On the Role of the Immunoproteasome in Protein Homeostasis
Abstract
1. Introduction
2. The Proteasome, a Key Player in Protein Homeostasis
3. Immunoproteasomes in Protein Homeostasis. Contradictory Results
4. The Role of the Immunoproteasome in Degrading Oxidized Proteins
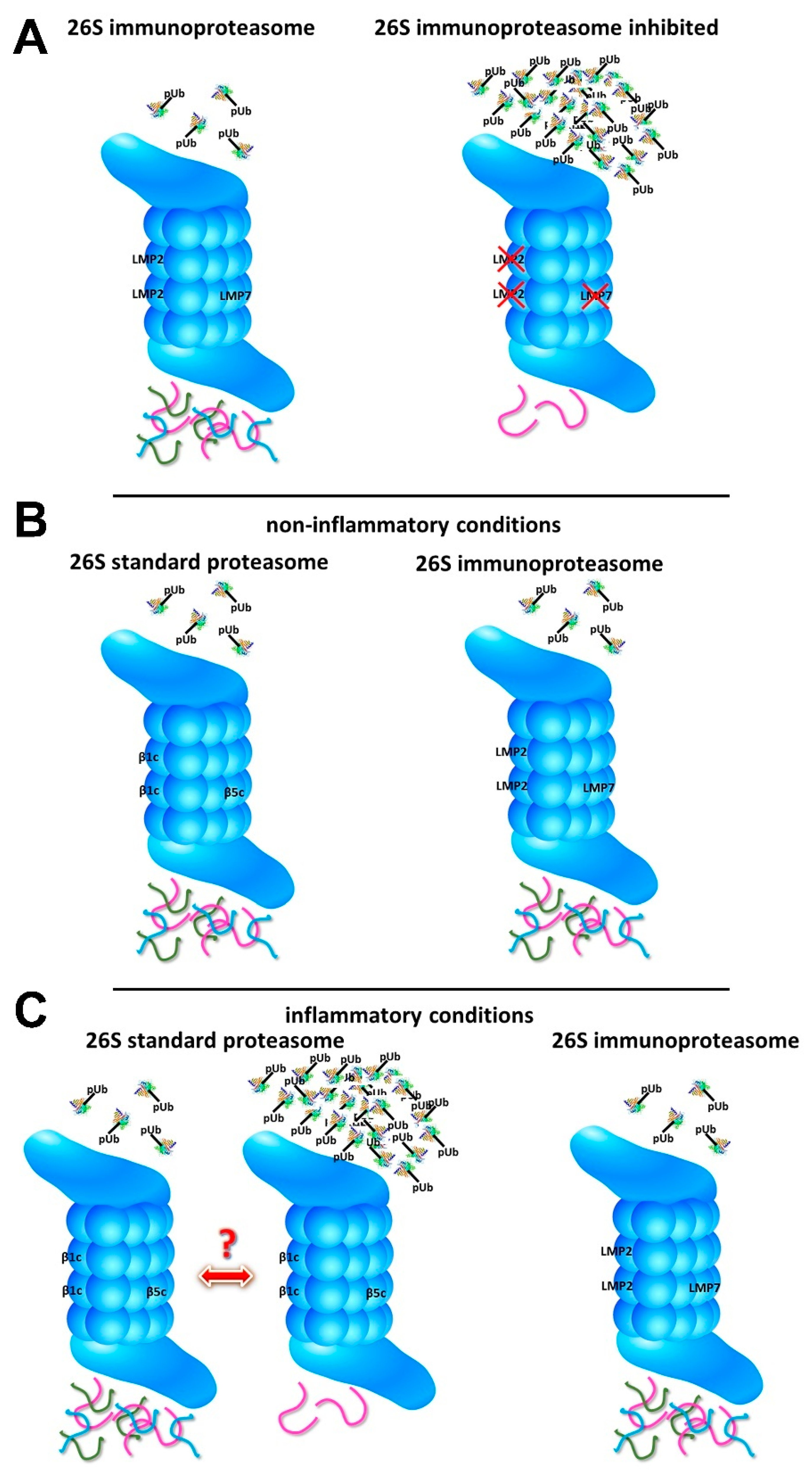
5. Immunoproteasome Inhibition and Protein Homeostasis
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, N.; Abi Habib, J.; Van den Eynde, B.J. Learning from the Proteasome How To Fine-Tune Cancer Immunotherapy. Trends Cancer 2017, 3, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Groettrup, M.; Kirk, C.J.; Basler, M. Proteasomes in immune cells: More than peptide producers? Nat. Rev. Immunol. 2010, 10, 73–78. [Google Scholar] [CrossRef]
- Chen, W.S.; Norbury, C.C.; Cho, Y.J.; Yewdell, J.W.; Bennink, J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001, 193, 1319–1326. [Google Scholar] [CrossRef]
- Basler, M.; Youhnovski, N.; Van Den Broek, M.; Przybylski, M.; Groettrup, M. Immunoproteasomes down-regulate presentation of a subdominant T cell epitope from lymphocytic choriomeningitis virus. J. Immunol. 2004, 173, 3925–3934. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Moebius, J.; Elenich, L.; Groettrup, M.; Monaco, J.J. An Altered T Cell Repertoire in MECL-1-Deficient Mice. J. Immunol. 2006, 176, 6665–6672. [Google Scholar] [CrossRef] [PubMed]
- Chou, B.; Hisaeda, H.; Shen, J.; Duan, X.; Imai, T.; Tu, L.; Murata, S.; Tanaka, K.; Himeno, K. Critical contribution of immunoproteasomes in the induction of protective immunity against Trypanosoma cruzi in mice vaccinated with a plasmid encoding a CTL epitope fused to green fluorescence protein. Microbes Infect. 2008, 10, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Ersching, J.; Vasconcelos, J.R.; Ferreira, C.P.; Caetano, B.C.; Machado, A.V.; Bruna-Romero, O.; Baron, M.A.; Ferreira, L.R.; Cunha-Neto, E.; Rock, K.L.; et al. The Combined Deficiency of Immunoproteasome Subunits Affects Both the Magnitude and Quality of Pathogen- and Genetic Vaccination-Induced CD8+ T Cell Responses to the Human Protozoan Parasite Trypanosoma cruzi. PLoS Pathog. 2016, 12, e1005593. [Google Scholar] [CrossRef]
- Basler, M.; Mundt, S.; Groettrup, M. The immunoproteasome subunit LMP7 is required in the murine thymus for filling up a hole in the T cell repertoire. Eur. J. Immunol. 2018, 48, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Moebius, J.; van den Broek, M.; Groettrup, M.; Basler, M. Immunoproteasomes are essential for survival and expansion of T cells in virus-infected mice. Eur. J. Immunol. 2010, 40, 3439–3449. [Google Scholar] [CrossRef] [PubMed]
- Caudill, C.M.; Jayarapu, K.; Elenich, L.; Monaco, J.J.; Colbert, R.A.; Griffin, T.A. T cells lacking immunoproteasome subunits MECL-1 and LMP7 hyperproliferate in response to polyclonal mitogens. J. Immunol. 2006, 176, 4075–4082. [Google Scholar] [CrossRef] [PubMed]
- Kalim, K.W.; Basler, M.; Kirk, C.J.; Groettrup, M. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. J. Immunol. 2012, 189, 4182–4193. [Google Scholar] [CrossRef]
- Vachharajani, N.; Joeris, T.; Luu, M.; Hartmann, S.; Pautz, S.; Jenike, E.; Pantazis, G.; Prinz, I.; Hofer, M.J.; Steinhoff, U.; et al. Prevention of colitis-associated cancer by selective targeting of immunoproteasome subunit LMP7. Oncotarget 2017, 8, 50447–50459. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Kammerl, I.E.; Vosyka, O.; Baumann, T.; Yu, Y.; Wu, Y.; Irmler, M.; Overkleeft, H.S.; Beckers, J.; Eickelberg, O.; et al. Immunoproteasome dysfunction augments alternative polarization of alveolar macrophages. Cell Death Differ. 2016, 23, 1026–1037. [Google Scholar] [CrossRef]
