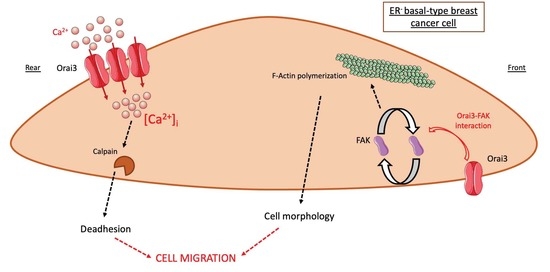
Orai3 Calcium Channel Regulates Breast Cancer Cell Migration through Calcium-Dependent and -Independent Mechanisms
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. Transient Transfections
2.3. Quantitative Real-Time PCR (qRT-PCR)
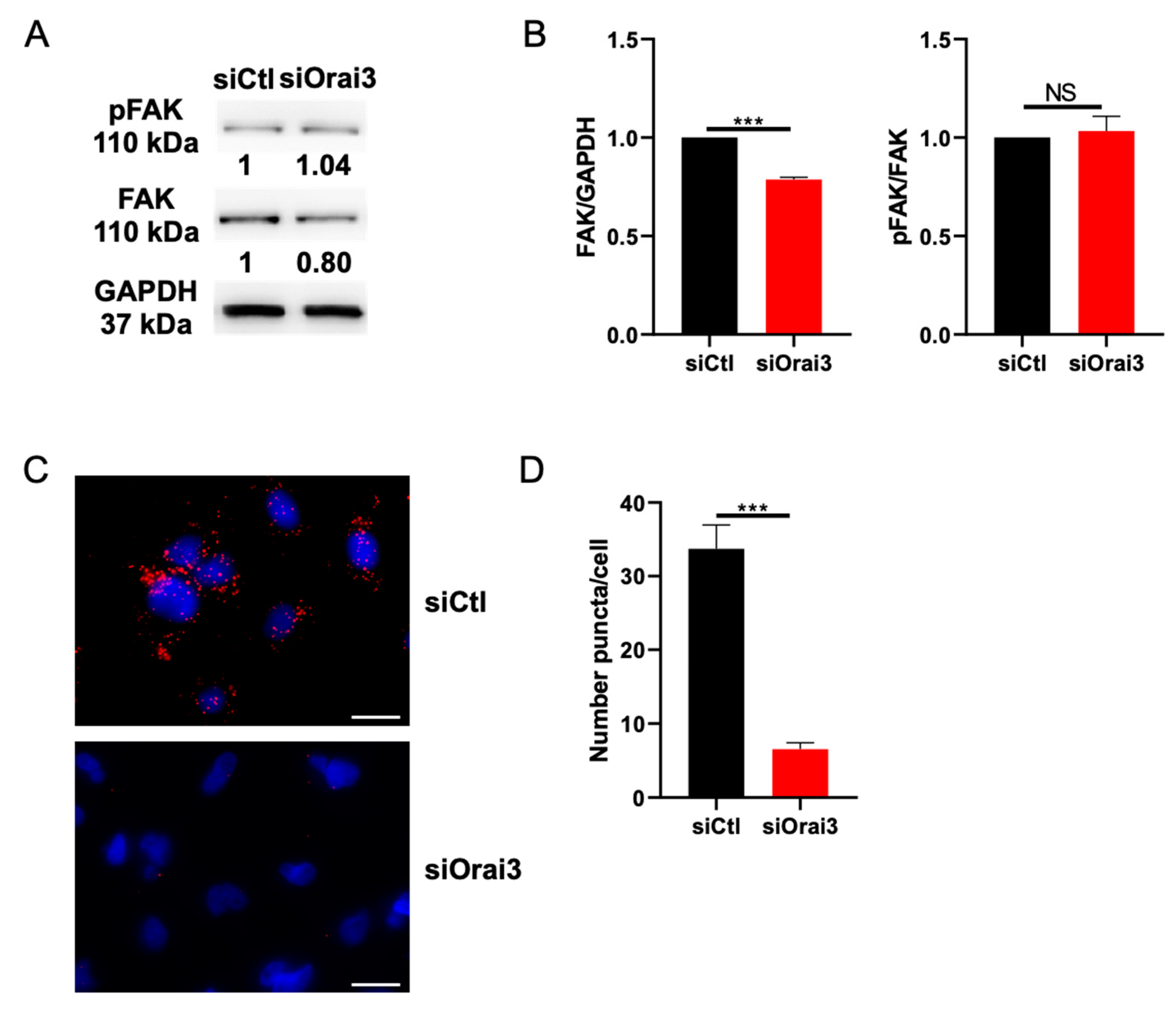
2.4. Western Blot Analysis
2.5. Cell Migration
2.6. Cell Adhesion Assays
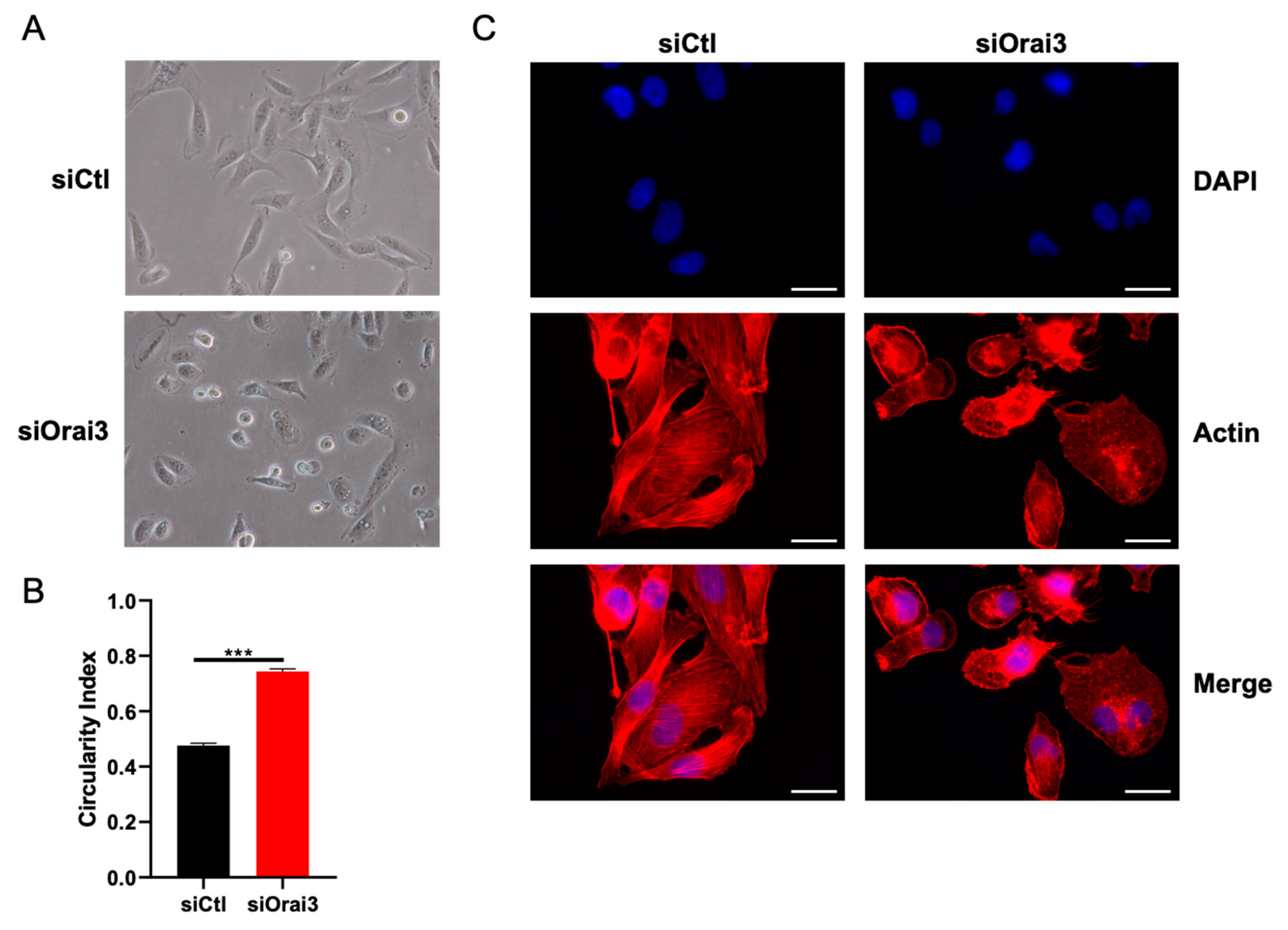
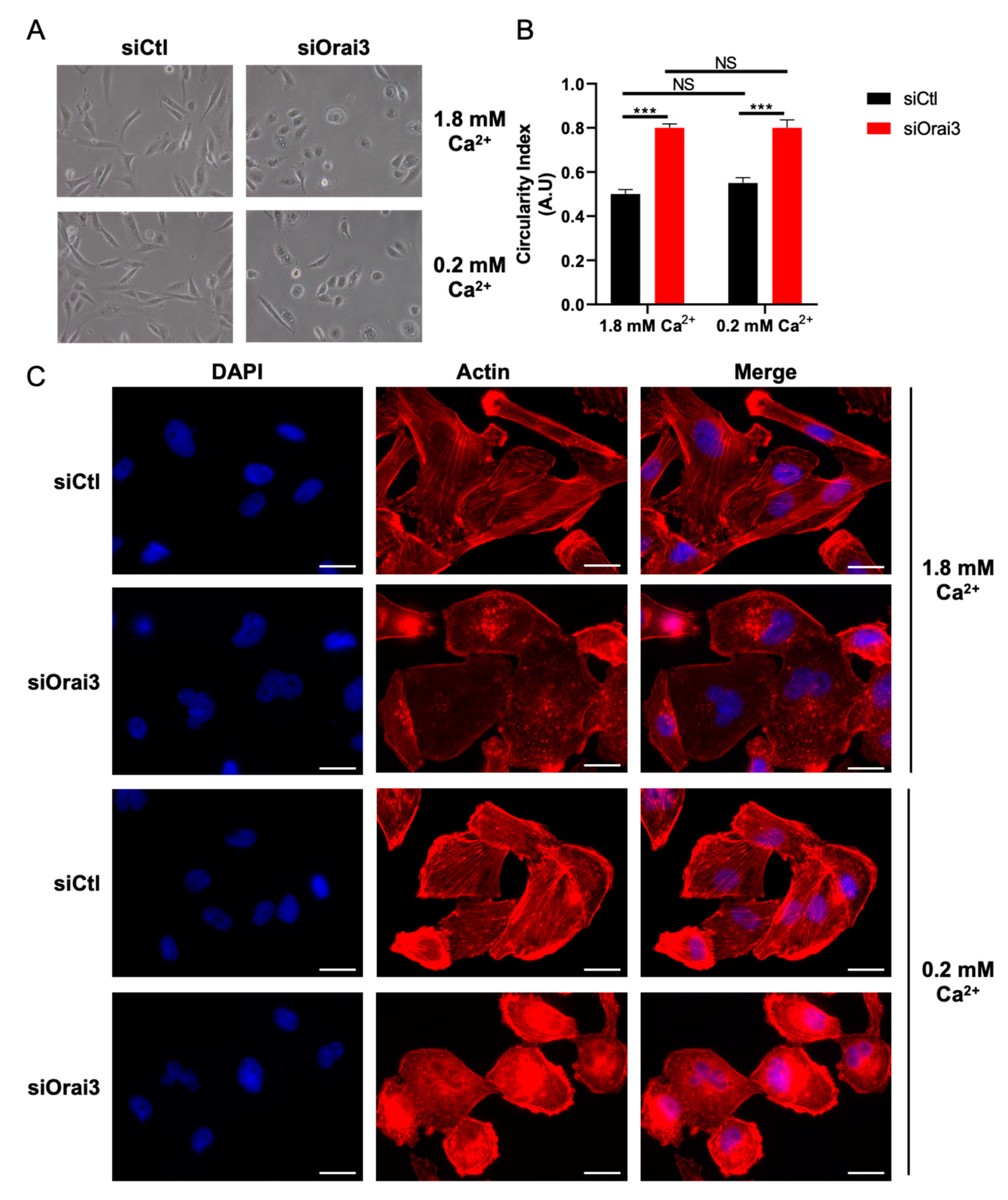
2.7. Cell Morphology
2.8. Immunofluorescence
2.9. Measurement of Calpain Activity
2.10. Calcium Imaging
2.11. Proximity Ligation Assay
2.12. Statistical Analysis
3. Results
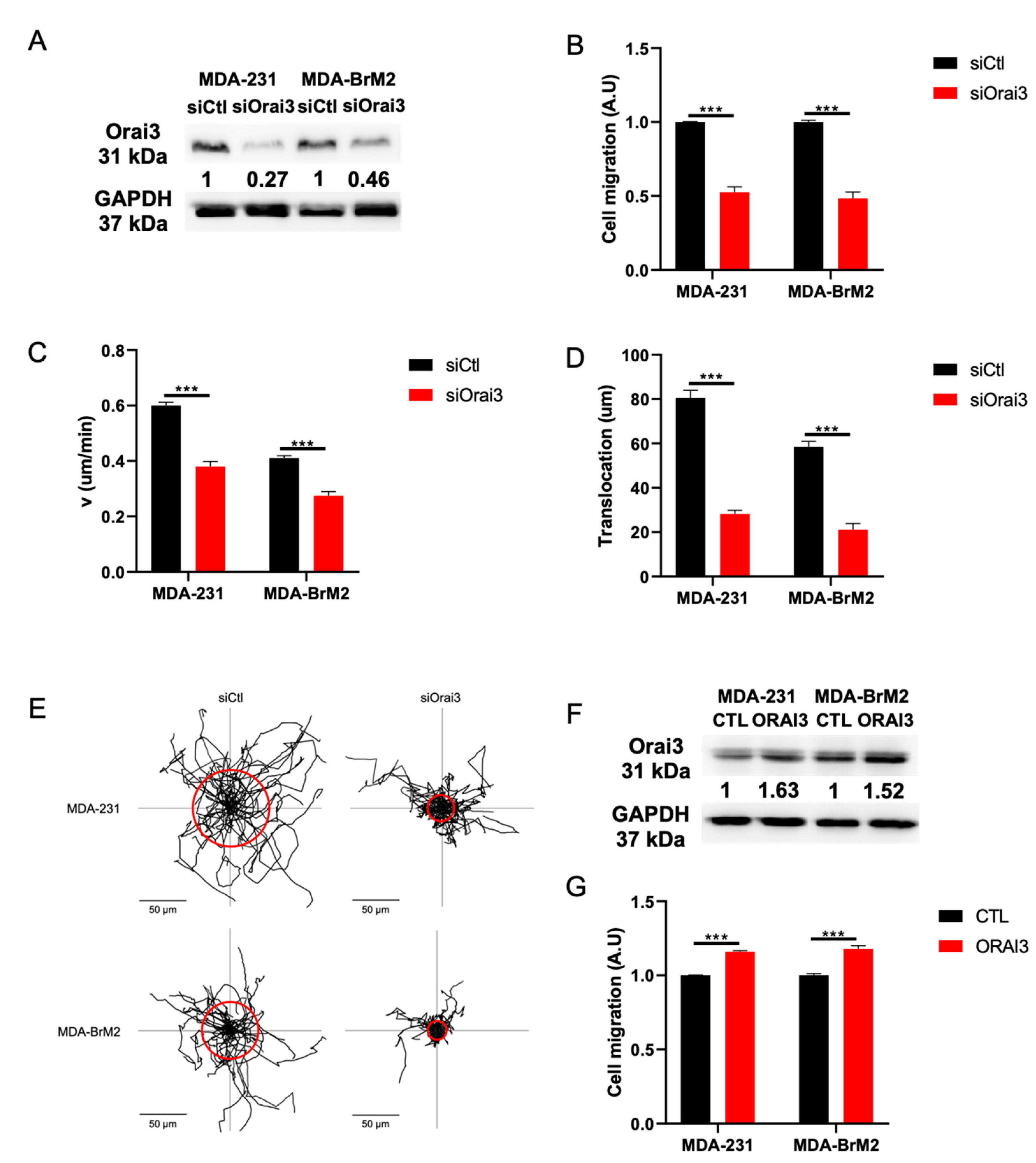
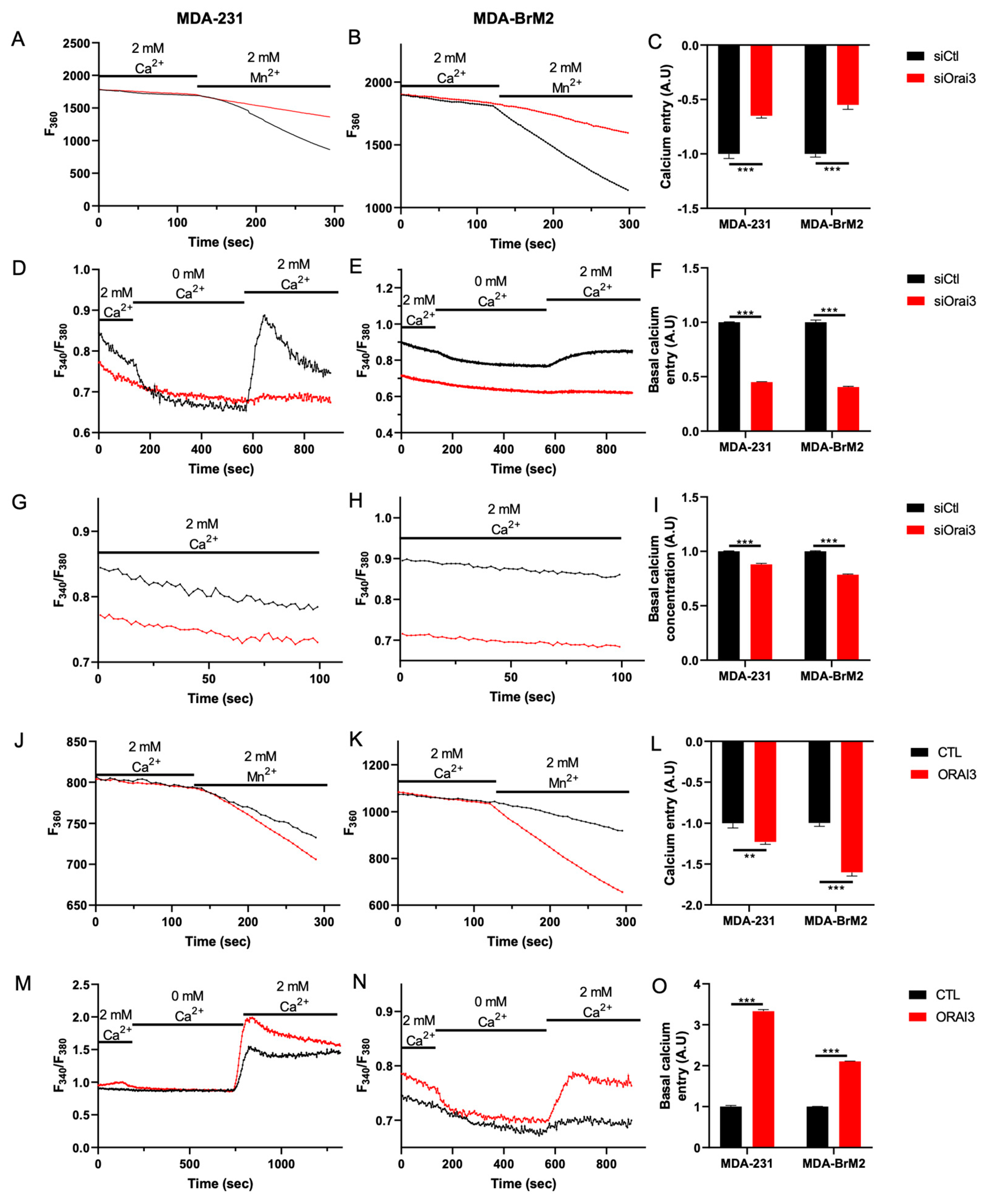
3.1. The Orai3 Ca2+ Channel Regulates Cell Migration and Is Activated at a Basal Level inMDA-231 and MDA-BrM2 Breast Cancer Cells
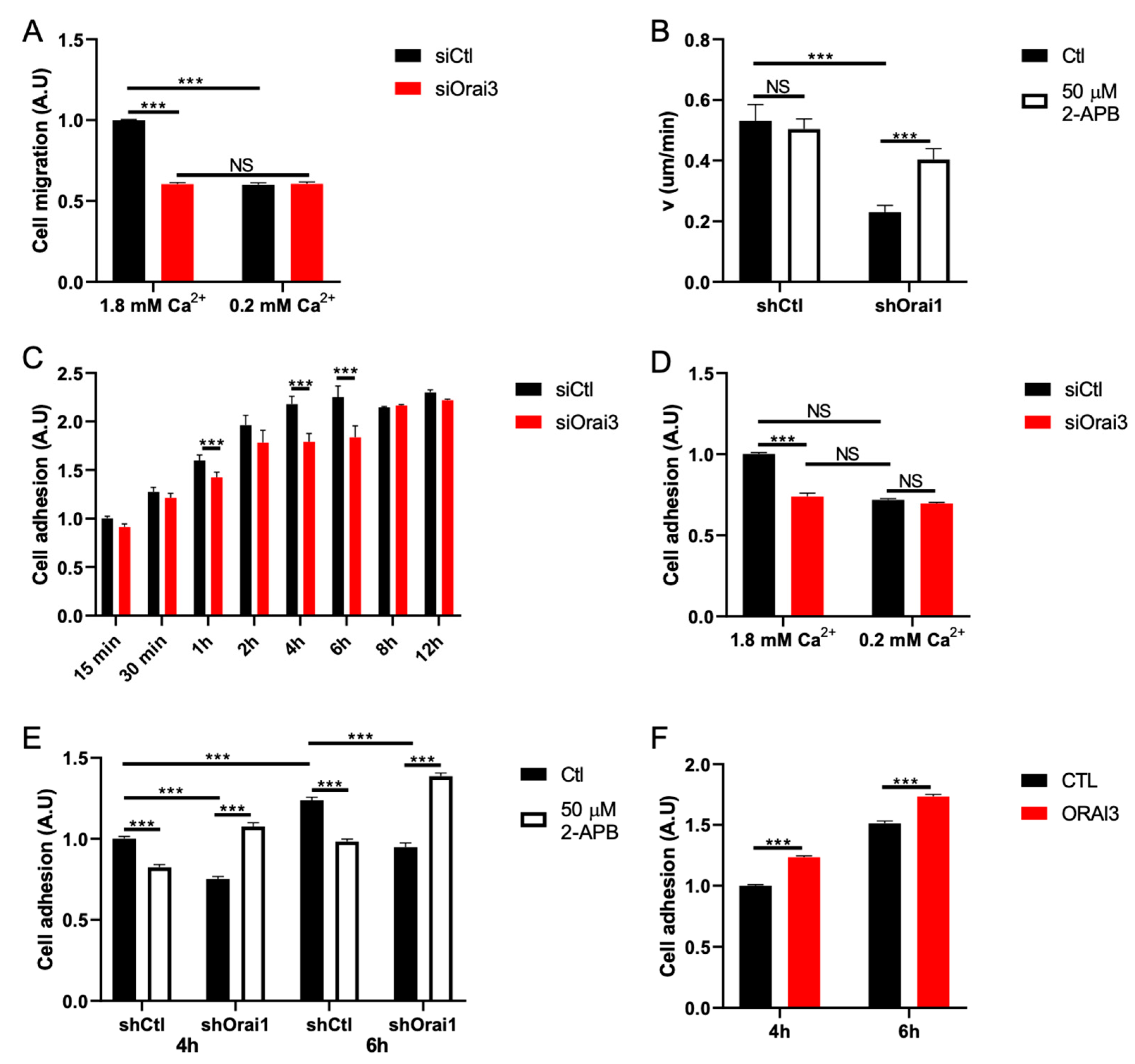
3.2. Orai3 Regulates Cell Migration in a Ca2+-Dependent Manner via a Modulation of Cell Adhesive Capacities
3.3. Orai3, by Regulating Ca2+ Entry, Controls Calpain Activity
3.4. Orai3 Maintains an Elongated Cell Morphology through a Ca2+-Independent Mechanism
3.5. Orai3 Interacts with FAK and Regulates Its Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Gupta, G.P.; Massague, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peart, O. Metastatic Breast Cancer. Radiol. Technol. 2017, 88, 519M–539M. [Google Scholar] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horwitz, R.; Webb, D. Cell migration. Curr. Biol. 2003, 13, R756–R759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, H.; Wyckoff, J.; Condeelis, J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005, 17, 559–564. [Google Scholar] [CrossRef]
- Ciobanasu, C.; Faivre, B.; Le Clainche, C. Actin dynamics associated with focal adhesions. Int. J. Cell Biol. 2012, 2012, 941292. [Google Scholar] [CrossRef] [Green Version]
