Inherited and Acquired Rhythm Disturbances in Sick Sinus Syndrome, Brugada Syndrome, and Atrial Fibrillation: Lessons from Preclinical Modeling
Abstract
1. Introduction
2. The Embryological Development, Anatomy, and Physiology of the Heart Rhythm
3. Heart Rhythm Diseases: Clinical Signs, Underlying Causes, and Modeling Approaches
4. Sick Sinus Syndrome (SSS): The Complex Modeling of a Multifactorial Disease
- -
- SSS due to genetic mutations
- -
- SSS due to aging
- -
- SSS due to inflammatory conditions and structural alterations
- -
- SSS due to drug or chemical cardiotoxicity
5. Brugada Syndrome: Modeling a Rhythm Disorder with Still Incompletely Defined Etiology
- -
- BrS due to SCN5A haploinsufficiency
- -
- BrS due to SCN5A point mutations
- -
- BrS due to mutations in other genes
- -
- BrS due to unknown gene mutations
- -
- BrS due to drug cardiotoxicity
- -
- BrS modeling in silico
6. Atrial Fibrillation: Modeling a Rhythm Disturbance Highly Prevalent and Often Untreatable
- -
- Modeling of AF-related KCNQ1 mutations
- -
- Modeling of AF-related NPPA mutations
- -
- Modeling of AF-related mutations of TBX5 and other transcription factor genes
- -
- Modeling of AF-related mutations in other genes or in case of unknown mutations
- -
- Other AF modeling modalities
- -
- Modeling AF through in silico reconstruction
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EHRA. White Book. Available online: https://www.escardio.org/static-file/Escardio/Subspecialty/EHRA/Publications/Documents/2017/ehra-white-book-2017.pdf (accessed on 15 April 2021).
- Gutiérrez, M.; Figueroa, F.; Rivero, S.; Salles, J.P.; Arnaiz, P.; Cruzat, L.; Jacobelli, S. Maternal connective tissue disease associated with congenital AV block. Rev. Med. Chil. 1989, 117, 789–793. [Google Scholar] [PubMed]
- Manolis, A.A.; Manolis, T.A.; Melita, H.; Manolis, A.S. Congenital heart block: Pace earlier (Childhood) than later (Adulthood). Trends Cardiovasc. Med. 2020, 30, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-F.; Chen, Y.-J.; Lin, Y.-J.; Chen, S.-A. Inflammation and the pathogenesis of atrial fibrillation. Nat. Rev. Cardiol. 2015, 12, 230–243. [Google Scholar] [CrossRef] [PubMed]
- His, W.J. Die Tätigkeit des embryonalen Herzens und deren Bedeutung für die Lehre von der Herzbewegung beim Erwachsenen. Arb. Medizinischen Klin. 1983, 1, 14–50. [Google Scholar]
- Keith, A.; Flack, M. The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J. Anat. Physiol. 1907, 41, 172–189. [Google Scholar]
- Tawara, S. Die topographie und histologie der bruckenfasern.Ein beitrag zur lehre von der bedeutung der Purkinjeschen faden. Zentralbl. Physiol. 1906, 19, 70–76. [Google Scholar]
- Kent, A.F.S. Observations on the auriculo-ventricular junction of the mammalian heart. Q. J. Exp. Physiol 1913, 7, 193–195. [Google Scholar] [CrossRef]
- James, T.N. Anatomy of the human sinus node. Anat. Rec. 1961, 141, 109–139. [Google Scholar] [CrossRef]
- James, T.N. Anatomy of the sinus node of the dog. Anat. Rec. 1962, 143, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Challice, C.E.; Virágh, S. Origin and early differentiation of the sinus node in the mouse embryo heart. Adv. Myocardiol. 1980, 1, 267–277. [Google Scholar] [PubMed]
- Virágh, S.; Challice, C.E. The development of the conduction system in the mouse embryo heart: III. The development of sinus muscle and sinoatrial node. Dev. Biol. 1980, 80, 28–45. [Google Scholar] [CrossRef]