- Liao, J.; Xie, Y.; Lin, Q.; Yang, X.; An, X.; Xia, Y.; Du, J.; Wang, F.; Li, H.H. Immunoproteasome subunit β5i regulates diet-induced atherosclerosis through altering MERTK-mediated efferocytosis in Apoe knockout mice. J. Pathol. 2020, 250, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Kremer, M.; Henn, A.; Kolb, C.; Basler, M.; Moebius, J.; Guillaume, B.; Leist, M.; Van den Eynde, B.J.; Groettrup, M. Reduced immunoproteasome formation and accumulation of immunoproteasomal precursors in the brains of lymphocytic choriomeningitis virus-infected mice. J. Immunol. 2010, 185, 5549–5560. [Google Scholar] [CrossRef] [PubMed]
- Mundt, S.; Engelhardt, B.; Kirk, C.J.; Groettrup, M.; Basler, M. Inhibition and deficiency of the immunoproteasome subunit LMP7 attenuates LCMV-induced meningitis. Eur. J. Immunol. 2016, 46, 104–113. [Google Scholar] [CrossRef]
- Keller, I.E.; Vosyka, O.; Takenaka, S.; Kloss, A.; Dahlmann, B.; Willems, L.I.; Verdoes, M.; Overkleeft, H.S.; Marcos, E.; Adnot, S.; et al. Regulation of immunoproteasome function in the lung. Sci. Rep. 2015, 5, 10230. [Google Scholar] [CrossRef] [PubMed]
- Kammerl, I.E.; Dann, A.; Mossina, A.; Brech, D.; Lukas, C.; Vosyka, O.; Nathan, P.; Conlon, T.M.; Wagner, D.E.; Overkleeft, H.S.; et al. Impairment of Immunoproteasome Function by Cigarette Smoke and in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2016, 193, 1230–1241. [Google Scholar] [CrossRef]
- Kammerl, I.E.; Hardy, S.; Flexeder, C.; Urmann, A.; Peierl, J.; Wang, Y.; Vosyka, O.; Frankenberger, M.; Milger, K.; Behr, J.; et al. Activation of immune cell proteasomes in peripheral blood of smokers and COPD patients—Implications for therapy. Eur. Respir. J. 2021, 58. [Google Scholar] [CrossRef]
- Desterke, C.; Turhan, A.G.; Bennaceur-Griscelli, A.; Griscelli, F. HLA-dependent heterogeneity and macrophage immunoproteasome activation during lung COVID-19 disease. J. Transl. Med. 2021, 19, 290. [Google Scholar] [CrossRef] [PubMed]
- DiazHernandez, M.; Hernandez, F.; MartinAparicio, E.; GomezRamos, P.; Moran, M.A.; Castano, J.G.; Ferrer, I.; Avila, J.; Lucas, J.J. Neuronal induction of the immunoproteasome in Huntington’s disease. J. Neurosci. 2003, 23, 11653–11661. [Google Scholar] [CrossRef]
- Puttaparthi, K.; Elliott, J.L. Non-neuronal induction of immunoproteasome subunits in an ALS model: Possible mediation by cytokines. Exp. Neurol. 2005, 196, 441–451. [Google Scholar] [CrossRef]
- Mishto, M.; Bellavista, E.; Santoro, A.; Stolzing, A.; Ligorio, C.; Nacmias, B.; Spazzafumo, L.; Chiappelli, M.; Licastro, F.; Sorbi, S.; et al. Immunoproteasome and LMP2 polymorphism in aged and Alzheimer’s disease brains. Neurobiol. Aging 2006, 27, 54–66. [Google Scholar] [CrossRef]
- Yeo, I.J.; Lee, M.J.; Baek, A.; Miller, Z.; Bhattarai, D.; Baek, Y.M.; Jeong, H.J.; Kim, Y.K.; Kim, D.E.; Hong, J.T.; et al. A dual inhibitor of the proteasome catalytic subunits LMP2 and Y attenuates disease progression in mouse models of Alzheimer’s disease. Sci. Rep. 2019, 9, 18393. [Google Scholar] [CrossRef]
- Orre, M.; Kamphuis, W.; Dooves, S.; Kooijman, L.; Chan, E.T.; Kirk, C.J.; Dimayuga Smith, V.; Koot, S.; Mamber, C.; Jansen, A.H.; et al. Reactive glia show increased immunoproteasome activity in Alzheimer’s disease. Brain 2013, 136, 1415–1431. [Google Scholar] [CrossRef]
- Wagner, L.K.; Gilling, K.E.; Schormann, E.; Kloetzel, P.M.; Heppner, F.L.; Kruger, E.; Prokop, S. Immunoproteasome deficiency alters microglial cytokine response and improves cognitive deficits in Alzheimer’s disease-like APPPS1 mice. Acta Neuropathol. Commun. 2017, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Ugras, S.; Daniels, M.J.; Fazelinia, H.; Gould, N.S.; Yocum, A.K.; Luk, K.C.; Luna, E.; Ding, H.; McKennan, C.; Seeholzer, S.; et al. Induction of the Immunoproteasome Subunit Lmp7 Links Proteostasis and Immunity in alpha-Synuclein Aggregation Disorders. EBioMedicine 2018, 31, 307–319. [Google Scholar] [CrossRef]