- Lebart, M.C.; Benyamin, Y. Calpain involvement in the remodeling of cytoskeletal anchorage complexes. FEBS J. 2006, 273, 3415–3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef] [Green Version]
- Leloup, L.; Wells, A. Calpains as potential anti-cancer targets. Expert Opin. Ther. Targets 2011, 15, 309–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuidema, A.; Wang, W.; Sonnenberg, A. Crosstalk between Cell Adhesion Complexes in Regulation of Mechanotransduction. Bioessays 2020, 42, e2000119. [Google Scholar] [CrossRef]
- Deliot, N.; Constantin, B. Plasma membrane calcium channels in cancer: Alterations and consequences for cell proliferation and migration. Biochim. Biophys. Acta 2015, 1848, 2512–2522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammad, A.S.; Machaca, K. Store Operated Calcium Entry in Cell Migration and Cancer Metastasis. Cells 2021, 10, 1246. [Google Scholar] [CrossRef] [PubMed]
- Pratt, S.J.P.; Hernandez-Ochoa, E.; Martin, S.S. Calcium signaling: Breast cancer’s approach to manipulation of cellular circuitry. Biophys. Rev. 2020, 12, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.C.; Kuo, G.H.; Chang, S.W.; Tsai, P.J. Ca2+ signaling in cytoskeletal reorganization, cell migration, and cancer metastasis. Biomed. Res. Int. 2015, 2015, 409245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.-H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Burstein, S.R.; Long, S.B. Structures reveal opening of the store-operated calcium channel Orai. eLife 2018, 7, e36758. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wu, G.; Yu, Y.; Chen, X.; Ji, R.; Lu, J.; Li, X.; Zhang, X.; Yang, X.; Shen, Y. Molecular understanding of calcium permeation through the open Orai channel. PLoS Biol. 2019, 17, e3000096. [Google Scholar] [CrossRef] [PubMed]