- Virágh, S.; Challice, C.E. The development of the conduction system in the mouse embryo heart. IV. Differentiation of the atrioventricular conduction system. Dev. Biol. 1982, 89, 25–40. [Google Scholar] [CrossRef]
- Viragh, S.; Challice, C.E. The development of the early atrioventricular conduction system in the embryonic heart. Can. J. Physiol. Pharmacol. 1983, 61, 775–792. [Google Scholar] [CrossRef] [PubMed]
- Szabóa, E.; Virágh, S.; Challice, C.E. The structure of the atrioventricular conducting system in the avian heart. Anat. Rec. 1986, 215, 1–9. [Google Scholar] [CrossRef]
- Moskowitz, I.P.G.; Pizard, A.; Patel, V.V.; Bruneau, B.G.; Kim, J.B.; Kupershmidt, S.; Roden, D.; Berul, C.I.; Seidman, C.E.; Seidman, J.G. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 2004, 131, 4107–4116. [Google Scholar] [CrossRef] [PubMed]
- Hoogaars, W.M.H.; Tessari, A.; Moorman, A.F.M.; De Boer, P.A.J.; Hagoort, J.; Soufan, A.T.; Campione, M.; Christoffels, V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004, 62, 489–499. [Google Scholar] [CrossRef]
- Mohan, R.A.; Bosada, F.M.; Weerd, J.H.V.; Van Duijvenboden, K.; Wang, J.; Mommersteeg, M.T.M.; Hooijkaas, I.B.; Wakker, V.; De Vries, C.D.G.; Coronel, R.; et al. T-box transcription factor 3 governs a transcriptional program for the function of the mouse atrioventricular conduction system. Proc. Natl. Acad. Sci. USA 2020, 117, 18617–18626. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, I.P.G.; Kim, J.B.; Moore, M.L.; Wolf, C.M.; Peterson, M.A.; Shendure, J.; Nobrega, M.A.; Yokota, Y.; Berul, C.; Izumo, S.; et al. A Molecular Pathway Including Id2, Tbx5, and Nkx2-5 Required for Cardiac Conduction System Development. Cell 2007, 129, 1365–1376. [Google Scholar] [CrossRef]
- Hoogaars, W.M.H.; Engel, A.; Brons, J.F.; Verkerk, A.O.; de Lange, F.J.; Wong, L.Y.E.; Bakker, M.L.; Clout, D.E.; Wakker, V.; Barnett, P.; et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007, 21, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.L.; Boukens, B.J.; Mommersteeg, M.T.M.; Brons, J.F.; Wakker, V.; Moorman, A.F.M.; Christoffels, V.M. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008, 102, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Horsthuis, T.; Buermans, H.P.J.; Brons, J.F.; Verkerk, A.O.; Bakker, M.L.; Wakker, V.; Clout, D.E.W.; Moorman, A.F.M.; ’t Hoen, P.A.C.; Christoffels, V.M. Gene expression profiling of the forming atrioventricular node using a novel Tbx3-based node-specific transgenic reporter. Circ. Res. 2009, 105, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Greener, I.D.; Monfredi, O.; Inada, S.; Chandler, N.J.; Tellez, J.O.; Atkinson, A.; Taube, M.A.; Billeter, R.; Anderson, R.H.; Efimov, I.R.; et al. Molecular architecture of the human specialised atrioventricular conduction axis. J. Mol. Cell. Cardiol. 2011, 50, 642–651. [Google Scholar] [CrossRef]
- Arnolds, D.E.; Liu, F.; Fahrenbach, J.P.; Kim, G.H.; Schillinger, K.J.; Smemo, S.; McNally, E.M.; Nobrega, M.A.; Patel, V.V.; Moskowitz, I.P. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Investig. 2012, 122, 2509–2518. [Google Scholar] [CrossRef]
