The NOTCH3 Downstream Target HEYL Is Required for Efficient Human Airway Basal Cell Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primary Human Bronchial Epithelial Cell (HBEC) Culture
2.2. Air–Liquid Interface (ALI) Culture
2.3. RNA Extraction, cDNA Synthesis and Quantitative PCR Analysis
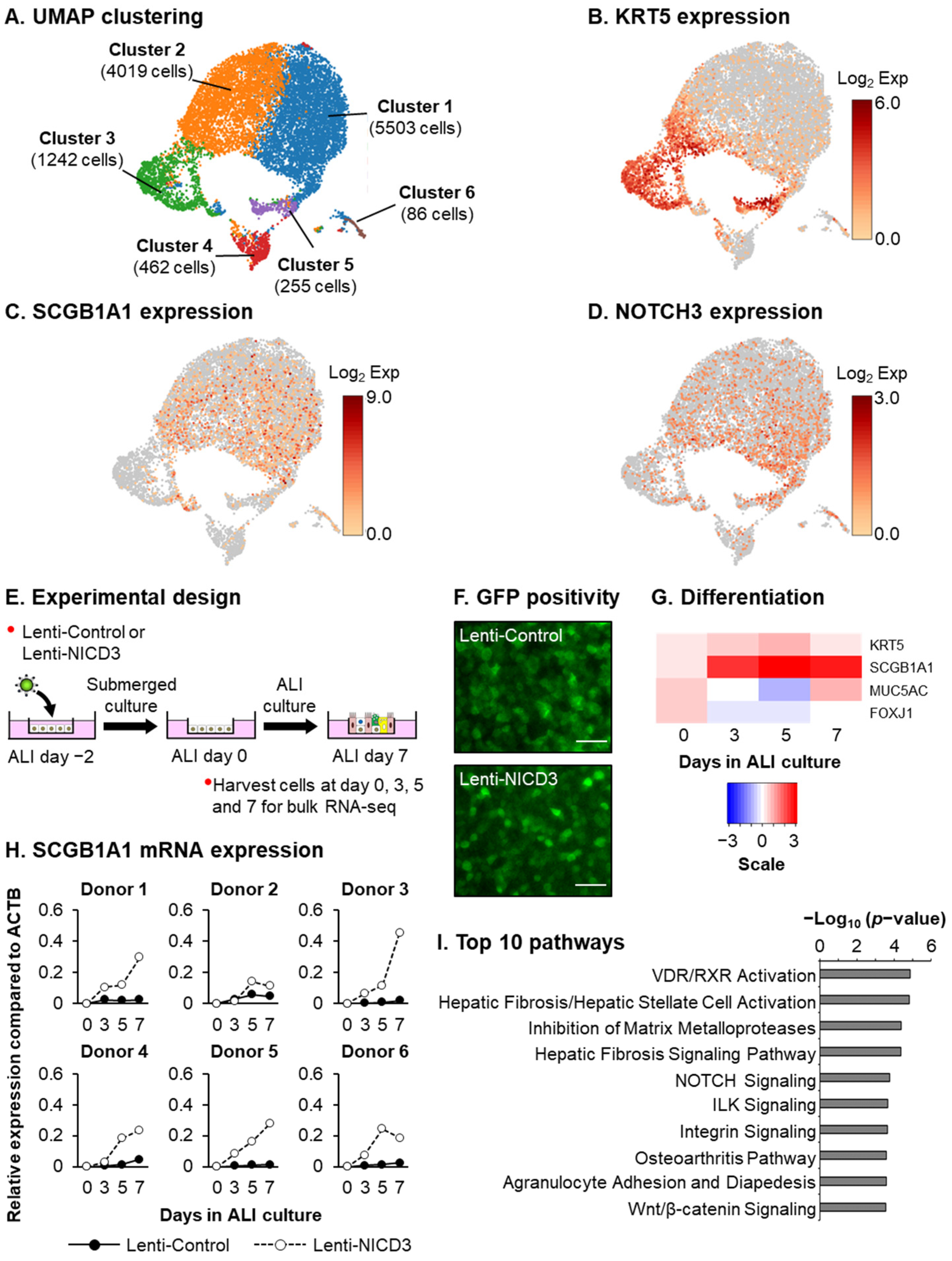
2.4. Single-Cell RNA Sequencing (scRNA-Seq)
2.5. Lentivirus-Based Overexpression of NICD3 and HEYL
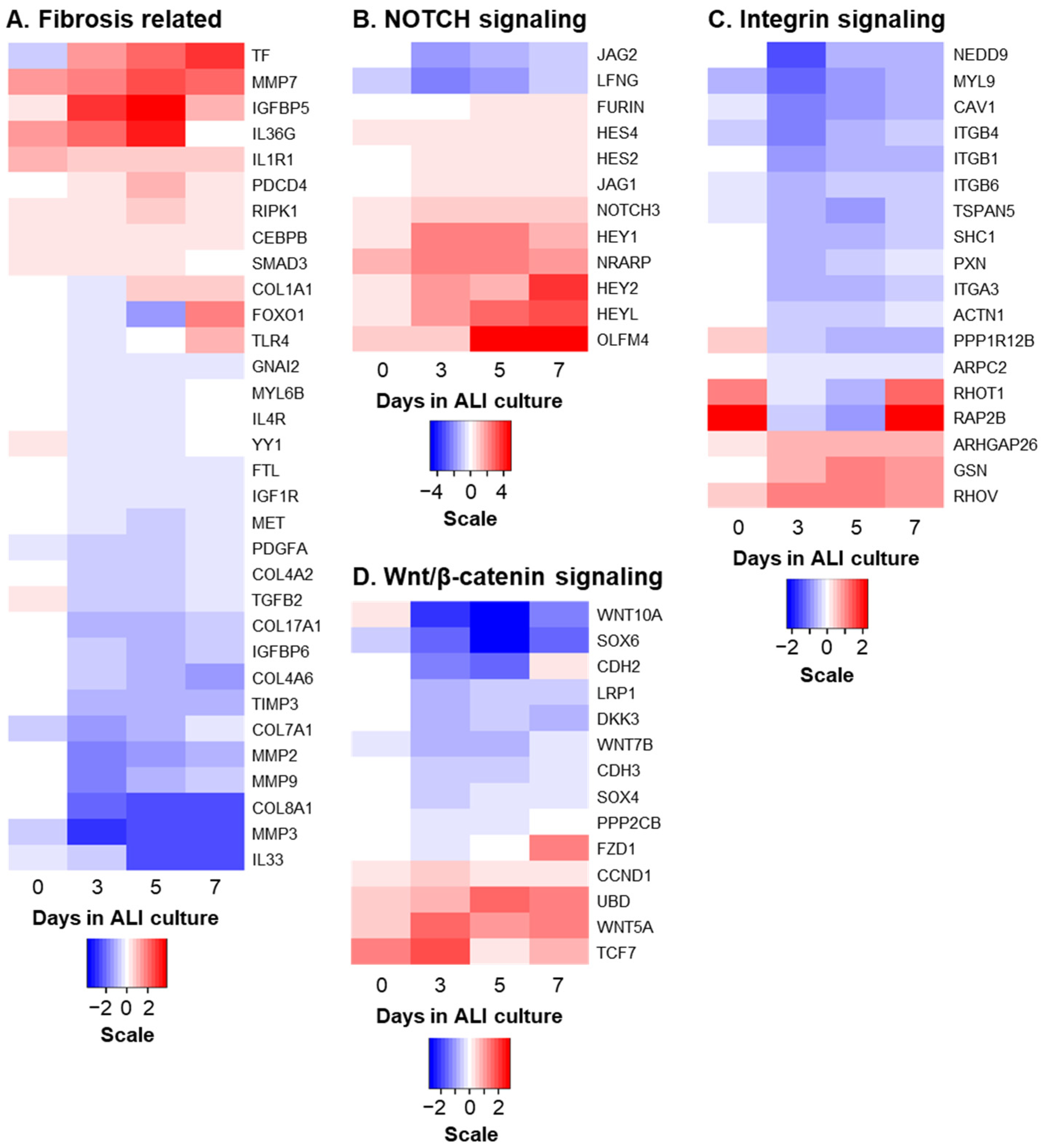
2.6. Bulk RNA Sequencing (Bulk RNA-Seq)
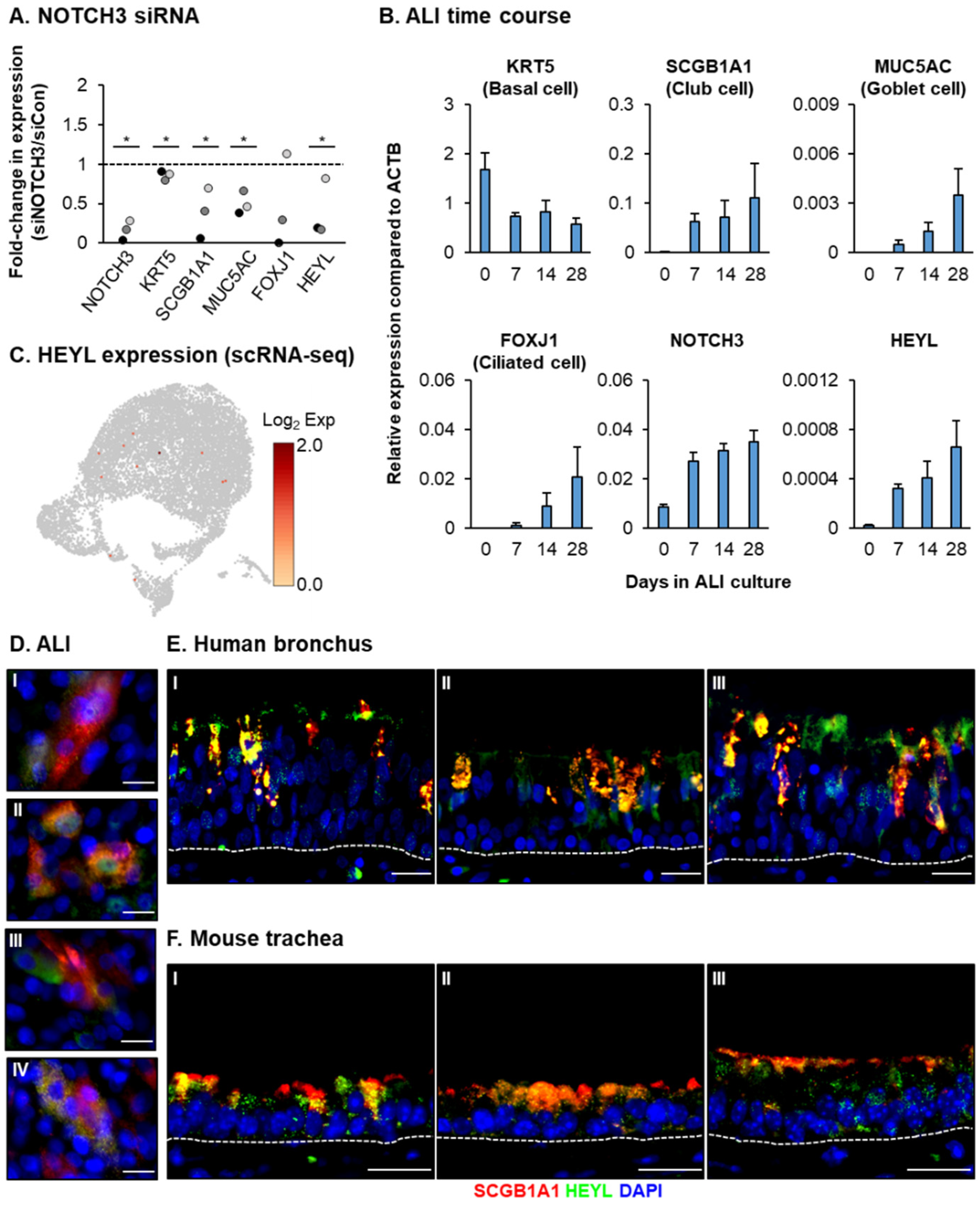
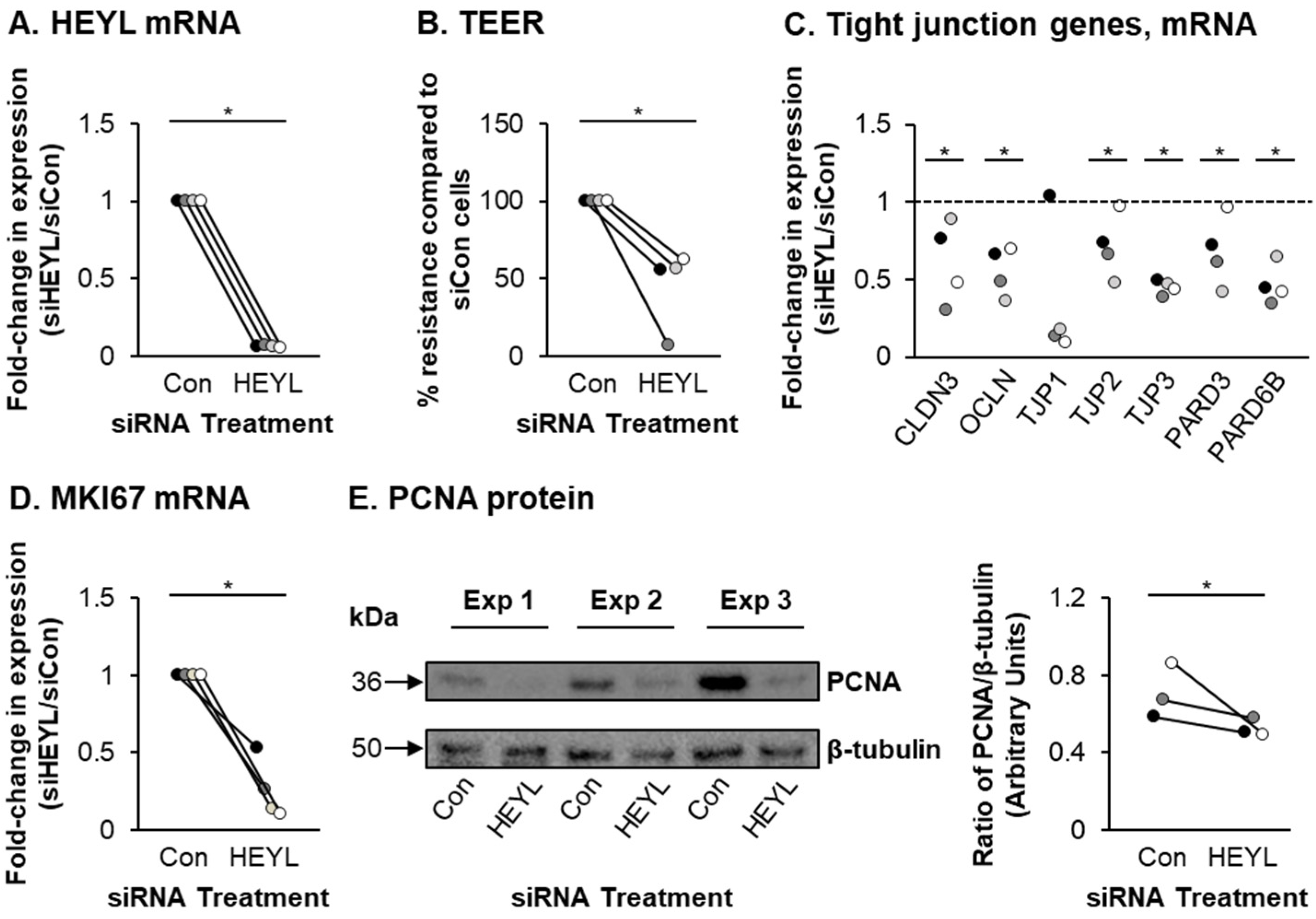
2.7. siRNA-Mediated Knockdown of NOTCH3 and HEYL
2.8. Immunofluorescence Staining
2.9. Mouse Trachea Collection
2.10. Transepithelial Electrical Resistance (TEER)
2.11. Western Blotting
2.12. Statistics
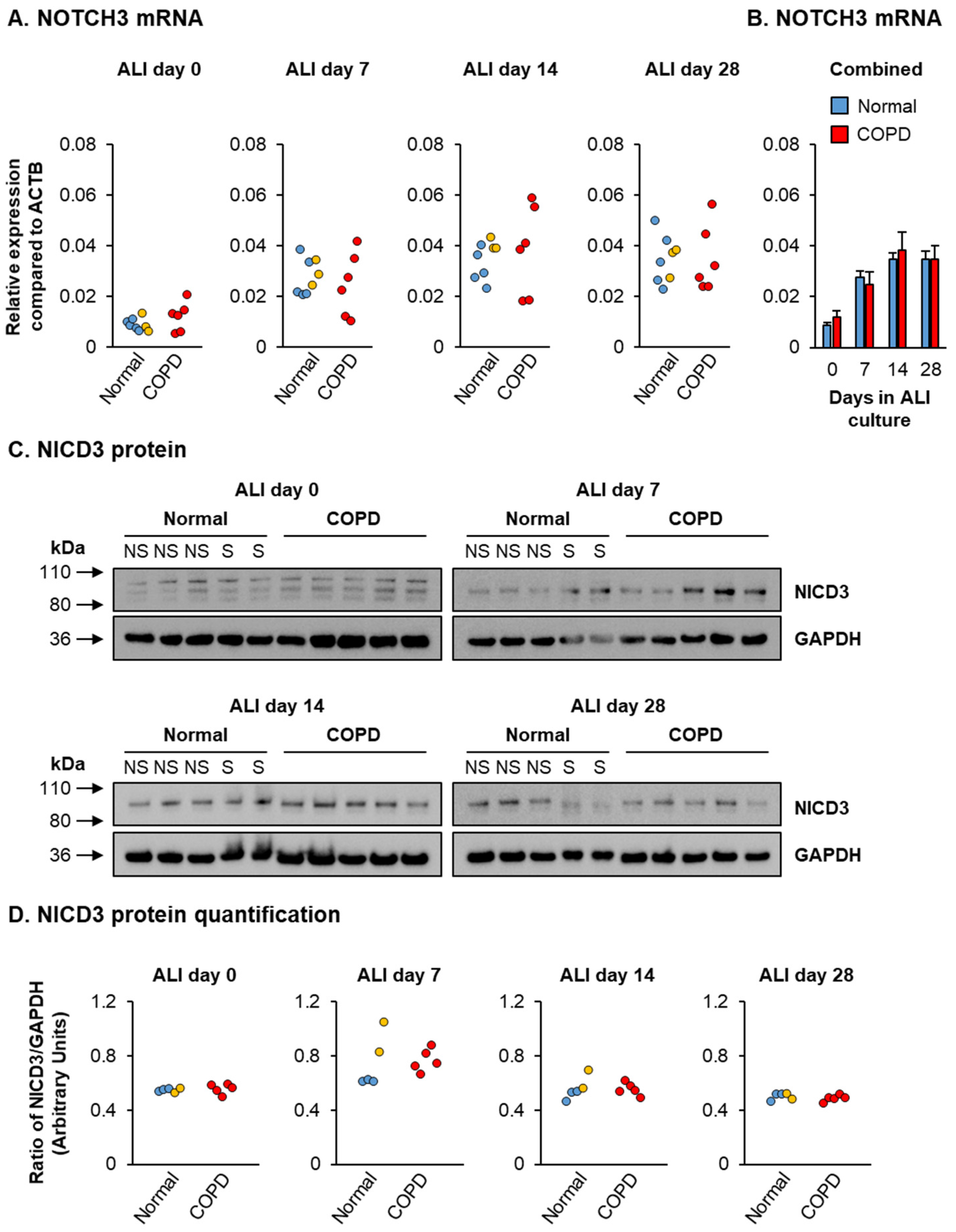
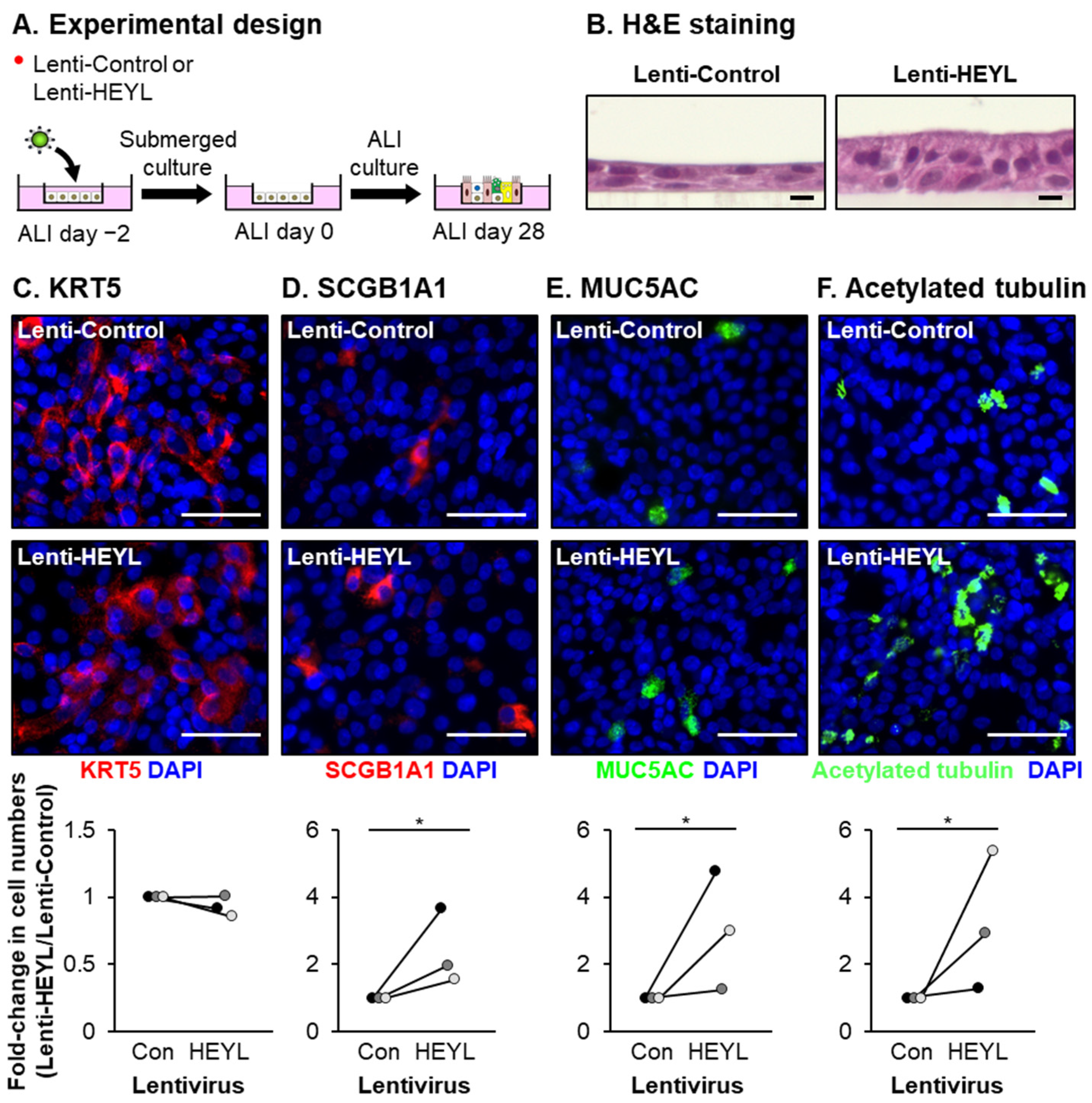
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogan, B.L.; Barkauskas, C.E.; Chapman, H.A.; Epstein, J.A.; Jain, R.; Hsia, C.C.; Niklason, L.; Calle, E.; Le, A.; Randell, S.H.; et al. Repair and Regeneration of the Respiratory System: Complexity, Plasticity, and Mechanisms of Lung Stem Cell Function. Cell Stem Cell 2014, 15, 123–138. [Google Scholar] [CrossRef] [Green Version]
- Rock, J.R.; Randell, S.H.; Hogan, B.L. Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis. Model Mech. 2010, 3, 545–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tata, P.R.; Rajagopal, J. Plasticity in the lung: Making and breaking cell identity. Development 2017, 144, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Whitsett, J.A.; Kalin, T.V.; Xu, Y.; Kalinichenko, V.V. Building and Regenerating the Lung Cell by Cell. Physiol. Rev. 2019, 99, 513–554. [Google Scholar] [CrossRef] [PubMed]
- Carraro, G.; Mulay, A.; Yao, C.; Mizuno, T.; Konda, B.; Petrov, M.; Lafkas, D.; Arron, J.R.; Hogaboam, C.M.; Chen, P.; et al. Single-Cell Reconstruction of Human Basal Cell Diversity in Normal and Idiopathic Pulmonary Fibrosis Lungs. Am. J. Respir. Crit. Care Med. 2020, 202, 1540–1550. [Google Scholar] [CrossRef]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Plasschaert, L.W.; Žilionis, R.; Choo-Wing, R.; Savova, V.; Knehr, J.; Roma, G.; Klein, A.M.; Jaffe, A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018, 560, 377–381. [Google Scholar] [CrossRef]
- García, S.R.; Deprez, M.; Lebrigand, K.; Cavard, A.; Paquet, A.; Arguel, M.-J.; Magnone, V.; Truchi, M.; Caballero, I.; Leroy, S.; et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 2019, 146, dev.177428. [Google Scholar] [CrossRef] [Green Version]