- Trebak, M.; Putney, J.W., Jr. ORAI Calcium Channels. Physiology 2017, 32, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Hammadi, M.; Chopin, V.; Matifat, F.; Dhennin-Duthille, I.; Chasseraud, M.; Sevestre, H.; Ouadid-Ahidouch, H. Human ether a-gogo K+ channel 1 (hEag1) regulates MDA-MB-231 breast cancer cell migration through Orai1-dependent calcium entry. J. Cell. Physiol. 2012, 227, 3837–3846. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, J.J.; Huang, X.Y. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faouzi, M.; Hague, F.; Potier, M.; Ahidouch, A.; Sevestre, H.; Ouadid-Ahidouch, H. Down-regulation of Orai3 arrests cell-cycle progression and induces apoptosis in breast cancer cells but not in normal breast epithelial cells. J. Cell. Physiol. 2011, 226, 542–551. [Google Scholar] [CrossRef]
- Faouzi, M.; Kischel, P.; Hague, F.; Ahidouch, A.; Benzerdjeb, N.; Sevestre, H.; Penner, R.; Ouadid-Ahidouch, H. ORAI3 silencing alters cell proliferation and cell cycle progression via c-myc pathway in breast cancer cells. Biochim. Biophys. Acta 2013, 1833, 752–760. [Google Scholar] [CrossRef] [Green Version]
- Hasna, J.; Hague, F.; Despoix, L.; Geerts, D.; Leroy, C.; Tulasne, D.; Ouadid-Ahidouch, H.; Kischel, P. Orai3 calcium channel and resistance to chemotherapy in breast cancer cells: The p53 connection. Cell Death Differ. 2018, 25, 693–707. [Google Scholar] [CrossRef]
- Motiani, R.K.; Zhang, X.; Harmon, K.E.; Keller, R.S.; Matrougui, K.; Bennett, J.A.; Trebak, M. Orai3 is an estrogen receptor α-regulated Ca2+ channel that promotes tumorigenesis. FASEB J. 2013, 27, 63–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bos, P.D.; Zhang, X.H.F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Badaoui, M.; Mimsy-Julienne, C.; Saby, C.; VAN Gulick, L.; Peretti, M.; Jeannesson, P.; Morjani, H.; Ouadid-Ahidouch, H. Collagen type 1 promotes survival of human breast cancer cells by overexpressing Kv10.1 potassium and Orai1 calcium channels through DDR1-dependent pathway. Oncotarget 2018, 9, 24653–24671. [Google Scholar] [CrossRef]