- Van Weerd, J.H.; Badi, I.; Van Den Boogaard, M.; Stefanovic, S.; Van De Werken, H.J.G.; Gomez-Velazquez, M.; Badia-Careaga, C.; Manzanares, M.; De Laat, W.; Barnett, P.; et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ. Res. 2014, 115, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Weigel, T.; Schmitz, T.; Pfister, T.; Gaetzner, S.; Jannasch, M.; Al-Hijailan, R.; Schürlein, S.; Suliman, S.; Mustafa, K.; Hansmann, J. A three-dimensional hybrid pacemaker electrode seamlessly integrates into engineered, functional human cardiac tissue in vitro. Sci. Rep. 2018, 8, 14545. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, R.J.; Hahurij, N.D.; Kuijper, S.; Just, S.; Wisse, L.J.; Deissler, K.; Maxelon, T.; Anastassiadis, K.; Spitzer, J.; Hardt, S.E.; et al. Targeted Mutation Reveals Essential Functions of the Homeodomain Transcription Factor Shox2 in Sinoatrial and Pacemaking Development. Circulation 2007, 115, 1830–1838. [Google Scholar] [CrossRef]
- Mommersteeg, M.T.M.; Hoogaars, W.M.H.; Prall, O.W.J.; De Gier-De Vries, C.; Wiese, C.; Clout, D.E.W.; Papaioannou, V.E.; Brown, N.A.; Harvey, R.P.; Moorman, A.F.M.; et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 2007, 100, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liang, X.; Najafi, N.; Cass, M.; Lin, L.; Cai, C.L.; Chen, J.; Evans, S.M. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 2007, 304, 286–296. [Google Scholar] [CrossRef]
- Espinoza-Lewis, R.A.; Yu, L.; He, F.; Liu, H.; Tang, R.; Shi, J.; Sun, X.; Martin, J.F.; Wang, D.; Yang, J.; et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009, 327, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Wiese, C.; Grieskamp, T.; Airik, R.; Mommersteeg, M.T.M.; Gardiwal, A.; de Gier-de Vries, C.; Schuster-Gossler, K.; Moorman, A.F.M.; Kispert, A.; Christoffels, V.M. Formation of the Sinus Node Head and Differentiation of Sinus Node Myocardium Are Independently Regulated by Tbx18 and Tbx3. Circ. Res. 2009, 104, 388–397. [Google Scholar] [CrossRef]
- Ammirabile, G.; Tessari, A.; Pignataro, V.; Szumska, D.; Sutera Sardo, F.; Benes, J.; Balistreri, M.; Bhattacharya, S.; Sedmera, D.; Campione, M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2012, 93, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, S.; Zhang, Q.; Zhuang, T.; Evans, S.M.; Liang, X.; Sun, Y. Expression of Isl1 during mouse development. Gene Expr. Patterns 2013, 13, 407–412. [Google Scholar] [CrossRef]
- Li, H.; Li, D.; Wang, Y.; Huang, Z.; Xu, J.; Yang, T.; Wang, L.; Tang, Q.; Cai, C.L.; Huang, H.; et al. Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development 2019, 146, dev178145. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, D.; Tamaddon, H.S.; Lo, C.W.; Taffet, S.M.; Delmar, M.; Morley, G.E.; Jalife, J. Null mutation of connexin43 causes slow propagation of ventricular activation in the late stages of mouse embryonic development. Circ. Res. 2001, 88, 1196–1202. [Google Scholar] [CrossRef]
- Alcoléa, S.; Jarry-Guichard, T.; De Bakker, J.; Gonzàlez, D.; Lamers, W.; Coppen, S.; Barrio, L.; Jongsma, H.; Gros, D.; Van Rijen, H. Replacement of Connexin40 by Connexin45 in the Mouse Impact on Cardiac Electrical Conduction. Circ. Res. 2004, 94, 100–109. [Google Scholar] [CrossRef]
