IL-1β and TNFα Cooperativity in Regulating IL-6 Expression in Adipocytes Depends on CREB Binding and H3K14 Acetylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Differentiation of 3T3-L1 Adipocytes
2.2. Differentiation of Human Adipocytes
2.3. Nile Red Staining of Lipids
2.4. Real-Time RT-PCR
2.5. ELISA
2.6. Confocal Microscopy
2.7. Chromatin Immunoprecipitation-qPCR
2.8. Statistical Analysis
3. Results
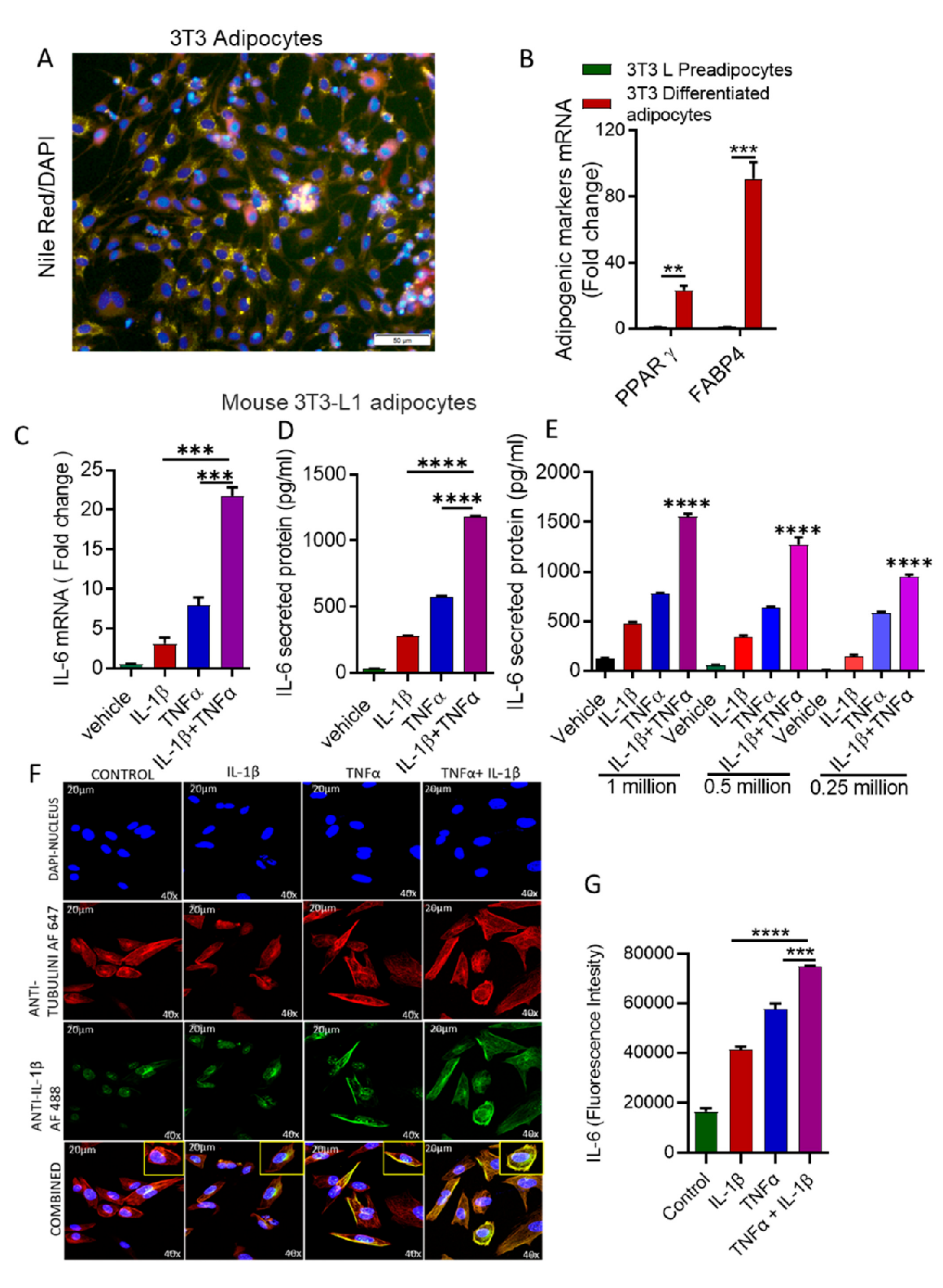
3.1. Stimulation with IL-1β and TNFα Increases IL-6 Expression in Mouse Adipocytes
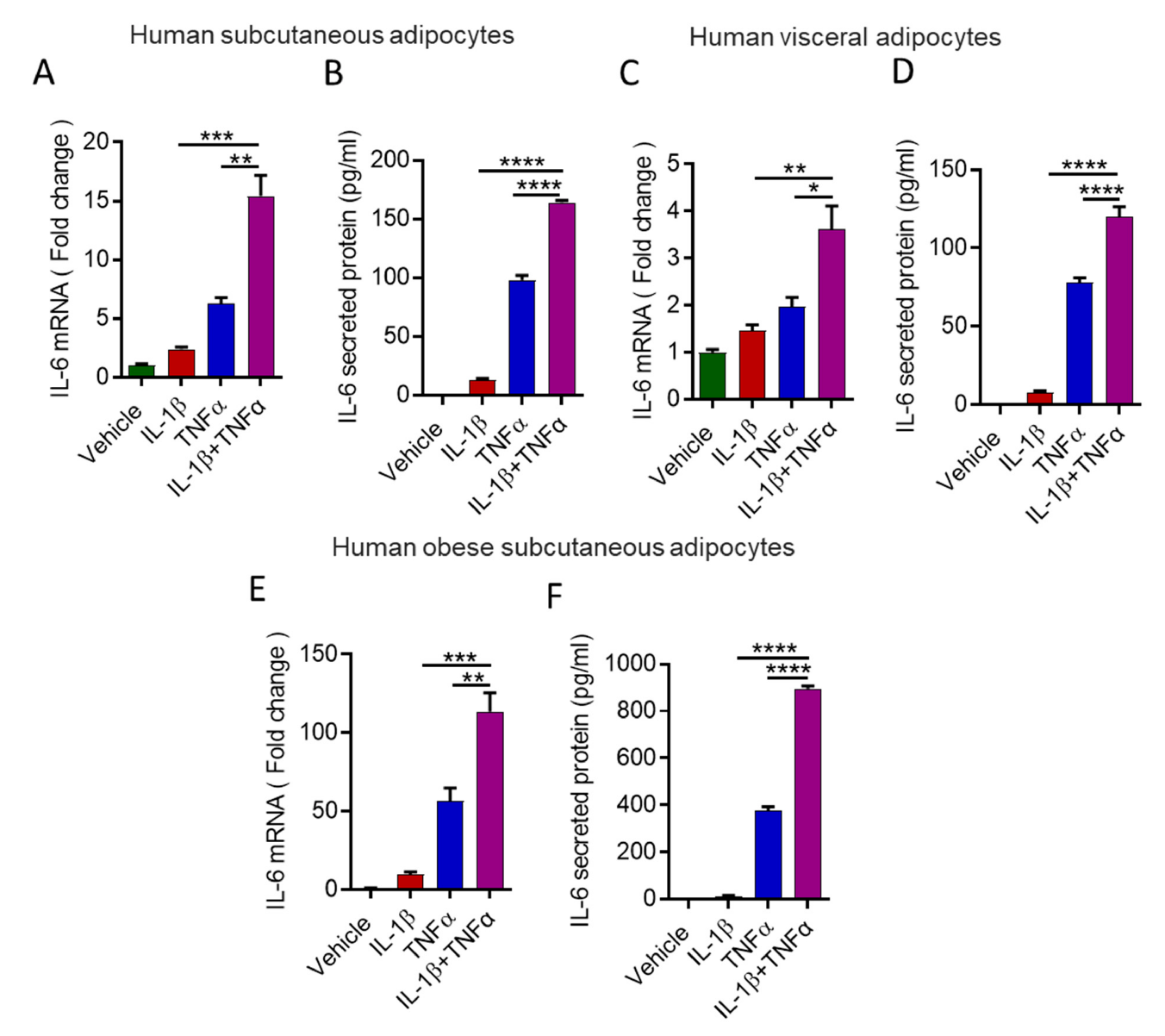
3.2. Stimulation with IL-1β and TNFα Increases IL-6 Expression in Human Primary Adipocytes
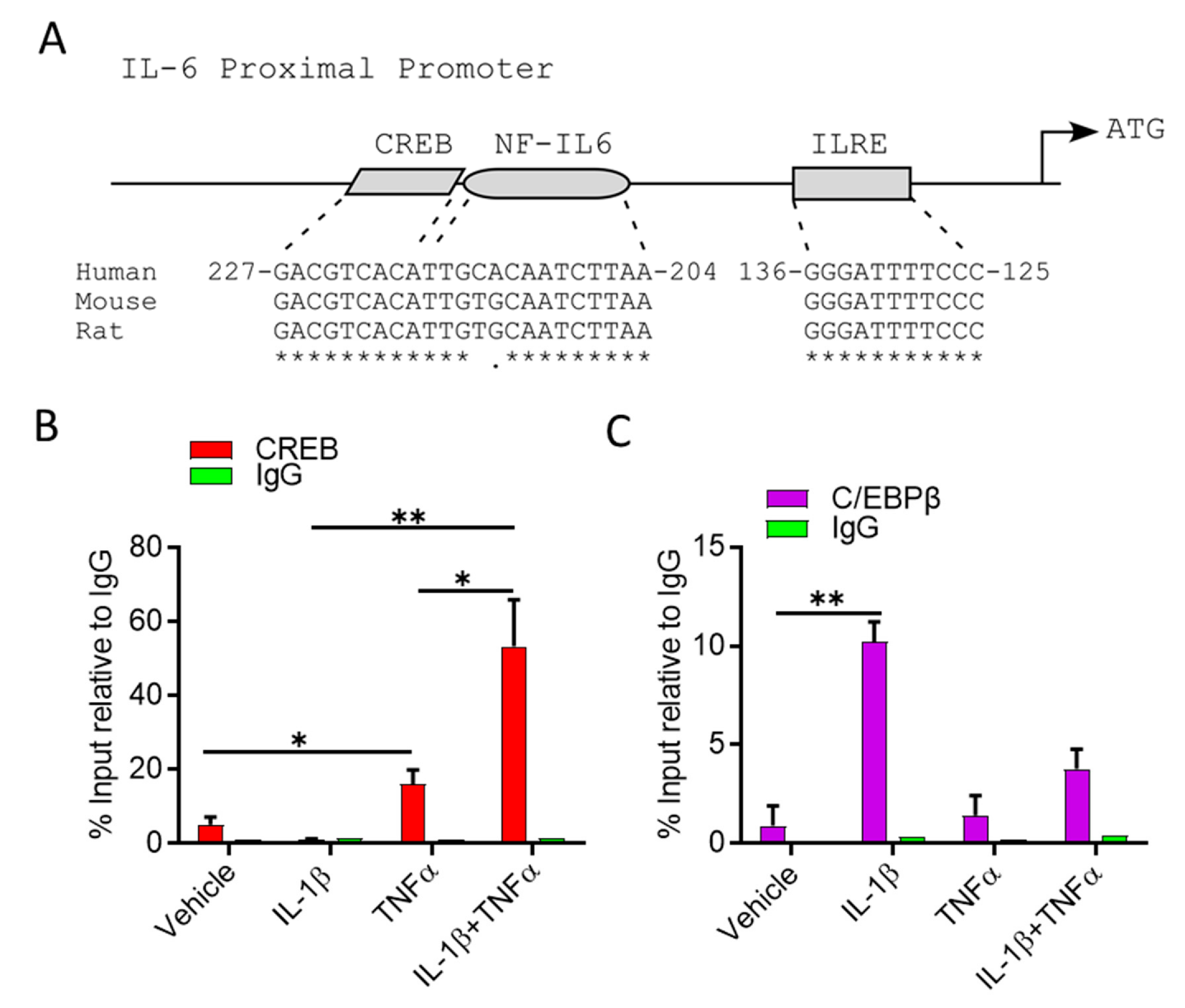
3.3. IL-1β/TNFα Stimulation Increases CREB Binding at IL-6 Promoter
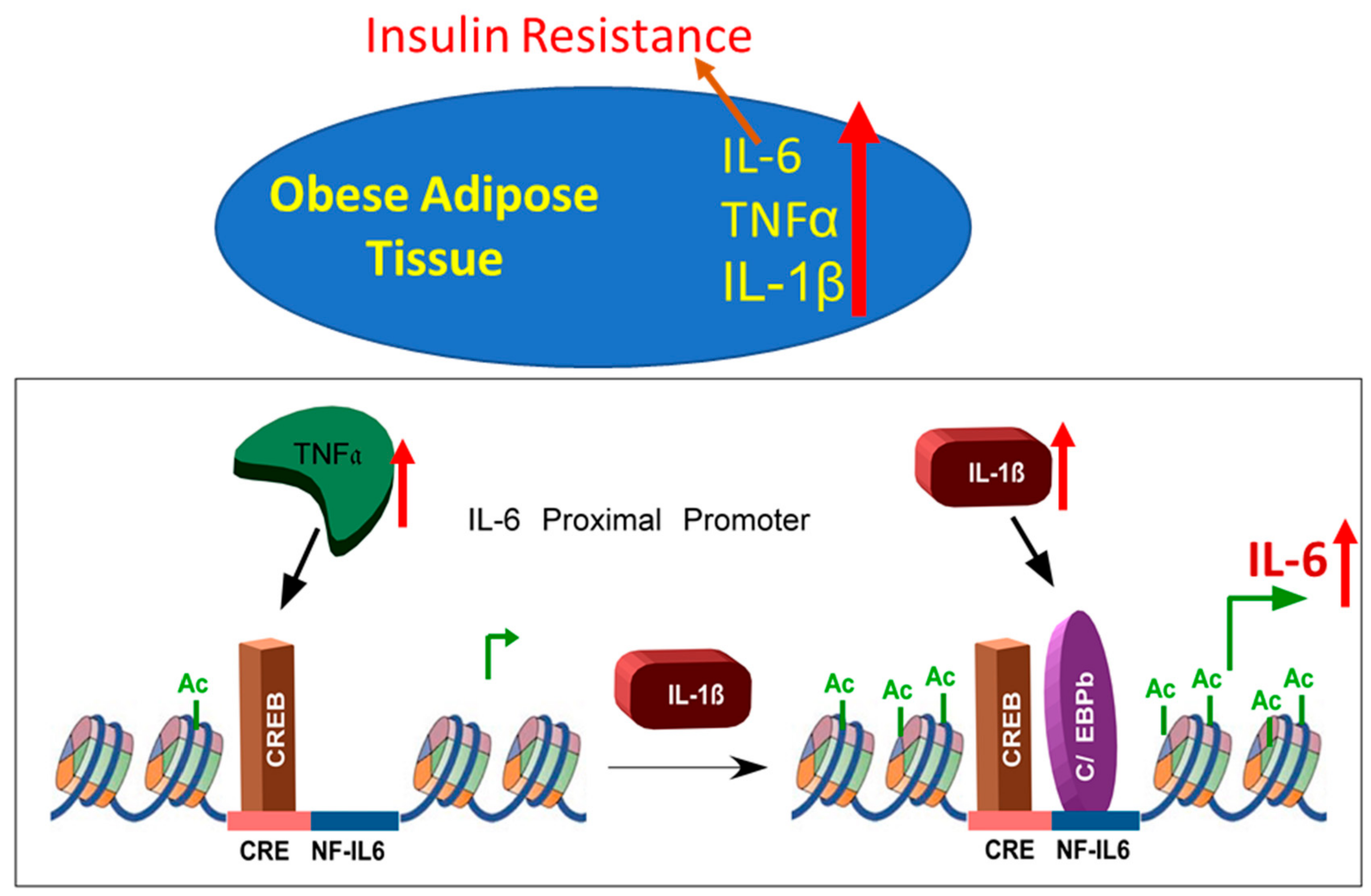
3.4. Cooperative Induction of IL-6 in Adipocyte Requires H3K14 Acetylation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the global burden of disease study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [Green Version]
- Donath, M.Y.; Shoelson, S.E. Type 2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 2011, 11, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Olefsky, J.M.; Glass, C.K. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 2010, 72, 219–246. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Kumari, M.; Wang, X.; Lantier, L.; Lyubetskaya, A.; Eguchi, J.; Kang, S.; Tenen, D.; Roh, H.C.; Kong, X.; Kazak, L.; et al. Irf3 promotes adipose inflammation and insulin resistance and represses browning. J. Clin. Investig. 2016, 126, 2839–2854. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Al-Roub, A.; Kochumon, S.; Akther, N.; Thomas, R.; Kumari, M.; Koshy, M.S.; Tiss, A.; Hannun, Y.A.; Tuomilehto, J.; et al. The synergy between palmitate and tnf-alpha for ccl2 production is dependent on the trif/irf3 pathway: Implications for metabolic inflammation. J. Immunol. 2018, 200, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Kumari, M.; Xiao, H.; Jacobs, C.; Kochumon, S.; Jedrychowski, M.; Chouchani, E.; Ahmad, R.; Rosen, E.D. Irf3 reduces adipose thermogenesis via isg15-mediated reprogramming of glycolysis. J. Clin. Investig. 2021, 131, e144888. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Murray, D.L.; Choy, L.N.; Spiegelman, B.M. Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 4854–4858. [Google Scholar] [CrossRef] [Green Version]
- Sindhu, S.; Kochumon, S.; Thomas, R.; Bennakhi, A.; Al-Mulla, F.; Ahmad, R. Enhanced adipose expression of interferon regulatory factor (irf)-5 associates with the signatures of metabolic inflammation in diabetic obese patients. Cells 2020, 9, 730. [Google Scholar] [CrossRef] [Green Version]