- Radoslavova, S.; Folcher, A.; Lefebvre, T.; Kondratska, K.; Guénin, S.; Dhennin-Duthille, I.; Gautier, M.; Prevarskaya, N.; Ouadid-Ahidouch, H. Orai1 Channel Regulates Human-Activated Pancreatic Stellate Cell Proliferation and TGFβ1 Secretion through the AKT Signaling Pathway. Cancers 2021, 13, 2395. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, P.; Klages, R.; Preuss, R.; Schwab, A. Anomalous dynamics of cell migration. Proc. Natl. Acad. Sci. USA 2008, 105, 459–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosser, B.G.; Powers, S.P.; Gores, G.J. Calpain activity increases in hepatocytes following addition of ATP. Demonstration by a novel fluorescent approach. J. Biol. Chem. 1993, 268, 23593–23600. [Google Scholar] [CrossRef]
- Schindl, R.; Bergsmann, J.; Frischauf, I.; Derler, I.; Fahrner, M.; Muik, M.; Fritsch, R.; Groschner, K.; Romanin, C. 2-aminoethoxydiphenyl borate alters selectivity of Orai3 channels by increasing their pore size. J. Biol. Chem. 2008, 283, 20261–20267. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Xin, P.; Yoast, R.E.; Emrich, S.M.; Johnson, M.T.; Pathak, T.; Benson, J.C.; Azimi, I.; Gill, D.L.; Monteith, G.R.; et al. Distinct pharmacological profiles of ORAI1, ORAI2, and ORAI3 channels. Cell Calcium 2020, 91, 102281. [Google Scholar] [CrossRef]
- De Pascalis, C.; Etienne-Manneville, S. Single and collective cell migration: The mechanics of adhesions. Mol. Biol. Cell 2017, 28, 1833–1846. [Google Scholar] [CrossRef]
- Huttenlocher, A.; Sandborg, R.R.; Horwitz, A.F. Adhesion in cell migration. Current opinion in cell biology 1995, 7, 697–706. [Google Scholar] [CrossRef]