- Krüger, O.; Maxeiner, S.; Kim, J.S.; van Rijen, H.V.M.; de Bakker, J.M.T.; Eckardt, D.; Tiemann, K.; Lewalter, T.; Ghanem, A.; Lüderitz, B.; et al. Cardiac morphogenetic defects and conduction abnormalities in mice homozygously deficient for connexin40 and heterozygously deficient for connexin45. J. Mol. Cell. Cardiol. 2006, 41, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Leaf, D.E.; Feig, J.E.; Vasquez, C.; Riva, P.L.; Yu, C.; Lader, J.M.; Kontogeorgis, A.; Baron, E.L.; Peters, N.S.; Fisher, E.A.; et al. Connexin40 imparts conduction heterogeneity to atrial tissue. Circ. Res. 2008, 103, 1001–1008. [Google Scholar] [CrossRef]
- Kontogeorgis, A.; Li, X.; Kang, E.Y.; Feig, J.E.; Ponzio, M.; Kang, G.; Kaba, R.A.; Wit, A.L.; Fisher, E.A.; Morley, G.E.; et al. Decreased connexin43 expression in the mouse heart potentiates pacing-induced remodeling of repolarizing currents. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H1905–H1916. [Google Scholar] [CrossRef] [PubMed]
- Benes, J.; Ammirabile, G.; Sankova, B.; Campione, M.; Krejci, E.; Kvasilova, A.; Sedmera, D. The role of connexin40 in developing atrial conduction. FEBS Lett. 2014, 588, 1465–1469. [Google Scholar] [CrossRef]
- George, S.A.; Sciuto, K.J.; Lin, J.; Salama, M.E.; Keener, J.P.; Gourdie, R.G.; Poelzing, S. Extracellular sodium and potassium levels modulate cardiac conduction in mice heterozygous null for the Connexin43 gene. Pflugers Arch. Eur. J. Physiol. 2015, 467, 2287–2297. [Google Scholar] [CrossRef] [PubMed]
- Poelmann, R.E.; Lie-Venema, H.; Gittenberger-De Groot, A.C. The role of the epicardium and neural crest: As extracardiac contributors to coronary vascular development. Texas Hear. Inst. J. 2002, 29, 255–261. [Google Scholar]
- St. Amand, T.R.; Lu, J.T.; Zamora, M.; Gu, Y.; Stricker, J.; Hoshijima, M.; Epstein, J.A.; Ross, J.J.; Ruiz-Lozano, P.; Chien, K.R. Distinct roles of HF-1b/Sp4 in ventricular and neural crest cells lineages affect cardiac conduction system development. Dev. Biol. 2006, 291, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Colbert, M.C.; Robbins, J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 2006, 98, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Pfeufer, A.; Van Noord, C.; Marciante, K.D.; Arking, D.E.; Larson, M.G.; Smith, A.V.; Tarasov, K.V.; Müller, M.; Sotoodehnia, N.; Sinner, M.F.; et al. Genome-wide association study of PR interval. Nat. Genet. 2010, 42, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Sotoodehnia, N.; Isaacs, A.; De Bakker, P.I.W.; DÖrr, M.; Newton-Cheh, C.; Nolte, I.M.; Van Der Harst, P.; Müller, M.; Eijgelsheim, M.; Alonso, A.; et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010, 42, 1068–1076. [Google Scholar] [CrossRef]
- Smith, J.G.; Magnani, J.W.; Palmer, C.; Meng, Y.A.; Soliman, E.Z.; Musani, S.K.; Kerr, K.F.; Schnabel, R.B.; Lubitz, S.A.; Sotoodehnia, N.; et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011, 7, e1001304. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Denny, J.C.; Zuvich, R.L.; Crawford, D.C.; Schildcrout, J.S.; Bastarache, L.; Ramirez, A.H.; Mosley, J.D.; Pulley, J.M.; Basford, M.A.; et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 2013, 127, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Wang, J.; Song, Y.; Yu, D.; Sun, C.; Liu, C.; Chen, F.; Zhang, Y.; Wang, F.; Harvey, R.P.; et al. A common Shox2–nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 2015, 142, 2521–2532. [Google Scholar]