- Sindhu, S.; Thomas, R.; Kochumon, S.; Wilson, A.; Abu-Farha, M.; Bennakhi, A.; Al-Mulla, F.; Ahmad, R. Increased adipose tissue expression of interferon regulatory factor (irf)-5 in obesity: Association with metabolic inflammation. Cells 2019, 8, 1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochumon, S.; Al-Rashed, F.; Abu-Farha, M.; Devarajan, S.; Tuomilehto, J.; Ahmad, R. Adipose tissue expression of ccl19 chemokine is positively associated with insulin resistance. Diabetes Metab. Res. Rev. 2019, 35, e3087. [Google Scholar] [CrossRef]
- Senn, J.J.; Klover, P.J.; Nowak, I.A.; Mooney, R.A. Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes 2002, 51, 3391–3399. [Google Scholar] [CrossRef] [Green Version]
- Kanemaki, T.; Kitade, H.; Kaibori, M.; Sakitani, K.; Hiramatsu, Y.; Kamiyama, Y.; Ito, S.; Okumura, T. Interleukin 1beta and interleukin 6, but not tumor necrosis factor alpha, inhibit insulin-stimulated glycogen synthesis in rat hepatocytes. Hepatology 1998, 27, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, C.; Papanicolaou, D.A.; Kyrou, I.; Defensor, R.; Mitsiadis, C.S.; Chrousos, G.P. Dose-dependent effects of recombinant human interleukin-6 on glucose regulation. J. Clin. Endocrinol. Metab. 1997, 82, 4167–4170. [Google Scholar] [CrossRef] [PubMed]
- Bastard, J.P.; Maachi, M.; Van Nhieu, J.T.; Jardel, C.; Bruckert, E.; Grimaldi, A.; Robert, J.J.; Capeau, J.; Hainque, B. Adipose tissue il-6 content correlates with resistance to insulin activation of glucose uptake both in vivo and in vitro. J. Clin. Endocrinol. Metab. 2002, 87, 2084–2089. [Google Scholar] [CrossRef]
- Kern, P.A.; Ranganathan, S.; Li, C.; Wood, L.; Ranganathan, G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E745–E751. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.D.; Manson, J.E.; Rifai, N.; Buring, J.E.; Ridker, P.M. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA 2001, 286, 327–334. [Google Scholar] [CrossRef]
- Mohamed-Ali, V.; Goodrick, S.; Rawesh, A.; Katz, D.R.; Miles, J.M.; Yudkin, J.S.; Klein, S.; Coppack, S.W. Subcutaneous adipose tissue releases interleukin-6, but not tumor necrosis factor-alpha, in vivo. J. Clin. Endocrinol. Metab. 1997, 82, 4196–4200. [Google Scholar] [PubMed] [Green Version]
- Stenlöf, K.; Wernstedt, I.; Fjällman, T.; Wallenius, V.; Wallenius, K.; Jansson, J.-O. Interleukin-6 levels in the central nervous system are negatively correlated with fat mass in overweight/obese subjects. J. Clin. Endocrinol. Metab. 2003, 88, 4379–4383. [Google Scholar]