- Franco, S.J.; Huttenlocher, A. Regulating cell migration: Calpains make the cut. J. Cell Sci. 2005, 118, 3829–3838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, A.; Huttenlocher, A.; Lauffenburger, D.A. Calpain proteases in cell adhesion and motility. Int. Rev. Cytol. 2005, 245, 1–16. [Google Scholar]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Vautrin-Glabik, A.; Botia, B.; Kischel, P.; Ouadid-Ahidouch, H.; Rodat-Despoix, L. IP3R3 silencing induced actin cytoskeletal reorganization through ARHGAP18/RhoA/mDia1/FAK pathway in breast cancer cell lines. Biochimica et biophysica acta. Mol. Cell Res. 2018, 1865, 945–958. [Google Scholar]
- Pardo-Pastor, C.; Rubio-Moscardo, F.; Vogel-González, M.; Serra, S.A.; Afthinos, A.; Mrkonjic, S.; Destaing, O.; Abenza, J.F.; Fernández-Fernández, J.M.; Trepat, X.; et al. Piezo2 channel regulates RhoA and actin cytoskeleton to promote cell mechanobiological responses. Proc. Natl. Acad. Sci. USA 2018, 115, 1925–1930. [Google Scholar] [CrossRef] [Green Version]
- Sieg, D.J.; Hauck, C.R.; Schlaepfer, D.D. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J. Cell Sci. 1999, 112, 2677–2691. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, M.A.; Serrels, B.; Fincham, V.J.; Frame, M.C.; Carragher, N.O. SRC-mediated phosphorylation of focal adhesion kinase couples actin and adhesion dynamics to survival signaling. Mol. Cell Biol. 2004, 24, 8113–8133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.F.; Wei, L.; Zhou, X.; Lu, Z.-Y.; Francis, K.; Hu, X.-Y.; Liu, Y.; Xiong, W.-C.; Zhang, X.; Banik, N.L.; et al. Formation of Kv2.1-FAK complex as a mechanism of FAK activation, cell polarization and enhanced motility. J. Cell. Physiol. 2008, 217, 544–557. [Google Scholar] [CrossRef] [Green Version]