- Zaragosi, L.; Deprez, M.; Barbry, P. Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract. Biochem. Soc. Trans. 2020, 48, 327–336. [Google Scholar] [CrossRef]
- Gamez, A.S.; Gras, D.; Petit, A.; Knabe, L.; Molinari, N.; Vachier, I.; Chanez, P.; Bourdin, A. Supplementing Defect in Club Cell Secretory Protein Attenuates Airway Inflammation in COPD. Chest 2015, 147, 1467–1476. [Google Scholar] [CrossRef]
- Pilette, C.; Godding, V.; Kiss, R.; Delos, M.; Verbeken, E.; Decaestecker, C.; De Paepe, K.; Vaerman, J.-P.; Decramer, M.; Sibille, Y. Reduced Epithelial Expression of Secretory Component in Small Airways Correlates with Airflow Obstruction in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2001, 163, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, P.; Ahmed, E.; Serre, I.; Knabe, L.; Bommart, S.; Suehs, C.; Vachier, I.; Berthet, J.P.; Romagnoli, M.; Vernisse, C.; et al. Club Cell Loss as a Feature of Bronchiolization in ILD. Front. Immunol. 2021, 12, 311. [Google Scholar] [CrossRef] [PubMed]
- Shijubo, N.; Itoh, Y.; Yamaguchi, T.; Imada, A.; Hirasawa, M.; Yamada, T.; Kawai, T.; Abe, S. Clara Cell Protein–positive Epithelial Cells Are Reduced in Small Airways of Asthmatics. Am. J. Respir. Crit. Care Med. 1999, 160, 930–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodas, M.; Moore, A.R.; Subramaniyan, B.; Georgescu, C.; Wren, J.D.; Freeman, W.M.; Brown, B.R.; Metcalf, J.P.; Walters, M.S. Cigarette Smoke Activates NOTCH3 to Promote Goblet Cell Differentiation in Human Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2021, 64, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Saganta, A.; Law, B.M.; Tata, P.R.; Villoria, J.; Saez, B.; Mou, H.; Zhao, R.; Rajagopal, J. Injury Induces Direct Lineage Segregation of Functionally Distinct Airway Basal Stem/Progenitor Cell Subpopulations. Cell Stem Cell 2015, 16, 184–197. [Google Scholar] [CrossRef] [Green Version]
- Boucherat, O.; Chakir, J.; Jeannotte, L. The loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in lung airways. Biol. Open 2012, 1, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Carrer, M.; Crosby, J.R.; Sun, G.; Zhao, C.; Damle, S.S.; Kuntz, S.G.; Monia, B.P.; Hart, C.E.; Grossman, T.R. Antisense Oligonucleotides Targeting Jagged 1 Reduce House Dust Mite–induced Goblet Cell Metaplasia in the Adult Murine Lung. Am. J. Respir. Cell Mol. Biol. 2020, 63, 46–56. [Google Scholar] [CrossRef]