- Seyerle, A.A.; Lin, H.J.; Gogarten, S.M.; Stilp, A.; Méndez Giráldez, R.; Soliman, E.; Baldassari, A.; Graff, M.; Heckbert, S.; Kerr, K.F.; et al. Genome-wide association study of PR interval in Hispanics/Latinos identifies novel locus at ID2. Heart 2018, 104, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Van Setten, J.; Brody, J.A.; Jamshidi, Y.; Swenson, B.R.; Butler, A.M.; Campbell, H.; Del Greco, F.M.; Evans, D.S.; Gibson, Q.; Gudbjartsson, D.F.; et al. PR interval genome-wide association meta-analysis identifies 50 loci associated with atrial and atrioventricular electrical activity. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- van Eif, V.W.W.; Protze, S.I.; Bosada, F.M.; Yuan, X.; Sinha, T.; van Duijvenboden, K.; Ernault, A.C.; Mohan, R.A.; Wakker, V.; de Gier-de Vries, C.; et al. Genome-Wide Analysis Identifies an Essential Human TBX3 Pacemaker Enhancer. Circ. Res. 2020, 127, 1522–1535. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bai, Y.; Li, N.; Ye, W.; Zhang, M.; Greene, S.B.; Tao, Y.; Chen, Y.; Wehrens, X.H.T.; Martin, J.F. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 9181–9186. [Google Scholar] [CrossRef] [PubMed]
- Benzoni, P.; Nava, L.; Giannetti, F.; Guerini, G.; Gualdoni, A.; Bazzini, C.; Milanesi, R.; Bucchi, A.; Baruscotti, M.; Barbuti, A. Dual role of miR-1 in the development and function of sinoatrial cells. J. Mol. Cell. Cardiol. 2021, 157, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Preussner, J.; Chen, X.; Guenther, S.; Yuan, X.; Yekelchyk, M.; Kuenne, C.; Looso, M.; Zhou, Y.; Teichmann, S.; et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Prins, B.P.; Mead, T.J.; Brody, J.A.; Sveinbjornsson, G.; Ntalla, I.; Bihlmeyer, N.A.; van den Berg, M.; Bork-Jensen, J.; Cappellani, S.; Van Duijvenboden, S.; et al. Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol. 2018, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Van Eif, V.W.W.; Stefanovic, S.; van Duijvenboden, K.; Bakker, M.; Wakker, V.; de Gier-de Vries, C.; Zaffran, S.; Verkerk, A.O.; Boukens, B.J.; Christoffels, V.M. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 2019, 146, dev173161. [Google Scholar] [CrossRef] [PubMed]
- Galang, G.; Mandla, R.; Ruan, H.; Jung, C.; Sinha, T.; Stone, N.R.; Wu, R.S.; Mannion, B.J.; Allu, P.K.R.; Chang, K.; et al. ATAC-Seq Reveals an Isl1 Enhancer That Regulates Sinoatrial Node Development and Function. Circ. Res. 2020, 127, 1502–1518. [Google Scholar] [CrossRef]
- Hoelscher, S.C.; Stich, T.; Diehm, A.; Lahm, H.; Dreßen, M.; Zhang, Z.; Neb, I.; Aherrahrou, Z.; Erdmann, J.; Schunkert, H.; et al. MiR-128a acts as a regulator in cardiac development by modulating differentiation of cardiac progenitor cell populations. Int. J. Mol. Sci. 2020, 21, 1158. [Google Scholar] [CrossRef] [PubMed]
- Yanni, J.; D’Souza, A.; Wang, Y.; Li, N.; Hansen, B.J.; Zakharkin, S.O.; Smith, M.; Hayward, C.; Whitson, B.A.; Mohler, P.J.; et al. Silencing miR-370-3p rescues funny current and sinus node function in heart failure. Sci. Rep. 2020, 10, 11279. [Google Scholar] [CrossRef] [PubMed]
- Petkova, M.; Atkinson, A.J.; Yanni, J.; Stuart, L.; Aminu, A.J.; Ivanova, A.D.; Pustovit, K.B.; Geragthy, C.; Feather, A.; Li, N.; et al. Identification of key small non-coding micrornas controlling pacemaker mechanisms in the human sinus node. J. Am. Heart Assoc. 2020, 9, e016590. [Google Scholar] [CrossRef]
- Yang, D.; Wan, X.; Dennis, A.T.; Bektik, E.; Wang, Z.; Costa, M.G.S.; Fagnen, C.; Vénien-Bryan, C.; Xu, X.; Gratz, D.H.; et al. MicroRNA Biophysically Modulates Cardiac Action Potential by Direct Binding to Ion Channel. Circulation 2021, 143, 1597–1613. [Google Scholar] [CrossRef]