- Bhattarai, D.; Lee, M.J.; Baek, A.; Yeo, I.J.; Miller, Z.; Baek, Y.M.; Lee, S.; Kim, D.E.; Hong, J.T.; Kim, K.B. LMP2 Inhibitors as a Potential Treatment for Alzheimer’s Disease. J. Med. Chem. 2020, 63, 3763–3783. [Google Scholar] [CrossRef] [PubMed]
- Bi, M.; Du, X.; Xiao, X.; Dai, Y.; Jiao, Q.; Chen, X.; Zhang, L.; Jiang, H. Deficient immunoproteasome assembly drives gain of alpha-synuclein pathology in Parkinson’s disease. Redox Biol. 2021, 47, 102167. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Dajee, M.; Moll, C.; Groettrup, M.; Kirk, C.J. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J. Immunol. 2010, 185, 634–641. [Google Scholar] [CrossRef]
- Zaiss, D.M.; Bekker, C.P.; Grone, A.; Lie, B.A.; Sijts, A.J. Proteasome immunosubunits protect against the development of CD8 T cell-mediated autoimmune diseases. J. Immunol. 2011, 187, 2302–2309. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, L.R.; Khare, V.; Small, J.S.; Koltun, W.A. Dextran sulfate sodium-induced colitis is associated with enhanced low molecular mass polypeptide 2 (LMP2) expression and is attenuated in LMP2 knockout mice. Dig. Dis. Sci. 2006, 51, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Koerner, J.; Brunner, T.; Groettrup, M. Inhibition and deficiency of the immunoproteasome subunit LMP7 suppress the development and progression of colorectal carcinoma in mice. Oncotarget 2017, 8, 50873–50888. [Google Scholar] [CrossRef] [PubMed]
- Bockstahler, M.; Fischer, A.; Goetzke, C.C.; Neumaier, H.L.; Sauter, M.; Kespohl, M.; Müller, A.M.; Meckes, C.; Salbach, C.; Schenk, M.; et al. Heart-Specific Immune Responses in an Animal Model of AutoimmuneRelated Myocarditis Mitigated by an Immunoproteasome Inhibitor and Genetic Ablation. Circulation 2020, 141, 1885–1902. [Google Scholar] [CrossRef] [PubMed]
- French, T.; Israel, N.; Dusedau, H.P.; Tersteegen, A.; Steffen, J.; Cammann, C.; Topfstedt, E.; Dieterich, D.; Schuler, T.; Seifert, U.; et al. The Immunoproteasome Subunits LMP2, LMP7 and MECL-1 Are Crucial Along the Induction of Cerebral Toxoplasmosis. Front. Immunol. 2021, 12, 619465. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Chama, L.L.; Ebstein, F.; Wiesrecker, B.; Wagh, P.R.; Hammer, E.; Weiss, F.U.; Junker, H.; Studencka-Turski, M.; Lerch, M.M.; Kruger, E.; et al. Immunoproteasome impairment via beta5i/LMP7-deletion leads to sustained pancreatic injury from experimental pancreatitis. J. Cell. Mol. Med. 2021, 25, 6786–6799. [Google Scholar] [CrossRef]
- Du, S.H.; Xiang, Y.J.; Liu, L.; Nie, M.; Hou, Y.; Wang, L.; Li, B.B.; Xu, M.; Teng, Q.L.; Peng, J.; et al. Co-Inhibition of the Immunoproteasome Subunits LMP2 and LMP7 Ameliorates Immune Thrombocytopenia. Front. Immunol. 2020, 11, 603278. [Google Scholar] [CrossRef]
- Basler, M.; Kirk, C.J.; Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 2013, 25, 74–80. [Google Scholar] [CrossRef]
- Hayashi, T.; Faustman, D. NOD mice are defective in proteasome production and activation of NF- kappa B. Mol. Cell. Biol. 1999, 19, 8646–8659. [Google Scholar] [CrossRef]
- Hayashi, T.; Faustman, D. Essential role of human leukocyte antigen-encoded proteasome subunits in NF-kappa B activation and prevention of tumor necrosis factor- alpha-induced apoptosis. J. Biol. Chem. 2000, 275, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Runnels, H.A.; Watkins, W.A.; Monaco, J.J. LMP2 expression and proteasome activity in NOD mice. Nat. Med. 2000, 6, 1064–1065, author reply 1065-1066. [Google Scholar] [CrossRef] [PubMed]