- Danahay, H.; Pessotti, A.D.; Coote, J.; Montgomery, B.E.; Xia, D.; Wilson, A.; Yang, H.; Wang, Z.; Bevan, L.; Thomas, C.; et al. Notch2 Is Required for Inflammatory Cytokine-Driven Goblet Cell Metaplasia in the Lung. Cell Rep. 2015, 10, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Gomi, K.; Arbelaez, V.; Crystal, R.G.; Walters, M.S. Activation of NOTCH1 or NOTCH3 Signaling Skews Human Airway Basal Cell Differentiation toward a Secretory Pathway. PLoS ONE 2015, 10, e0116507. [Google Scholar] [CrossRef] [Green Version]
- Gomi, K.; Staudt, M.R.; Salit, J.; Kaner, R.J.; Heldrich, J.; Rogalski, A.M.; Arbelaez, V.; Crystal, R.G.; Walters, M.S. JAG1-Mediated Notch Signaling Regulates Secretory Cell Differentiation of the Human Airway Epithelium. Stem Cell Rev. Rep. 2016, 12, 454–463. [Google Scholar] [CrossRef] [Green Version]
- Guseh, J.; Bores, S.A.; Stanger, B.Z.; Zhou, Q.; Anderson, W.; Melton, D.A.; Rajagopal, J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 2009, 136, 1751–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Udaka, N.; Yazawa, T.; Okudela, K.; Hayashi, H.; Sudo, T.; Guillemot, F.; Kageyama, R.; Kitamura, H. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 2000, 127, 3913–3921. [Google Scholar] [CrossRef]
- Jing, Y.; Gimenes, J.A.; Mishra, R.; Pham, D.; Comstock, A.T.; Yu, D.; Sajjan, U. NOTCH3 contributes to rhinovirus-induced goblet cell hyperplasia in COPD airway epithelial cells. Thorax 2018, 74, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Kiyokawa, H.; Morimoto, M. Notch signaling in the mammalian respiratory system, specifically the trachea and lungs, in development, homeostasis, regeneration, and disease. Dev. Growth Differ. 2020, 62, 67–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafkas, D.; Shelton, A.; Chiu, C.; Boenig, G.D.L.; Chen, Y.; Stawicki, S.S.; Siltanen, C.; Reichelt, M.; Zhou, M.; Wu, X.; et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 2015, 528, 127–131. [Google Scholar] [CrossRef]
- Marcet, B.; Chevalier, B.; Luxardi, G.; Coraux, C.; Zaragosi, L.-E.; Cibois, M.; Robbe-Sermesant, K.; Jolly, T.; Cardinaud, B.; Moreilhon, C.; et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011, 13, 693–699. [Google Scholar] [CrossRef]
- Mori, M.; Mahoney, J.E.; Stupnikov, M.R.; Paez-Cortez, J.R.; Szymaniak, A.D.; Varelas, X.; Herrick, D.; Schwob, J.; Zhang, H.; Cardoso, W.V. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development 2015, 142, 258–267. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, M.; Liu, Z.; Cheng, H.T.; Winters, N.; Bader, D.; Kopan, R. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J. Cell Sci. 2010, 123, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, M.; Nishinakamura, R.; Saga, Y.; Kopan, R. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development 2012, 139, 4365–4373. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Saganta, A.; Law, B.M.; Gonzalez-Celeiro, M.; Vinarsky, V.; Rajagopal, J. Ciliated Cells of Pseudostratified Airway Epithelium Do Not Become Mucous Cells after Ovalbumin Challenge. Am. J. Respir. Cell Mol. Biol. 2013, 48, 364–373. [Google Scholar] [CrossRef] [Green Version]