- Chalmers, S.B.; Monteith, G.R. ORAI channels and cancer. Cell Calcium 2018, 74, 160–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallinger, M.; Berlansky, S.; Frischauf, I. Orai channels: Key players in Ca2+ homeostasis. Curr. Opin. Physiol. 2020, 17, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Fiorio Pla, A.; Kondratska, K.; Prevarskaya, N. STIM and ORAI proteins: Crucial roles in hallmarks of cancer. Am. J. Physiol. Cell Physiol. 2016, 310, C509–C519. [Google Scholar] [CrossRef]
- Motiani, R.K.; Abdullaev, I.F.; Trebak, M. A novel native store-operated calcium channel encoded by Orai3: Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. The J. Biol. Chem. 2010, 285, 19173–19183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, I.; Milevskiy, M.J.G.; Chalmers, S.B.; Yapa, K.T.D.S.; Robitaille, M.; Henry, C.; Baillie, G.J.; Thompson, E.W.; Roberts-Thomson, S.J.; Monteith, G.R. ORAI1 and ORAI3 in Breast Cancer Molecular Subtypes and the Identification of ORAI3 as a Hypoxia Sensitive Gene and a Regulator of Hypoxia Responses. Cancers 2019, 11, 208. [Google Scholar] [CrossRef] [Green Version]
- Maziveyi, M.; Alahari, S.K. Cell matrix adhesions in cancer: The proteins that form the glue. Oncotarget 2017, 8, 48471–48487. [Google Scholar] [CrossRef] [Green Version]
- Greenwood, J.A.; Murphy-Ullrich, J.E. Signaling of de-adhesion in cellular regulation and motility. Microsc. Res. Tech. 1998, 43, 420–432. [Google Scholar] [CrossRef]
- Croce, K.; Flaumenhaft, R.; Rivers, M.; Furie, B.; Furie, B.C.; Herman, I.M.; Potter, D.A. Inhibition of calpain blocks platelet secretion, aggregation, and spreading. J. Biol. Chem. 1999, 274, 36321–36327. [Google Scholar] [CrossRef] [Green Version]
- Huttenlocher, A.; Palecek, S.P.; Lu, Q.; Zhang, W.; Mellgren, R.L.; Lauffenburger, D.A.; Ginsberg, M.H.; Horwitz, A.F. Regulation of cell migration by the calcium-dependent protease calpain. J. Biol. Chem. 1997, 272, 32719–32722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potter, D.A.; Tirnauer, J.S.; Janssen, R.; Croall, D.E.; Hughes, C.N.; Fiacco, K.A.; Mier, J.W.; Maki, M.; Herman, I.M. Calpain regulates actin remodeling during cell spreading. J. Cell Biol. 1998, 141, 647–662. [Google Scholar] [CrossRef]