- Ahima, R.S.; Flier, J.S. Adipose tissue as an endocrine organ. Trends Endocrinol. Metab. 2000, 11, 327–332. [Google Scholar] [CrossRef]
- Haider, M.; Al-Rashed, F.; Albaqsumi, Z.; Alobaid, K.; Alqabandi, R.; Al-Mulla, F.; Ahmad, R. Candida albicans induces foaming and inflammation in macrophages through fabp4: Its implication for atherosclerosis. Biomedicines 2021, 9, 1567. [Google Scholar] [CrossRef]
- Al-Roub, A.; Akhter, N.; Al-Sayyar, A.; Wilson, A.; Thomas, R.; Kochumon, S.; Al-Rashed, F.; Al-Mulla, F.; Sindhu, S.; Ahmad, R. Short chain fatty acid acetate increases tnfα-induced mcp-1 production in monocytic cells via acsl1/mapk/nf-κb axis. Int. J. Mol. Sci. 2021, 22, 7683. [Google Scholar] [CrossRef]
- Al Madhoun, A.; Haddad, D.; Al Tarrah, M.; Jacob, S.; Al-Ali, W.; Nizam, R.; Miranda, L.; Al-Rashed, F.; Sindhu, S.; Ahmad, R.; et al. Microarray analysis reveals onc201 mediated differential mechanisms of chop gene regulation in metastatic and nonmetastatic colorectal cancer cells. Sci. Rep. 2021, 11, 11893. [Google Scholar] [CrossRef]
- Sindhu, S.; Kochumon, S.; Shenouda, S.; Wilson, A.; Al-Mulla, F.; Ahmad, R. The cooperative induction of ccl4 in human monocytic cells by tnf-α and palmitate requires myd88 and involves mapk/nf-κb signaling pathways. Int. J. Mol. Sci. 2019, 20, 4658. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.; Al-Rashed, F.; Akhter, N.; Al-Mulla, F.; Ahmad, R. Acsl1 regulates tnfα-induced gm-csf production by breast cancer mda-mb-231 cells. Biomolecules 2019, 9, 555. [Google Scholar] [CrossRef] [Green Version]
- Kochumon, S.; Al-Sayyar, A.; Jacob, T.; Hasan, A.; Al-Mulla, F.; Sindhu, S.; Ahmad, R. Tnf-α increases ip-10 expression in mcf-7 breast cancer cells via activation of the jnk/c-jun pathways. Biomolecules 2021, 11, 1355. [Google Scholar] [CrossRef] [PubMed]
- Akhter, N.; Wilson, A.; Thomas, R.; Al-Rashed, F.; Kochumon, S.; Al-Roub, A.; Arefanian, H.; Al-Madhoun, A.; Al-Mulla, F.; Ahmad, R.; et al. Ros/tnf-α crosstalk triggers the expression of il-8 and mcp-1 in human monocytic thp-1 cells via the nf-κb and erk1/2 mediated signaling. Int. J. Mol. Sci. 2021, 22, 10519. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashed, F.; Thomas, R.; Al-Roub, A.; Al-Mulla, F.; Ahmad, R. Lps induces gm-csf production by breast cancer mda-mb-231 cells via long-chain acyl-coa synthetase 1. Molecules 2020, 25, 4709. [Google Scholar] [CrossRef]