- Pardo-Saganta, A.; Tata, P.R.; Law, B.M.; Saez, B.; Chow, R.; Prabhu, M.; Gridley, T.; Rajagopal, J. Parent stem cells can serve as niches for their daughter cells. Nat. Cell Biol. 2015, 523, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Bisht, B.; Darmawan, D.O.; Chiou, R.; Ha, V.L.; Wallace, W.D.; Chon, A.T.; Hegab, A.E.; Grogan, T.; Elashoff, D.A.; et al. Dynamic Changes in Intracellular ROS Levels Regulate Airway Basal Stem Cell Homeostasis through Nrf2-Dependent Notch Signaling. Cell Stem Cell 2014, 15, 199–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, A.T.; Nichol, K.S.; Veerati, P.C.; Moheimani, F.; Kicic, A.; Stick, S.; Bartlett, N.; Grainge, C.L.; Wark, P.A.B.; Hansbro, P.; et al. Blocking Notch3 Signaling Abolishes MUC5AC Production in Airway Epithelial Cells from Individuals with Asthma. Am. J. Respir. Cell Mol. Biol. 2020, 62, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Gao, X.; Xue, Y.; Randell, S.H.; Kong, Y.-Y.; Hogan, B.L. Notch-Dependent Differentiation of Adult Airway Basal Stem Cells. Cell Stem Cell 2011, 8, 639–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stupnikov, M.R.; Yang, Y.; Mori, M.; Lu, J.; Cardoso, W.V. Jagged and Delta-like ligands control distinct events during airway progenitor cell differentiation. Elife 2019, 8, e50487. [Google Scholar] [CrossRef] [PubMed]
- Tata, P.R.; Mou, H.; Pardo-Saganta, A.; Zhao, R.; Prabhu, M.; Law, B.M.; Vinarsky, V.; Cho, J.; Breton, S.; Sahay, A.; et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nat. Cell Biol. 2013, 503, 218–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tata, P.R.; Pardo-Saganta, A.; Prabhu, M.; Vinarsky, V.; Law, B.M.; Fontaine, B.A.; Tager, A.M.; Rajagopal, J. Airway-Specific Inducible Transgene Expression Using Aerosolized Doxycycline. Am. J. Respir. Cell Mol. Biol. 2013, 49, 1048–1056. [Google Scholar] [CrossRef] [Green Version]
- Tsao, P.-N.; Matsuoka, C.; Wei, S.-C.; Sato, A.; Sato, S.; Hasegawa, K.; Chen, H.-K.; Ling, T.-Y.; Mori, M.; Cardoso, W.V.; et al. Epithelial Notch signaling regulates lung alveolar morphogenesis and airway epithelial integrity. Proc. Natl. Acad. Sci. USA 2016, 113, 8242–8247. [Google Scholar] [CrossRef] [Green Version]