- Schmitteckert, S.; Griesbeck, A.; Sumer, S.; Jauch, A.; Rolletschek, A.; Niesler, B.; Rappold, G.A.; Hoffmann, S. Murine transgenic embryonic stem cell lines for the investigation of sinoatrial node-related molecular pathways. Stem Cell Res. 2017, 25, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Han, P.; Kim, E.H.; Mak, J.; Zhang, R.; Torrente, A.G.; Goldhaber, J.I.; Marbán, E.; Cho, H.C. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem Cells 2020, 38, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Munshi, N.V. Gene regulatory networks in cardiac conduction system development. Circ. Res. 2012, 110, 1525–1537. [Google Scholar] [CrossRef]
- Kawashima, T.; Sasaki, H. A macroscopic anatomical investigation of atrioventricular bundle locational variation relative to the membranous part of the ventricular septum in elderly human hearts. Surg. Radiol. Anat. 2005, 27, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Sasaki, H. Gross anatomy of the human cardiac conduction system with comparative morphological and developmental implications for human application. Ann. Anat. 2011, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.H.; Yanni, J.; Boyett, M.R.; Chandler, N.J.; Dobrzynski, H. The anatomy of the cardiac conduction system. Clin. Anat. 2009, 22, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Dobrzynski, H.; Anderson, R.H.; Atkinson, A.; Borbas, Z.; D’Souza, A.; Fraser, J.F.; Inada, S.; Logantha, S.J.R.J.; Monfredi, O.; Morris, G.M.; et al. Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol. Ther. 2013, 139, 260–288. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.H.; Boyett, M.R.; Dobrzynski, H.; Moorman, A.F.M. The anatomy of the conduction system: Implications for the clinical cardiologist. J. Cardiovasc. Transl. Res. 2013, 6, 187–196. [Google Scholar] [CrossRef]
- Shinohara, G.; Morita, K.; Hoshino, M.; Ko, Y.; Tsukube, T.; Kaneko, Y.; Morishita, H.; Oshima, Y.; Matsuhisa, H.; Iwaki, R.; et al. Three Dimensional Visualization of Human Cardiac Conduction Tissue in Whole Heart Specimens by High-Resolution Phase-Contrast CT Imaging Using Synchrotron Radiation. World J. Pediatr. Congenit. Heart Surg. 2016, 7, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, R.S.; Atkinson, A.; Kottas, P.; Perde, F.; Jafarzadeh, F.; Bateman, M.; Iaizzo, P.A.; Zhao, J.; Zhang, H.; Anderson, R.H.; et al. High resolution 3-Dimensional imaging of the human cardiac conduction system from microanatomy to mathematical modeling. Sci. Rep. 2017, 7, 7188. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, R.S.; Jones, C.B.; Guerrero, R.; Zhao, J.; Anderson, R.H.; Jarvis, J.C. High-Resolution Contrast-Enhanced Micro-Computed Tomography to Identify the Cardiac Conduction System in Congenitally Malformed Hearts: Valuable Insight From a Hospital Archive. JACC Cardiovasc. Imaging 2018, 11, 1706–1712. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Sato, F. First in situ 3D visualization of the human cardiac conduction system and its transformation associated with heart contour and inclination. Sci. Rep. 2021, 11, 8636. [Google Scholar] [CrossRef]
- Padala, S.K.; Cabrera, J.A.; Ellenbogen, K.A. Anatomy of the cardiac conduction system. PACE Pacing Clin. Electrophysiol. 2021, 44, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Sommer, J.R.; Johnson, E.A. Cardiac muscle. A comparative study of Purkinje fibers and ventricular fibers. J. Cell Biol. 1968, 36, 497–526. [Google Scholar] [CrossRef] [PubMed]