- Kochumon, S.; Arefanian, H.; Azim, R.; Shenouda, S.; Jacob, T.; Abu Khalaf, N.; Al-Rashed, F.; Hasan, A.; Sindhu, S.; Al-Mulla, F.; et al. Stearic acid and tnf-α co-operatively potentiate mip-1α production in monocytic cells via myd88 independent tlr4/tbk/irf3 signaling pathway. Biomedicines 2020, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashed, F.; Sindhu, S.; Arefanian, H.; Al Madhoun, A.; Kochumon, S.; Thomas, R.; Al-Kandari, S.; Alghaith, A.; Jacob, T.; Al-Mulla, F.; et al. Repetitive intermittent hyperglycemia drives the m1 polarization and inflammatory responses in thp-1 macrophages through the mechanism involving the tlr4-irf5 pathway. Cells 2020, 9, 1892. [Google Scholar] [CrossRef]
- Sindhu, S.; Akhter, N.; Wilson, A.; Thomas, R.; Arefanian, H.; Al Madhoun, A.; Al-Mulla, F.; Ahmad, R. Mip-1α expression induced by co-stimulation of human monocytic cells with palmitate and tnf-α involves the tlr4-irf3 pathway and is amplified by oxidative stress. Cells 2020, 9, 1799. [Google Scholar] [CrossRef]
- Kochumon, S.; Arefanian, H.; Sindhu, S.; Shenouda, S.; Thomas, R.; Al-Mulla, F.; Tuomilehto, J.; Ahmad, R. Adipose tissue steroid receptor rna activator 1 (sra1) expression is associated with obesity, insulin resistance, and inflammation. Cells 2021, 10, 2602. [Google Scholar] [CrossRef]
- Al-Rashed, F.; Ahmad, Z.; Snider, A.J.; Thomas, R.; Kochumon, S.; Melhem, M.; Sindhu, S.; Obeid, L.M.; Al-Mulla, F.; Hannun, Y.A.; et al. Ceramide kinase regulates tnf-α-induced immune responses in human monocytic cells. Sci. Rep. 2021, 11, 8259. [Google Scholar] [CrossRef]
- Al-Rashed, F.; Ahmad, Z.; Thomas, R.; Melhem, M.; Snider, A.J.; Obeid, L.M.; Al-Mulla, F.; Hannun, Y.A.; Ahmad, R. Neutral sphingomyelinase 2 regulates inflammatory responses in monocytes/macrophages induced by tnf-α. Sci. Rep. 2020, 10, 16802. [Google Scholar] [CrossRef]
- Al Madhoun, A.S.; Voronova, A.; Ryan, T.; Zakariyah, A.; McIntire, C.; Gibson, L.; Shelton, M.; Ruel, M.; Skerjanc, I.S. Testosterone enhances cardiomyogenesis in stem cells and recruits the androgen receptor to the mef2c and hcn4 genes. J. Mol. Cell. Cardiol. 2013, 60, 164–171. [Google Scholar] [CrossRef]
- Al Madhoun, A.S.; Mehta, V.; Li, G.; Figeys, D.; Wiper-Bergeron, N.; Skerjanc, I.S. Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of mef2c. EMBO J. 2011, 30, 2477–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunout, D.; Muñoz, C.; López, M.; de la Maza, M.P.; Schlesinger, L.; Hirsch, S.; Pettermann, M. Interleukin 1 and tumor necrosis factor in obese alcoholics compared with normal-weight patients. Am. J. Clin. Nutr. 1996, 63, 373–376. [Google Scholar] [CrossRef] [Green Version]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Investig. 1995, 95, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Gustin, J.A.; Pincheira, R.; Mayo, L.D.; Ozes, O.N.; Kessler, K.M.; Baerwald, M.R.; Korgaonkar, C.K.; Donner, D.B. Tumor necrosis factor activates cre-binding protein through a p38 mapk/msk1 signaling pathway in endothelial cells. Am. J. Physiol. Cell Physiol. 2004, 286, C547–C555. [Google Scholar] [CrossRef] [PubMed]