- Tsao, P.-N.; Vasconcelos, M.; Izvolsky, K.I.; Qian, J.; Lu, J.; Cardoso, W.V. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 2009, 136, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Tsao, P.-N.; Wei, S.-C.; Wu, M.-F.; Huang, M.-T.; Lin, H.-Y.; Lee, M.-C.; Lin, K.; Wang, I.-J.; Kaartinen, V.; Yang, L.-T.; et al. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development 2011, 138, 3533–3543. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Li, A.; Borok, Z.; Li, C.; Minoo, P. NOTCH1 Is Required for Regeneration of Clara Cells during Repair of Airway Injury. Stem Cells 2012, 30, 946–955. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Moghal, N.; Egan, S.E. Notch Signaling in Lung Development and Disease. Adv. Exp. Med. Biol. 2012, 727, 89–98. [Google Scholar] [CrossRef]
- Zhang, S.; Loch, A.J.; Radtke, F.; Egan, S.E.; Xu, K. Jagged1 is the major regulator of notch-dependent cell fate in proximal airways. Dev. Dyn. 2013, 242, 678–686. [Google Scholar] [CrossRef] [Green Version]
- Zuo, W.; Zhang, T.; Wu, D.Z.; Guan, S.P.; Liew, A.-A.; Yamamoto, Y.; Wang, X.; Lim, S.J.; Vincent, M.; Lessard, M.; et al. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nat. Cell Biol. 2015, 517, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Giuranno, L.; Roig, E.M.; Wansleeben, C.; Berg, A.V.D.; Groot, A.J.; Dubois, L.; Vooijs, M. NOTCH inhibition promotes bronchial stem cell renewal and epithelial barrier integrity after irradiation. Stem Cells Transl. Med. 2020, 9, 799–812. [Google Scholar] [CrossRef] [Green Version]
- Giuranno, L.; Wansleeben, C.; Iannone, R.; Arathoon, L.; Hounjet, J.; Groot, A.J.; Vooijs, M. NOTCH signaling promotes the survival of irradiated basal airway stem cells. Am. J. Physiol. Cell. Mol. Physiol. 2019, 317, L414–L423. [Google Scholar] [CrossRef] [PubMed]
- Kovall, R.A.; Gebelein, B.; Sprinzak, D.; Kopan, R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev. Cell 2017, 41, 228–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [Green Version]
- Hendrix, A.Y.; Kheradmand, F. The Role of Matrix Metalloproteinases in Development, Repair, and Destruction of the Lungs. Prog. Mol. Biol. Transl. Sci. 2017, 148, 1–29. [Google Scholar] [CrossRef]
- Teoh, C.M.; Tan, S.S.L.; Tran, T. Integrins as Therapeutic Targets for Respiratory Diseases. Curr. Mol. Med. 2015, 15, 714–734. [Google Scholar] [CrossRef]
- Wang, Z.; Li, W.; Guo, Q.; Wang, Y.; Ma, L.; Zhang, X. Insulin-Like Growth Factor-1 Signaling in Lung Development and Inflammatory Lung Diseases. BioMed Res. Int. 2018, 2018, 6057589. [Google Scholar] [CrossRef] [Green Version]