- Lakatta, E.G.; DiFrancesco, D. What keeps us ticking: A funny current, a calcium clock, or both? J. Mol. Cell. Cardiol. 2009, 47, 157–170. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. A Brief History of Pacemaking. Front. Physiol. 2020, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- Mesirca, P.; Torrente, A.G.; Mangoni, M.E. Functional role of voltage gated Ca2+ channels in heart automaticity. Front. Physiol. 2015, 6, 19. [Google Scholar] [CrossRef]
- Baudot, M.; Torre, E.; Bidaud, I.; Louradour, J.; Torrente, A.G.; Fossier, L.; Talssi, L.; Nargeot, J.; Barrère-Lemaire, S.; Mesirca, P.; et al. Concomitant genetic ablation of L-type Cav1.3 (α1D) and T-type Cav3.1 (α1G) Ca2+ channels disrupts heart automaticity. Sci. Rep. 2020, 10, 1–19. [Google Scholar] [CrossRef]
- Baruscotti, M.; Barbuti, A.; Bucchi, A. The cardiac pacemaker current. J. Mol. Cell. Cardiol. 2010, 48, 55–64. [Google Scholar] [CrossRef]
- Vetulli, H.M.; Elizari, M.V.; Naccarelli, G.V.; Gonzalez, M.D. Cardiac automaticity: Basic concepts and clinical observations. J. Interv. Card. Electrophysiol. 2018, 52, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Davis, L.M.; Westphale, E.M.; Beyer, E.C.; Saffitz, J.E. Expression of multiple gap junction proteins in human fetal and infant hearts. Pediatr. Res. 1994, 36, 561–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davis, L.M.; Rodefeld, M.E.; Green, K.; Beyer, E.C.; Saffitz, J.E. Gap Junction Protein Phenotypes of the Human Heart and Conduction System. J. Cardiovasc. Electrophysiol. 1995, 6, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Meijler, F.L.; Janse, M.J. Morphology and electrophysiology of the mammalian atrioventricular node. Physiol. Rev. 1988, 68, 608–647. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, A.R.; Dubé, B. Propagation in the AV node: A model based on a simplified two-dimensional structure and a bidomain tissue representation. Med. Biol. Eng. Comput. 1993, 31, 545–556. [Google Scholar] [CrossRef]
- Choi, B.R.; Salama, G. Optical mapping of atrioventricular node reveals a conduction barrier between atrial and nodal cells. Am. J. Physiol. Heart Circ. Physiol. 1998, 274, H829–H845. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Kaza, A.K.; Hitchcock, R.W.; Sachse, F.B. Identification of nodal tissue in the living heart using rapid scanning fiber-optics confocal microscopy and extracellular fluorophores. Circ. Cardiovasc. Imaging 2013, 6, 739–746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xia, R.; Vlcek, J.; Bauer, J.; Kääb, S.; Ishikawa-Ankerhold, H.; van den Heuvel, D.A.; Schulz, C.; Massberg, S.; Clauss, S. Whole-mount immunofluorescence staining, confocal imaging and 3d reconstruction of the sinoatrial and atrioventricular node in the mouse. J. Vis. Exp. 2020, 2020, 166. [Google Scholar] [CrossRef] [PubMed]
- Miquerol, L.; Meysen, S.; Mangoni, M.; Bois, P.; Van Rijen, H.V.M.; Abran, P.; Jongsma, H.; Nargeot, J.; Gros, D. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc. Res. 2004, 63, 77–86. [Google Scholar] [CrossRef]