- Hungness, E.; Pritts, T.; Luo, G.; Hershko, D.; Robb, B.; Hasselgren, P. Il-1β activates c/ebp-β and c/ebp-δ in human enterocytes through a mitogen-activated protein-kinase signaling pathway. Int. J. Biochem. Cell Biol. 2002, 34, 382–395. [Google Scholar] [CrossRef]
- Robinson, K.F.; Narasipura, S.D.; Wallace, J.; Ritz, E.M.; Al-Harthi, L. Β-catenin and tcfs/lef signaling discordantly regulate il-6 expression in astrocytes. Cell Commun. Signal. 2020, 18, 93. [Google Scholar] [CrossRef]
- Hu, L.; Yu, Y.; Huang, H.; Fan, H.; Hu, L.; Yin, C.; Li, K.; Fulton, D.J.; Chen, F. Epigenetic regulation of interleukin 6 by histone acetylation in macrophages and its role in paraquat-induced pulmonary fibrosis. Front. Immunol. 2016, 7, 696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voronova, A.; Fischer, A.; Ryan, T.; Al Madhoun, A.; Skerjanc, I.S. Ascl1/mash1 is a novel target of gli2 during gli2-induced neurogenesis in p19 ec cells. PLoS ONE 2011, 6, e19174. [Google Scholar] [CrossRef] [Green Version]
- Eliseeva, E.D.; Valkov, V.; Jung, M.; Jung, M.O. Characterization of novel inhibitors of histone acetyltransferases. Mol. Cancer Ther. 2007, 6, 2391–2398. [Google Scholar] [CrossRef] [Green Version]
- Reuter, S.; Gupta, S.C.; Park, B.; Goel, A.; Aggarwal, B.B. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011, 6, 93–108. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Lu, J.; Zhang, Y.; Cheng, C.; Li, L.; Han, L.; Huang, B. Hdac inhibitors tsa and sodium butyrate enhanced the human il-5 expression by altering histone acetylation status at its promoter region. Immunol. Lett. 2007, 108, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Rosas, F.; Hernández-Oliveras, A.; Flores-Peredo, L.; Rodríguez, G.; Zarain-Herzberg, Á.; Caba, M.; Santiago-García, J. Histone deacetylase inhibitors induce the expression of tumor suppressor genes per1 and per2 in human gastric cancer cells. Oncol. Lett. 2018, 16, 1981–1990. [Google Scholar] [PubMed]
- Nish, S.A.; Schenten, D.; Wunderlich, F.T.; Pope, S.D.; Gao, Y.; Hoshi, N.; Yu, S.; Yan, X.; Lee, H.K.; Pasman, L.; et al. T cell-intrinsic role of il-6 signaling in primary and memory responses. eLife 2014, 3, e01949. [Google Scholar] [CrossRef]
- El-Mikkawy, D.M.E.; El-Sadek, M.A.; El-Badawy, M.A.; Samaha, D. Circulating level of interleukin-6 in relation to body mass indices and lipid profile in egyptian adults with overweight and obesity. Egypt. Rheumatol. Rehabil. 2020, 47, 7. [Google Scholar] [CrossRef]