- Kessler, B.M.; Lennon-Dumenil, A.M.; Shinohara, M.L.; Lipes, M.A.; Ploegh, H.L. LMP2 expression and proteasome activity in NOD mice. Nat. Med. 2000, 6, 1065–1066. [Google Scholar] [CrossRef] [PubMed]
- Visekruna, A.; Joeris, T.; Seidel, D.; Kroesen, A.; Loddenkemper, C.; Zeitz, M.; Kaufmann, S.H.; Schmidt-Ullrich, R.; Steinhoff, U. Proteasome-mediated degradation of IkappaBalpha and processing of p105 in Crohn disease and ulcerative colitis. J. Clin. Investig. 2006, 116, 3195–3203. [Google Scholar] [CrossRef] [PubMed]
- Bitzer, A.; Basler, M.; Krappmann, D.; Groettrup, M. Immunoproteasome subunit deficiency has no influence on the canonical pathway of NF-kappaB activation. Mol. Immunol. 2017, 83, 147–153. [Google Scholar] [CrossRef]
- Muchamuel, T.; Basler, M.; Aujay, M.A.; Suzuki, E.; Kalim, K.W.; Lauer, C.; Sylvain, C.; Ring, E.R.; Shields, J.; Jiang, J.; et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 2009, 15, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Mundt, S.; Muchamuel, T.; Moll, C.; Jiang, J.; Groettrup, M.; Kirk, C.J. Inhibition of the immunoproteasome ameliorates experimental autoimmune encephalomyelitis. EMBO Mol. Med. 2014, 6, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Maurits, E.; de Bruin, G.; Koerner, J.; Overkleeft, H.S.; Groettrup, M. Amelioration of autoimmunity with an inhibitor selectively targeting all active centres of the immunoproteasome. Br. J. Pharmacol. 2018, 175, 38–52. [Google Scholar] [CrossRef]
- Basler, M.; Mundt, S.; Bitzer, A.; Schmidt, C.; Groettrup, M. The immunoproteasome: A novel drug target for autoimmune diseases. Clin. Exp. Rheumatol. 2015, 33, 74–79. [Google Scholar]
- Basler, M.; Groettrup, M. Recent insights how combined inhibition of immuno/proteasome subunits enables therapeutic efficacy. Genes Immun. 2020, 21, 273–287. [Google Scholar] [CrossRef]
- Tubio-Santamaria, N.; Ebstein, F.; Heidel, F.H.; Kruger, E. Immunoproteasome Function in Normal and Malignant Hematopoiesis. Cells 2021, 10, 1577. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Groll, M. A Nut for Every Bolt: Subunit-Selective Inhibitors of the Immunoproteasome and Their Therapeutic Potential. Cells 2021, 10, 1929. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Hohn, A.; Tramutola, A.; Cascella, R. Proteostasis Failure in Neurodegenerative Diseases: Focus on Oxidative Stress. Oxid. Med. Cell Longev. 2020, 2020, 5497046. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Lemberg, M.K.; Strisovsky, K. Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol. Cell 2021, 81, 2507–2519. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Zhang, D.; Hannink, M.; Arvisais, E.; Kaufman, R.J.; Diehl, J.A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 2003, 23, 7198–7209. [Google Scholar] [CrossRef]
- Schubert, U.; Anton, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef]
- Kincaid, E.Z.; Che, J.W.; York, I.; Escobar, H.; Reyes-Vargas, E.; Delgado, J.C.; Welsh, R.M.; Karow, M.L.; Murphy, A.J.; Valenzuela, D.M.; et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2012, 13, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Christ, M.; Goebel, H.; Groettrup, M. Immunoproteasome Upregulation Is Not Required to Control Protein Homeostasis during Viral Infection. J. Immunol. 2021, 206, 1697–1708. [Google Scholar] [CrossRef]
- Hensley, S.E.; Zanker, D.; Dolan, B.P.; David, A.; Hickman, H.D.; Embry, A.C.; Skon, C.N.; Grebe, K.M.; Griffin, T.A.; Chen, W.; et al. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J. Immunol. 2010, 184, 4115–4122. [Google Scholar] [CrossRef]