- Baarsma, H.A.; Konigshoff, M. ‘WNT-er is coming’: WNT signalling in chronic lung diseases. Thorax 2017, 72, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Staudt, M.; Buro-Auriemma, L.J.; Walters, M.S.; Salit, J.; Vincent, T.; Shaykhiev, R.; Mezey, J.G.; Tilley, A.E.; Kaner, R.J.; Ho, M.W.Y.; et al. Airway Basal Stem/Progenitor Cells Have Diminished Capacity to Regenerate Airway Epithelium in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2014, 190, 955–958. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, M.; Miller, Y.E.; Nakachi, I.; Kwon, J.B.; Barón, A.E.; Brantley, A.E.; Merrick, D.T.; Franklin, W.A.; Keith, R.L.; Vandivier, R.W. Exhaustion of Airway Basal Progenitor Cells in Early and Established Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2018, 197, 885–896. [Google Scholar] [CrossRef]
- Gohy, S.; Carlier, F.M.; Fregimilicka, C.; Detry, B.; Lecocq, M.; Ladjemi, M.Z.; Verleden, S.; Hoton, D.; Weynand, B.; Bouzin, C.; et al. Altered generation of ciliated cells in chronic obstructive pulmonary disease. Sci. Rep. 2019, 9, 17963. [Google Scholar] [CrossRef] [PubMed]
- Rao, W.; Wang, S.; Duleba, M.; Niroula, S.; Goller, K.; Xie, J.; Mahalingam, R.; Neupane, R.; Liew, A.A.; Vincent, M.; et al. Regenerative Metaplastic Clones in COPD Lung Drive Inflammation and Fibrosis. Cell 2020, 181, 848–864.e18. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Horowitz, J.C.; Naba, A.; Ambalavanan, N.; Atabai, K.; Balestrini, J.; Bitterman, P.B.; Corley, R.A.; Ding, B.-S.; Engler, A.J.; et al. Extracellular matrix in lung development, homeostasis and disease. Matrix Biol. 2018, 73, 77–104. [Google Scholar] [CrossRef] [Green Version]
- Tschumperlin, D.J. Matrix, Mesenchyme, and Mechanotransduction. Ann. Am. Thorac. Soc. 2015, 12, S24–S29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, J.K.; Mauad, T.; Tjin, G.; Karlsson, J.C.; Westergren-Thorsson, G. The extracellular matrix—The under-recognized element in lung disease? J. Pathol. 2016, 240, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Vera, L.; Garcia-Olloqui, P.; Petri, E.; Viñado, A.C.; Valera, P.S.; Blasco-Iturri, Z.; Calvo, I.A.; Cenzano, I.; Ruppert, C.; Zulueta, J.J.; et al. Notch3 Deficiency Attenuates Pulmonary Fibrosis and Impedes Lung-Function Decline. Am. J. Respir. Cell Mol. Biol. 2021, 64, 465–476. [Google Scholar] [CrossRef]
- McCauley, K.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Ng-Blichfeldt, J.P.; Ota, C.; Ciminieri, C.; Ren, W.; Hiemstra, P.S.; Stolk, J.; Gosens, R.; Königshoff, M. Wnt/beta-catenin signaling is critical for regenerative potential of distal lung epithelial progenitor cells in homeostasis and emphysema. Stem Cells 2020, 38, 1467–1478. [Google Scholar] [CrossRef] [PubMed]