- Alzamil, H. Elevated serum tnf-alpha is related to obesity in type 2 diabetes mellitus and is associated with glycemic control and insulin resistance. J. Obes. 2020, 2020, 5076858. [Google Scholar] [CrossRef] [Green Version]
- Jager, J.; Grémeaux, T.; Cormont, M.; Le Marchand-Brustel, Y.; Tanti, J.F. Interleukin-1beta-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology 2007, 148, 241–251. [Google Scholar] [CrossRef]
- Ruscitti, P.; Ursini, F.; Cipriani, P.; Greco, M.; Alvaro, S.; Vasiliki, L.; Di Benedetto, P.; Carubbi, F.; Berardicurti, O.; Gulletta, E.; et al. Il-1 inhibition improves insulin resistance and adipokines in rheumatoid arthritis patients with comorbid type 2 diabetes: An observational study. Medicine 2019, 98, e14587. [Google Scholar] [CrossRef] [PubMed]
- Um, J.Y.; Chung, H.S.; Song, M.Y.; Shin, H.D.; Kim, H.M. Association of interleukin-1beta gene polymorphism with body mass index in women. Clin. Chem. 2004, 50, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Lee, O.Y.; Park, Y.; Seo, M.W.; Lee, D.S. Il-1beta induces il-6 production and increases invasiveness and estrogen-independent growth in a tg2-dependent manner in human breast cancer cells. BMC Cancer 2016, 16, 724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandere-Grzybowska, K.; Letourneau, R.; Kempuraj, D.; Donelan, J.; Poplawski, S.; Boucher, W.; Athanassiou, A.; Theoharides, T.C. Il-1 induces vesicular secretion of il-6 without degranulation from human mast cells. J. Immunol. 2003, 171, 4830–4836. [Google Scholar] [CrossRef] [Green Version]
- Tosato, G.; Jones, K.D. Interleukin-1 induces interleukin-6 production in peripheral blood monocytes. Blood 1990, 75, 1305–1310. [Google Scholar] [CrossRef] [Green Version]
- De Cesaris, P.; Starace, D.; Riccioli, A.; Padula, F.; Filippini, A.; Ziparo, E. Tumor necrosis factor-alpha induces interleukin-6 production and integrin ligand expression by distinct transduction pathways. J. Biol. Chem. 1998, 273, 7566–7571. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, K.; Matsushima-Nishiwaki, R.; Yamaguchi, S.; Iida, H.; Dohi, S.; Kozawa, O. Mechanisms of tumor necrosis factor-α-induced interleukin-6 synthesis in glioma cells. J. Neuroinflamm. 2010, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Confalone, E.; D’Alessio, G.; Furia, A. Il-6 induction by tnfα and il-1β in an osteoblast-like cell line. Int. J. Biomed. Sci. 2010, 6, 135–140. [Google Scholar] [PubMed]