- De Verteuil, D.A.; Rouette, A.; Hardy, M.P.; Lavallee, S.; Trofimov, A.; Gaucher, E.; Perreault, C. Immunoproteasomes shape the transcriptome and regulate the function of dendritic cells. J. Immunol. 2014, 193, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Hewing, B.; Ludwig, A.; Dan, C.; Potzsch, M.; Hannemann, C.; Petry, A.; Lauer, D.; Gorlach, A.; Kaschina, E.; Muller, D.N.; et al. Immunoproteasome subunit ss5i/LMP7-deficiency in atherosclerosis. Sci. Rep. 2017, 7, 13342. [Google Scholar] [CrossRef] [PubMed]
- Seifert, U.; Bialy, L.P.; Ebstein, F.; Bech-Otschir, D.; Voigt, A.; Schroter, F.; Prozorovski, T.; Lange, N.; Steffen, J.; Rieger, M.; et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010, 142, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef] [PubMed]
- Pierre, P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol. Rev. 2005, 207, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Szeto, J.; Kaniuk, N.A.; Canadien, V.; Nisman, R.; Mizushima, N.; Yoshimori, T.; Bazett-Jones, D.P.; Brumell, J.H. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2006, 2, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Nathan, J.A.; Spinnenhirn, V.; Schmidtke, G.; Basler, M.; Groettrup, M.; Goldberg, A.L. Immuno- and constitutive proteasomes do not differ in their abilities to degrade ubiquitinated proteins. Cell 2013, 152, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P.; Hilton, C.; Kisselev, A.F.; Rock, K.L.; Goldberg, A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001, 20, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Abi Habib, J.; De Plaen, E.; Stroobant, V.; Zivkovic, D.; Bousquet, M.P.; Guillaume, B.; Wahni, K.; Messens, J.; Busse, A.; Vigneron, N.; et al. Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci. Rep. 2020, 10, 15765. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; van den Broek, M.; Schwarz, K.; de Giuli, R.; Diener, P.A.; Groettrup, M. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 2001, 167, 6859–6868. [Google Scholar] [CrossRef] [PubMed]
- Barton, L.F.; Cruz, M.; Rangwala, R.; Deepe, G.S.; Monaco, J.J. Regulation of immunoproteasome subunit expression in vivo following pathogenic fungal infection. J. Immunol. 2002, 169, 3046–3052. [Google Scholar] [CrossRef]
- Strehl, B.; Joeris, T.; Rieger, M.; Visekruna, A.; Textoris-Taube, K.; Kaufmann, S.H.; Kloetzel, P.M.; Kuckelkorn, U.; Steinhoff, U. Immunoproteasomes are essential for clearance of Listeria monocytogenes in nonlymphoid tissues but not for induction of bacteria-specific CD8+ T cells. J. Immunol. 2006, 177, 6238–6244. [Google Scholar] [CrossRef]
- Mao, X.; Green, J.M.; Safer, B.; Lindsten, T.; Frederickson, R.M.; Miyamoto, S.; Sonenberg, N.; Thompson, C.B. Regulation of translation initiation factor gene expression during human T cell activation. J. Biol. Chem. 1992, 267, 20444–20450. [Google Scholar] [CrossRef]
- Kleijn, M.; Proud, C.G. The regulation of protein synthesis and translation factors by CD3 and CD28 in human primary T lymphocytes. BMC Biochem. 2002, 3, 11. [Google Scholar] [CrossRef][Green Version]
- Opitz, E.; Koch, A.; Klingel, K.; Schmidt, F.; Prokop, S.; Rahnefeld, A.; Sauter, M.; Heppner, F.L.; Volker, U.; Kandolf, R.; et al. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011, 7, e1002233. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, C.; Morgand, E.; Benhammadi, M.; Rouette, A.; Hardy, M.P.; Gaboury, L.; Perreault, C. Immunoproteasomes Control the Homeostasis of Medullary Thymic Epithelial Cells by Alleviating Proteotoxic Stress. Cell Rep. 2017, 21, 2558–2570. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.K.; Jameson, S.C.; Hogquist, K.A. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003, 21, 139–176. [Google Scholar] [CrossRef]
- Nil, A.; Firat, E.; Sobek, V.; Eichmann, K.; Niedermann, G. Expression of housekeeping and immunoproteasome subunit genes is differentially regulated in positively and negatively selecting thymic stroma subsets. Eur. J. Immunol. 2004, 34, 2681–2689. [Google Scholar] [CrossRef]