- Carlier, F.M.; Dupasquier, S.; Ambroise, J.; Detry, B.; Lecocq, M.; Biétry–Claudet, C.; Boukala, Y.; Gala, J.-L.; Bouzin, C.; Verleden, S.E.; et al. Canonical WNT pathway is activated in the airway epithelium in chronic obstructive pulmonary disease. EBioMedicine 2020, 61, 103034. [Google Scholar] [CrossRef] [PubMed]
- Aros, C.J.; Paul, M.; Pantoja, C.J.; Bisht, B.; Meneses, K.; Vijayaraj, P.; Sandlin, J.M.; France, B.; Tse, J.A.; Chen, M.W.; et al. High-Throughput Drug Screening Identifies a Potent Wnt Inhibitor that Promotes Airway Basal Stem Cell Homeostasis. Cell Rep. 2020, 30, 2055–2064.e5. [Google Scholar] [CrossRef] [Green Version]
- Aros, C.J.; Vijayaraj, P.; Pantoja, C.J.; Bisht, B.; Meneses, L.K.; Sandlin, J.M.; Tse, J.A.; Chen, M.W.; Purkayastha, A.; Shia, D.W.; et al. Distinct Spatiotemporally Dynamic Wnt-Secreting Niches Regulate Proximal Airway Regeneration and Aging. Cell Stem Cell 2020, 27, 413–429.e4. [Google Scholar] [CrossRef]
- Aros, C.J.; Pantoja, C.J.; Gomperts, B.N. Wnt signaling in lung development, regeneration, and disease progression. Commun. Biol. 2021, 4, 601. [Google Scholar] [CrossRef]
- Hirano, K.; Namihira, M. LSD1 Mediates Neuronal Differentiation of Human Fetal Neural Stem Cells by Controlling the Expression of a Novel Target Gene, HEYL. Stem Cells 2016, 34, 1872–1882. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Diehl, A.; Nguyen, N.K.; Korangath, P.; Teo, W.; Cho, S.; Kominsky, S.; Huso, D.L.; Feigenbaum, L.; Rein, A.; et al. The Notch pathway inhibits TGFbeta signaling in breast cancer through HEYL-mediated crosstalk. Cancer Res. 2014, 74, 6509–6518. [Google Scholar] [CrossRef] [Green Version]
- Kuo, K.K.; Jian, S.F.; Li, Y.J.; Wan, S.W.; Weng, C.C.; Fang, K.; Wu, D.C.; Cheng, K.H. Epigenetic inactivation of transforming growth factor-beta1 target gene HEYL, a novel tumor suppressor, is involved in the P53-induced apoptotic pathway in hepatocellular carcinoma. Hepatol. Res. 2015, 45, 782–793. [Google Scholar] [CrossRef]
- Lavery, D.N.; Villaronga, M.A.; Walker, M.M.; Patel, A.; Belandia, B.; Bevan, C.L. Repression of Androgen Receptor Activity by HEYL, a Third Member of the Hairy/Enhancer-of-split-related Family of Notch Effectors. J. Biol. Chem. 2011, 286, 17796–17808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalali, A.; Bassuk, A.G.; Kan, L.; Israsena, N.; Mukhopadhyay, A.; McGuire, T.; Kessler, J.A. HeyL promotes neuronal differentiation of neural progenitor cells. J. Neurosci. Res. 2011, 89, 299–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rea, M.; Eckstein, M.; Eleazer, R.; Smith, C.; Fondufe-Mittendorf, Y.N. Genome-wide DNA methylation reprogramming in response to inorganic arsenic links inhibition of CTCF binding, DNMT expression and cellular transformation. Sci. Rep. 2017, 7, 41474. [Google Scholar] [CrossRef] [PubMed]