- Chantome, A.; Potier-Cartereau, M.; Clarysse, L.; Fromont, G.; Lambot, S.M.; Guéguinou, M.; Pages, J.-C.; Collin, C.; Oullier, T.; Girault, A.; et al. Pivotal role of the lipid Raft SK3-Orai1 complex in human cancer cell migration and bone metastases. Cancer Res. 2013, 73, 4852–4861. [Google Scholar] [CrossRef] [Green Version]
- Gueguinou, M.; Crottès, D.; Chantôme, A.; Rapetti-Mauss, R.; Potier-Cartereau, M.; Clarysse, L.; Girault, A.; Fourbon, Y.; Jézéquel, P.; Guérin-Charbonnel, C.; et al. The SigmaR1 chaperone drives breast and colorectal cancer cell migration by tuning SK3-dependent Ca2+ homeostasis. Oncogene 2017, 36, 3640–3647. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K.; Guilluy, C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridley, A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 2015, 36, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Ballestrem, C.; Kam, Z.; Geiger, B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003, 116, 4605–4613. [Google Scholar] [CrossRef] [Green Version]
- Roa-Espitia, A.L.; Hernández-Rendón, E.R.; Baltiérrez-Hoyos, R.; Muñoz-Gotera, R.J.; Cote-Vélez, A.; Jiménez, I.; González-Márquez, H.; Hernández-González, E.O. Focal adhesion kinase is required for actin polymerization and remodeling of the cytoskeleton during sperm capacitation. Biol. Open 2016, 5, 1189–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salgado-Lucio, M.L.; Ramírez-Ramírez, D.; Jorge-Cruz, C.Y.; Roa-Espitia, A.L.; Hernández-González, E.O. FAK regulates actin polymerization during sperm capacitation via the ERK2/GEF-H1/RhoA signaling pathway. J. Cell Sci. 2020, 133, jcs239186. [Google Scholar] [CrossRef]