- Tian, Q.; Miyazaki, R.; Ichiki, T.; Imayama, I.; Inanaga, K.; Ohtsubo, H.; Yano, K.; Takeda, K.; Sunagawa, K. Inhibition of tumor necrosis factor-α–induced interleukin-6 expression by telmisartan through cross-talk of peroxisome proliferator-activated receptor-γ with nuclear factor κb and ccaat/enhancer-binding protein-β. Hypertension 2009, 53, 798–804. [Google Scholar] [CrossRef]
- Popko, K.; Gorska, E.; Stelmaszczyk-Emmel, A.; Plywaczewski, R.; Stoklosa, A.; Gorecka, D.; Pyrzak, B.; Demkow, U. Proinflammatory cytokines il-6 and tnf-α and the development of inflammation in obese subjects. Eur. J. Med. Res. 2010, 15, 120. [Google Scholar] [CrossRef] [PubMed]
- Song, K.S.; Lee, W.J.; Chung, K.C.; Koo, J.S.; Yang, E.J.; Choi, J.Y.; Yoon, J.H. Interleukin-1 beta and tumor necrosis factor-alpha induce muc5ac overexpression through a mechanism involving erk/p38 mitogen-activated protein kinases-msk1-creb activation in human airway epithelial cells. J. Biol. Chem. 2003, 278, 23243–23250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.C.; Chang, H.-h.; Lee, M.-Y.; Lin, C.-C.; Yeh, H.-W.; Yang, T.-t.; Lin, P.-S.; Tseng, W.Y.; Jeng, J.-H. Prostaglandin f(2alpha)-induced interleukin-8 production in human dental pulp cells is associated with mek/erk signaling. J. Endod. 2009, 35, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Giltiay, N.V.; Karakashian, A.A.; Alimov, A.P.; Ligthle, S.; Nikolova-Karakashian, M.N. Ceramide- and erk-dependent pathway for the activation of ccaat/enhancer binding protein by interleukin-1beta in hepatocytes. J. Lipid Res. 2005, 46, 2497–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Hodge, D.R.; Wang, L.; Yang, X.; Zhang, X.; Farrar, W.L. Co-operative functions between nuclear factors nfkappab and ccat/enhancer-binding protein-beta (c/ebp-beta) regulate the il-6 promoter in autocrine human prostate cancer cells. Prostate 2004, 61, 354–370. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Lin, J.X.; Vilcek, J. Interleukin-6 induction by tumor necrosis factor and interleukin-1 in human fibroblasts involves activation of a nuclear factor binding to a kappa b-like sequence. Mol. Cell. Biol. 1990, 10, 3818–3823. [Google Scholar]
- Zhang, Y.; Broser, M.; Rom, W.N. Activation of the interleukin 6 gene by mycobacterium tuberculosis or lipopolysaccharide is mediated by nuclear factors nf-il6 and nf-kappa b. Proc. Natl. Acad. Sci. USA 1994, 91, 2225–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, H.; Mitomo, K.; Watanabe, T.; Okamoto, S.; Yamamoto, K. Involvement of a nf-kappa b-like transcription factor in the activation of the interleukin-6 gene by inflammatory lymphokines. Mol. Cell. Biol. 1990, 10, 561–568. [Google Scholar]