| Phenotype | Batchnumber | Age (Years) | Gender | Race | Smoker |
|---|---|---|---|---|---|
| Normal | |||||
| Donor 1 | 501936 | 42 | Female | Hispanic | No |
| Donor 2 | 544414 | 48 | Male | Caucasian | No |
| Donor 3 | 619261 | 53 | Male | Caucasian | No |
| Donor 4 | 543643 | 57 | Female | Caucasian | No |
| Donor 5 | 613375 | 65 | Female | Black | No |
| Donor 6 | 529235 | 67 | Female | Black | No |
| Donor 7 | 608196 | 67 | Male | Caucasian | No |
| Donor 8 | 420927 | 69 | Female | Hispanic | No |
| Donor 9 | 444771 | 69 | Male | Black | No |
| Donor 10 | 619260 | 65 | Female | Caucasian | Yes |
| Donor 11 | 625963 | 65 | Female | Caucasian | Yes |
| Donor 12 | 508777 | 66 | Male | Caucasian | Yes |
| COPD | |||||
| Donor 1 | 436083 | 59 | Male | Caucasian | Yes |
| Donor 2 | 636518 | 62 | Female | Black | Yes |
| Donor 3 | 440551 | 63 | Male | Black | Yes |
| Donor 4 | 430905 | 66 | Male | Caucasian | Yes |
| Donor 5 | 636518 | 62 | Female | Black | Yes |
| Donor 6 | 18TL186386 | 69 | Female | Hispanic | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodas, M.; Subramaniyan, B.; Moore, A.R.; Metcalf, J.P.; Ocañas, S.R.; Freeman, W.M.; Georgescu, C.; Wren, J.D.; Walters, M.S. The NOTCH3 Downstream Target HEYL Is Required for Efficient Human Airway Basal Cell Differentiation. Cells 2021, 10, 3215. https://doi.org/10.3390/cells10113215
Bodas M, Subramaniyan B, Moore AR, Metcalf JP, Ocañas SR, Freeman WM, Georgescu C, Wren JD, Walters MS. The NOTCH3 Downstream Target HEYL Is Required for Efficient Human Airway Basal Cell Differentiation. Cells. 2021; 10(11):3215. https://doi.org/10.3390/cells10113215
Chicago/Turabian StyleBodas, Manish, Bharathiraja Subramaniyan, Andrew R. Moore, Jordan P. Metcalf, Sarah R. Ocañas, Willard M. Freeman, Constantin Georgescu, Jonathan D. Wren, and Matthew S. Walters. 2021. "The NOTCH3 Downstream Target HEYL Is Required for Efficient Human Airway Basal Cell Differentiation" Cells 10, no. 11: 3215. https://doi.org/10.3390/cells10113215
APA StyleBodas, M., Subramaniyan, B., Moore, A. R., Metcalf, J. P., Ocañas, S. R., Freeman, W. M., Georgescu, C., Wren, J. D., & Walters, M. S. (2021). The NOTCH3 Downstream Target HEYL Is Required for Efficient Human Airway Basal Cell Differentiation. Cells, 10(11), 3215. https://doi.org/10.3390/cells10113215