- Halangk, W.; Lerch, M.M.; Brandt-Nedelev, B.; Roth, W.; Ruthenbuerger, M.; Reinheckel, T.; Domschke, W.; Lippert, H.; Peters, C.; Deussing, J. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J. Clin. Investig. 2000, 106, 773–781. [Google Scholar] [CrossRef]
- Lukas, J.; Pospech, J.; Oppermann, C.; Hund, C.; Iwanov, K.; Pantoom, S.; Petters, J.; Frech, M.; Seemann, S.; Thiel, F.G.; et al. Role of endoplasmic reticulum stress and protein misfolding in disorders of the liver and pancreas. Adv. Med. Sci. 2019, 64, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.J.; Lee, H.S. Experimental models of pancreatitis. Clin. Endosc. 2014, 47, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Iles, K.E.; Forman, H.J. Macrophage signaling and respiratory burst. Immunol. Res. 2002, 26, 95–105. [Google Scholar] [CrossRef]
- Kuwano, Y.; Kawahara, T.; Yamamoto, H.; Teshima-Kondo, S.; Tominaga, K.; Masuda, K.; Kishi, K.; Morita, K.; Rokutan, K. Interferon-gamma activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006, 290, C433–C443. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie 2001, 83, 301–310. [Google Scholar] [CrossRef]
- Inai, Y.; Nishikimi, M. Increased degradation of oxidized proteins in yeast defective in 26 S proteasome assembly. Arch. Biochem. Biophys. 2002, 404, 279–284. [Google Scholar] [CrossRef]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Sun, H.; Murray, K.K.; Costa, J.; Williams, T.D.; Bigelow, D.J.; Squier, T.C. Selective degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 2001, 276, 937–943. [Google Scholar] [CrossRef]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef]
- Kotamraju, S.; Matalon, S.; Matsunaga, T.; Shang, T.; Hickman-Davis, J.M.; Kalyanaraman, B. Upregulation of immunoproteasomes by nitric oxide: Potential antioxidative mechanism in endothelial cells. Free Radic. Biol. Med. 2006, 40, 1034–1044. [Google Scholar] [CrossRef]
- Kwak, M.K.; Wakabayashi, N.; Greenlaw, J.L.; Yamamoto, M.; Kensler, T.W. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 2003, 23, 8786–8794. [Google Scholar] [CrossRef]
- Cenci, S.; Oliva, L.; Cerruti, F.; Milan, E.; Bianchi, G.; Raule, M.; Mezghrani, A.; Pasqualetto, E.; Sitia, R.; Cascio, P. Pivotal Advance: Protein synthesis modulates responsiveness of differentiating and malignant plasma cells to proteasome inhibitors. J. Leukoc. Biol. 2012, 92, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Weyburne, E.S.; Wilkins, O.M.; Sha, Z.; Williams, D.A.; Pletnev, A.A.; de Bruin, G.; Overkleeft, H.S.; Goldberg, A.L.; Cole, M.D.; Kisselev, A.F. Inhibition of the Proteasome beta2 Site Sensitizes Triple-Negative Breast Cancer Cells to beta5 Inhibitors and Suppresses Nrf1 Activation. Cell Chem. Biol. 2017, 24, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Pawar, A.; Basler, M.; Goebel, H.; Alvarez Salinas, G.O.; Groettrup, M.; Bottcher, T. Competitive Metabolite Profiling of Natural Products Reveals Subunit Specific Inhibitors of the 20S Proteasome. ACS Cent. Sci. 2020, 6, 241–246. [Google Scholar] [CrossRef]
- Basler, M.; Lindstrom, M.M.; LaStant, J.J.; Bradshaw, J.M.; Owens, T.D.; Schmidt, C.; Maurits, E.; Tsu, C.; Overkleeft, H.S.; Kirk, C.J.; et al. Co-inhibition of immunoproteasome subunits LMP2 and LMP7 is required to block autoimmunity. EMBO Rep. 2018, 19, e46512. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.W.B.; Lowe, E.; Anderl, J.L.; Fan, A.; Muchamuel, T.; Bowers, S.; Moebius, D.; Kirk, C.; McMinn, D.L. A required immunoproteasome subunit inhibition profile for anti-inflammatory efficacy and clinical candidate KZR-616 ((2S,3R)-N-((S)-3-(cyclopent-1-en-1-yl)-1-((R)-2-methyloxiran-2-yl)-1-oxopropan-2-yl)-3-hydroxy-3-(4-methoxyphenyl)-2-((S)-2-(2-morpholinoacetamido)propanamido)pr openamide). J. Med. Chem. 2018, 61, 11127–11143. [Google Scholar] [CrossRef]
