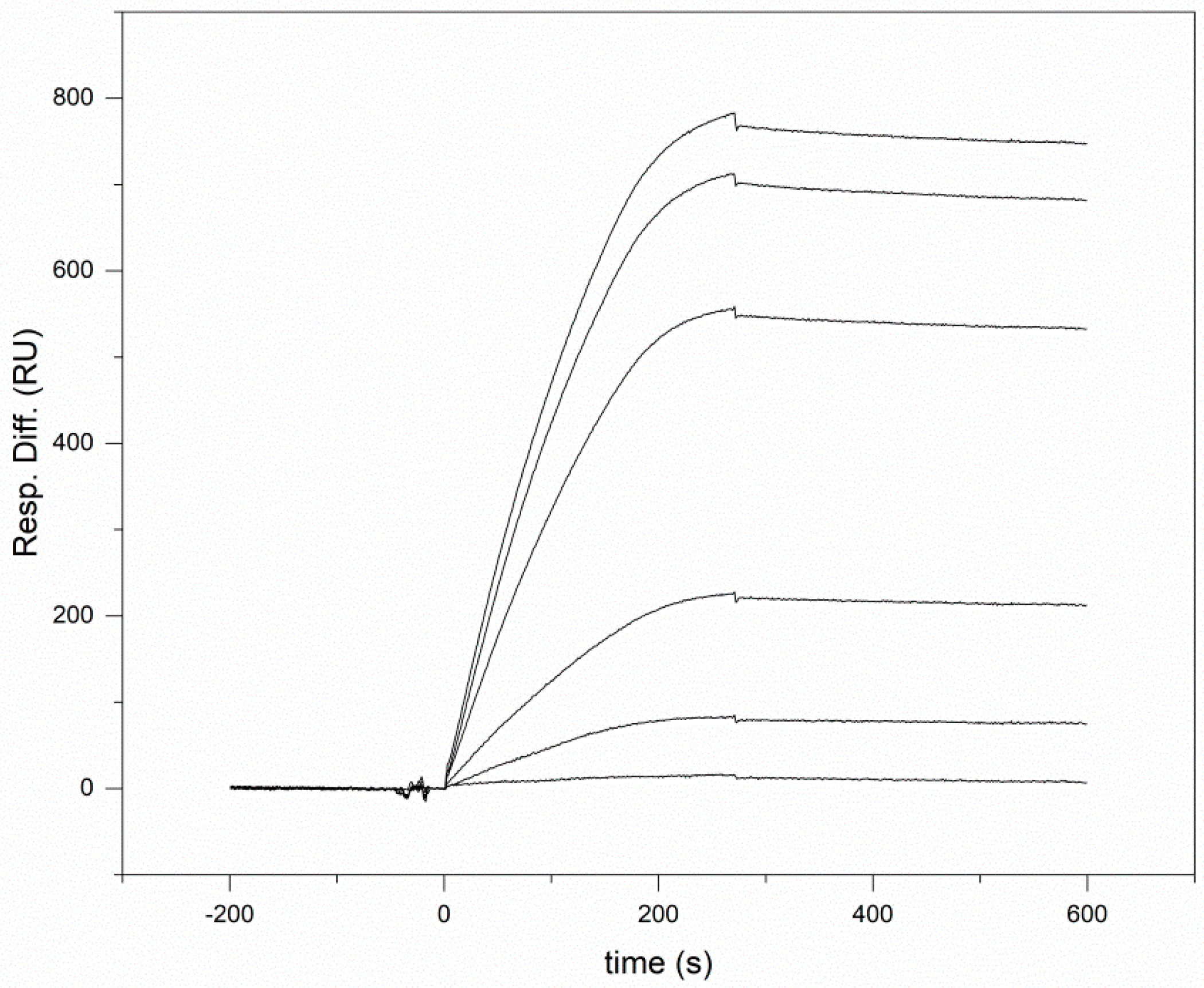

| Interacting Proteins | Kd (nM) | ka (1/Ms) | kd (1/s) |
|---|---|---|---|
| Shh—WIF1 | 2.06 ± 0.9 | 6.85 ± 0.6 × 104 | 1.41 ± 0.5 ×10−4 |
| Shh—WIF domain of WIF1 | 1.18 ± 0.3 | 4.26 ± 0.2 × 104 | 5.03 ± 1.3 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerekes, K.; Trexler, M.; Bányai, L.; Patthy, L. Wnt Inhibitory Factor 1 Binds to and Inhibits the Activity of Sonic Hedgehog. Cells 2021, 10, 3496. https://doi.org/10.3390/cells10123496
Kerekes K, Trexler M, Bányai L, Patthy L. Wnt Inhibitory Factor 1 Binds to and Inhibits the Activity of Sonic Hedgehog. Cells. 2021; 10(12):3496. https://doi.org/10.3390/cells10123496
Chicago/Turabian StyleKerekes, Krisztina, Mária Trexler, László Bányai, and László Patthy. 2021. "Wnt Inhibitory Factor 1 Binds to and Inhibits the Activity of Sonic Hedgehog" Cells 10, no. 12: 3496. https://doi.org/10.3390/cells10123496
APA StyleKerekes, K., Trexler, M., Bányai, L., & Patthy, L. (2021). Wnt Inhibitory Factor 1 Binds to and Inhibits the Activity of Sonic Hedgehog. Cells, 10(12), 3496. https://doi.org/10.3390/cells10123496