- Matsusaka, T.; Fujikawa, K.; Nishio, Y.; Mukaida, N.; Matsushima, K.; Kishimoto, T.; Akira, S. Transcription factors nf-il6 and nf-kappa b synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 1993, 90, 10193–10197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndlovu, M.N.; Van Lint, C.; Van Wesemael, K.; Callebert, P.; Chalbos, D.; Haegeman, G.; Vanden Berghe, W. Hyperactivated nf-κb and ap-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol. Cell. Biol. 2009, 29, 5488–5504. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Hodge, D.R.; Wang, L.; Yang, X.; Zhang, X.; Farrar, W.L. Nf-kappab activates il-6 expression through cooperation with c-jun and il6-ap1 site, but is independent of its il6-nfkappab regulatory site in autocrine human multiple myeloma cells. Cancer Biol. Ther. 2004, 3, 1007–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Islam, R.; Dash, D.; Singh, R. Intranasal curcumin and sodium butyrate modulates airway inflammation and fibrosis via hdac inhibition in allergic asthma. Cytokine 2021, 149, 155720. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Cao, Q.; Reilly, C.M.; Young, N.L.; Garcia, B.A.; Mishra, N. Histone deacetylase 9 deficiency protects against effector t cell-mediated systemic autoimmunity. J. Biol. Chem. 2011, 286, 28833–28843. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Roub, A.; Al Madhoun, A.; Akhter, N.; Thomas, R.; Miranda, L.; Jacob, T.; Al-Ozairi, E.; Al-Mulla, F.; Sindhu, S.; Ahmad, R. IL-1β and TNFα Cooperativity in Regulating IL-6 Expression in Adipocytes Depends on CREB Binding and H3K14 Acetylation. Cells 2021, 10, 3228. https://doi.org/10.3390/cells10113228
Al-Roub A, Al Madhoun A, Akhter N, Thomas R, Miranda L, Jacob T, Al-Ozairi E, Al-Mulla F, Sindhu S, Ahmad R. IL-1β and TNFα Cooperativity in Regulating IL-6 Expression in Adipocytes Depends on CREB Binding and H3K14 Acetylation. Cells. 2021; 10(11):3228. https://doi.org/10.3390/cells10113228
Chicago/Turabian StyleAl-Roub, Areej, Ashraf Al Madhoun, Nadeem Akhter, Reeby Thomas, Lavina Miranda, Texy Jacob, Ebaa Al-Ozairi, Fahd Al-Mulla, Sardar Sindhu, and Rasheed Ahmad. 2021. "IL-1β and TNFα Cooperativity in Regulating IL-6 Expression in Adipocytes Depends on CREB Binding and H3K14 Acetylation" Cells 10, no. 11: 3228. https://doi.org/10.3390/cells10113228
APA StyleAl-Roub, A., Al Madhoun, A., Akhter, N., Thomas, R., Miranda, L., Jacob, T., Al-Ozairi, E., Al-Mulla, F., Sindhu, S., & Ahmad, R. (2021). IL-1β and TNFα Cooperativity in Regulating IL-6 Expression in Adipocytes Depends on CREB Binding and H3K14 Acetylation. Cells, 10(11), 3228. https://doi.org/10.3390/cells10113228