- Serrels, B.; Serrels, A.; Brunton, V.G.; Holt, M.; McLean, G.W.; Gray, C.H.; Jones, G.E.; Frame, M.C. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat. Cell Biol. 2007, 9, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Mruk, D.D.; Cheng, C.Y. Focal adhesion kinase is a regulator of F-actin dynamics: New insights from studies in the testis. Spermatogenesis 2013, 3, e25385. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, A.; Hofmann, G.; Pillozzi, S.; Guasti, L.; Crociani, O.; Cilia, E.; Di Stefano, P.; Degani, S.; Balzi, S.; Olivotto, M.; et al. Human ether-a-go-go-related gene 1 channels are physically linked to β1 integrins and modulate adhesion-dependent signaling. Mol. Biol. Cell 2005, 16, 2972–2983. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamlali, M.; Kouba, S.; Rodat-Despoix, L.; Todesca, L.M.; Pethö, Z.; Schwab, A.; Ouadid-Ahidouch, H. Orai3 Calcium Channel Regulates Breast Cancer Cell Migration through Calcium-Dependent and -Independent Mechanisms. Cells 2021, 10, 3487. https://doi.org/10.3390/cells10123487
Chamlali M, Kouba S, Rodat-Despoix L, Todesca LM, Pethö Z, Schwab A, Ouadid-Ahidouch H. Orai3 Calcium Channel Regulates Breast Cancer Cell Migration through Calcium-Dependent and -Independent Mechanisms. Cells. 2021; 10(12):3487. https://doi.org/10.3390/cells10123487
Chicago/Turabian StyleChamlali, Mohamed, Sana Kouba, Lise Rodat-Despoix, Luca Matteo Todesca, Zoltán Pethö, Albrecht Schwab, and Halima Ouadid-Ahidouch. 2021. "Orai3 Calcium Channel Regulates Breast Cancer Cell Migration through Calcium-Dependent and -Independent Mechanisms" Cells 10, no. 12: 3487. https://doi.org/10.3390/cells10123487
APA StyleChamlali, M., Kouba, S., Rodat-Despoix, L., Todesca, L. M., Pethö, Z., Schwab, A., & Ouadid-Ahidouch, H. (2021). Orai3 Calcium Channel Regulates Breast Cancer Cell Migration through Calcium-Dependent and -Independent Mechanisms. Cells, 10(12), 3487. https://doi.org/10.3390/cells10123487