- Basler, M.; Claus, M.; Klawitter, M.; Goebel, H.; Groettrup, M. Immunoproteasome Inhibition Selectively Kills Human CD14(+) Monocytes and as a Result Dampens IL-23 Secretion. J. Immunol. 2019, 203, 1776–1785. [Google Scholar] [CrossRef]
- Ichikawa, H.T.; Conley, T.; Muchamuel, T.; Jiang, J.; Lee, S.; Owen, T.; Barnard, J.; Nevarez, S.; Goldman, B.I.; Kirk, C.J.; et al. Novel proteasome inhibitors have a beneficial effect in murine lupus via the dual inhibition of type i interferon and autoantibody secreting cells. Arthritis Rheum. 2012, 64, 493–503. [Google Scholar] [CrossRef]
- Li, J.; Basler, M.; Alvarez, G.; Brunner, T.; Kirk, C.J.; Groettrup, M. Immunoproteasome inhibition prevents chronic antibody-mediated allograft rejection in renal transplantation. Kidney Int. 2018, 93, 670–680. [Google Scholar] [CrossRef]
- Li, J.; Koerner, J.; Basler, M.; Brunner, T.; Kirk, C.J.; Groettrup, M. Immunoproteasome inhibition induces plasma cell apoptosis and preserves kidney allografts by activating the unfolded protein response and suppressing plasma cell survival factors. Kidney Int. 2019, 95, 611–623. [Google Scholar] [CrossRef]
- Basler, M.; Li, J.; Groettrup, M. On the role of the immunoproteasome in transplant rejection. Immunogenetics 2019, 71, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Berger, T.; Groettrup, M.; Basler, M. Immunoproteasome Inhibition Impairs T and B Cell Activation by Restraining ERK Signaling and Proteostasis. Front. Immunol. 2018, 9, 2386. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.B.; La Greca, F.; Arastu-Kapur, S.; Caiazza, F.; Cimermancic, P.; Buchholz, T.J.; Anderl, J.L.; Ravalin, M.; Bohn, M.F.; Sali, A.; et al. Immunoproteasome functions explained by divergence in cleavage specificity and regulation. Elife 2017, 6, e27364. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Beck, U.; Kirk, C.J.; Groettrup, M. The antiviral immune response in mice devoid of immunoproteasome activity. J. Immunol. 2011, 187, 5548–5557. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, G.; Schregle, R.; Alvarez, G.; Huber, E.M.; Groettrup, M. The 20S immunoproteasome and constitutive proteasome bind with the same affinity to PA28alphabeta and equally degrade FAT10. Mol. Immunol. 2019, 113, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Lesne, J.; Locard-Paulet, M.; Parra, J.; Zivkovic, D.; Menneteau, T.; Bousquet, M.P.; Burlet-Schiltz, O.; Marcoux, J. Conformational maps of human 20S proteasomes reveal PA28- and immuno-dependent inter-ring crosstalks. Nat. Commun. 2020, 11, 6140. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Xu, C.; Chen, K.; Zhao, Q.; Wang, S.; Yin, Y.; Peng, C.; Ding, Z.; Cong, Y. Cryo-EM of mammalian PA28alphabeta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28alphabeta. Nat. Commun. 2021, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.A.M.; Bashore, C.; Dong, K.C.; Martin, A. The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 2019, 177, 286–298.e215. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Weng, T.; Hu, S.; Yuan, Z.; Xiong, H.; Huang, B.; Cai, Y.; Li, L.; Fu, X. IFN-gamma-induced ER stress impairs autophagy and triggers apoptosis in lung cancer cells. Oncoimmunology 2021, 10, 1962591. [Google Scholar] [CrossRef] [PubMed]
- Heinemeyer, W.; Gruhler, A.; Mohrle, V.; Mahe, Y.; Wolf, D.H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 1993, 268, 5115–5120. [Google Scholar] [CrossRef]
- Arendt, C.S.; Hochstrasser, M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl. Acad. Sci. USA 1997, 94, 7156–7161. [Google Scholar] [CrossRef]
- Besse, A.; Besse, L.; Kraus, M.; Mendez-Lopez, M.; Bader, J.; Xin, B.T.; de Bruin, G.; Maurits, E.; Overkleeft, H.S.; Driessen, C. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell Chem. Biol. 2019, 26, 340–351.e343. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Callard, A.; Goldberg, A.L. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 2006, 281, 8582–8590. [Google Scholar] [CrossRef] [PubMed]
- Oberdorf, J.; Carlson, E.J.; Skach, W.R. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry 2001, 40, 13397–13405. [Google Scholar] [CrossRef] [PubMed]
| Effect on Protein Homeostasis | Inflammatory Condition | Cell Type, Organ, Proteasome Type | Effect on Protein Degradation | Reference |
|---|---|---|---|---|
| no | naïve mice | spleen of triple KO mice vs. wild-type mice | no accumulation of ubiquitylated proteins | [61] |
| no | unstimulated or LPS stimulated | LMP−/− vs. wild-type B cells | no accumulation of ubiquitylated proteins | [63] |
| no | immature DCs, LPS-matured DCs | LMP7−/−MECL-1−/− vs. wild-type DCs | no accumulation of ubiquitylated proteins | [64] |
| no | unstimulated or IFN-γ stimulated | LMP7−/− vs. wild-type macrophages | no accumulation of ubiquitylated proteins | [65] |
| yes | IFN-γ stimulated | LMP7−/− vs. wild-type MEFs | accumulation of poly-ubiquitylated proteins | [66] |
| ALIS formation | ||||
| LPS injection | liver of LPS stimulated LMP7−/− mice vs. wild-type mice | accumulation of poly-ubiquitylated proteins | ||
| experimental autoimmune encephalomyelitis | brain of diseased LMP7−/− mice vs. wild-type mice | accumulation of poly-ubiquitylated proteins | ||
| no | IFN-γ stimulated | LMP7−/− vs. wild-type MEFs | no accumulation of poly-ubiquitylated proteins | [70] |
| no ALIS formation | ||||
| purified 26S standard proteasomes or 26S immunoproteasomes | no difference of 26S standard or 26S immunoproteasomes in degrading Ub5DHFR | |||
| no | purified 26S proteasome of cells expressing standard proteasomes, intermediate proteasomes, or immunoproteasomes | efficiency to degrade ubiquitylated proteins is similar between different types of proteasomes | [72] | |
| no | naïve mice | spleen or liver of LMP7−/− mice vs. wild-type mice | no accumulation of poly-ubiquitylated proteins | [62] |
| LCMV-infected mice | spleen or liver of diseased LMP7−/− mice vs. wild-type mice | no accumulation of poly-ubiquitylated proteins on d3, 5, 7 post infection | ||
| yes | IFN-γ stimulated | primary cardiomyocytes or B cell depleted splenocytes of LMP7−/− mice vs. wild-type mice | accumulation of poly-ubiquitylated proteins | [78] |
| CVB3-infected mice | cardiac tissue of diseased LMP7−/− mice vs. wild-type mice | accumulation of poly-ubiquitylated proteins in cardiac tissues of diseased mice | ||
| yes | unstimulated | mTECs of LMP7−/−/MECL-1−/− mice vs. wild-type mice | induction of UPR | [79] |
| yes | acute pancreatitis | pancreas of diseased LMP7−/− mice vs. wild-type mice | accumulation of poly-ubiquitylated proteins in pancreas | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basler, M.; Groettrup, M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells 2021, 10, 3216. https://doi.org/10.3390/cells10113216
Basler M, Groettrup M. On the Role of the Immunoproteasome in Protein Homeostasis. Cells. 2021; 10(11):3216. https://doi.org/10.3390/cells10113216
Chicago/Turabian StyleBasler, Michael, and Marcus Groettrup. 2021. "On the Role of the Immunoproteasome in Protein Homeostasis" Cells 10, no. 11: 3216. https://doi.org/10.3390/cells10113216
APA StyleBasler, M., & Groettrup, M. (2021). On the Role of the Immunoproteasome in Protein Homeostasis. Cells, 10(11), 3216. https://doi.org/10.3390/cells10113216