- Atkinson, A.; Inada, S.; Li, J.; Tellez, J.O.; Yanni, J.; Sleiman, R.; Allah, E.A.; Anderson, R.H.; Zhang, H.; Boyett, M.R.; et al. Anatomical and molecular mapping of the left and right ventricular His-Purkinje conduction networks. J. Mol. Cell. Cardiol. 2011, 51, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, M.J.A.; Ten Velde, I.; Wessels, A.; Oosthoek, P.W.; Gros, D.; Jongsma, H.J.; Moorman, A.F.M.; Lamers, W.H. Differential connexin distribution accommodates cardiac function in different species. Microsc. Res. Tech. 1995, 31, 420–436. [Google Scholar] [CrossRef]
- Olejnickova, V.; Kocka, M.; Kvasilova, A.; Kolesova, H.; Dziacky, A.; Gidor, T.; Gidor, L.; Sankova, B.; Gregorovicova, M.; Gourdie, R.G.; et al. Gap junctional communication via connexin43 between purkinje fibers and working myocytes explains the epicardial activation pattern in the postnatal mouse left ventricle. Int. J. Mol. Sci. 2021, 22, 2475. [Google Scholar] [CrossRef]
- Vassalle, M.; Bocchi, L. Differences in ionic currents between canine myocardial and Purkinje cells. Physiol. Rep. 2013, 1, e00036. [Google Scholar] [CrossRef] [PubMed]
- Boyden, P.A.; Pu, J.; Pinto, J.; Ter Keurs, H.E.D.J. Ca2+ transients and Ca2+ waves in purkinje cells: Role in action potential initiation. Circ. Res. 2000, 86, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, A.; Ter Keurs, H.E.; Franzini-Armstrong, C. T-tubule profiles in Purkinje fibres of mammalian myocardium. J. Muscle Res. Cell Motil. 2007, 28, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Einthoven, W. The different forms of the human electrocardiogram and their signification. Lancet 1912, 179, 853–861. [Google Scholar] [CrossRef]
- Buxton, A.E.; Calkins, H.; Callans, D.J.; DiMarco, J.P.; Fisher, J.D.; Greene, H.L.; Haines, D.E.; Hayes, D.L.; Heidenreich, P.A.; Miller, J.M.; et al. ACC/AHA/HRS 2006 key data elements and definitions for electrophysiological studies and procedures: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards (ACC/AHA/HRS Writing Committee to Develop Data Standards on Electrophysiology). Circulation 2006, 114, 2360–2396. [Google Scholar]
- Kléber, A.G.; Rudy, Y. Basic Mechanisms of Cardiac Impulse Propagation and Associated Arrhythmias. Physiol. Rev. 2004, 84, 431–488. [Google Scholar] [CrossRef]
- Prinzen, F.W.; Strik, M.; Regoli, F.; Auricchio, A. Basic physiology and hemodynamics of cardiac pacing. In Clinical Cardiac Pacing, Defibrillation, and Resynchronization Therapy; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 203–233. ISBN 9781437716160. [Google Scholar]
- Lederer, W.J. Cardiac electrophysiology and the electrocardiogram. In Medical Physiology; Boron, W.E., Ed.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 504–528. [Google Scholar]
- Corrado, D.; Biffi, A.; Basso, C.; Pelliccia, A.; Thiene, G. 12-lead ECG in the athlete: Physiological versus pathological abnormalities. Br. J. Sports Med. 2009, 43, 669–676. [Google Scholar] [CrossRef]
- Shiroma, E.J.; Lee, I.M. Physical activity and cardiovascular health: Lessons learned from epidemiological studies across age, Gender, and race/ethnicity. Circulation 2010, 122, 743–752. [Google Scholar] [CrossRef]
- Franklin, B.A.; Thompson, C.P.D.; Al-Zaiti, S.S.; Albert, C.M.; Hivert, M.F.; Levine, B.D.; Lobelo, F.; Madan, K.; Sharrief, A.Z.; Eijsvogels, T.M.H. Exercise-related acute cardiovascular events and potential deleterious adaptations following long-term exercise training: Placing the risks into perspective-an update: A scientific statement from the American Heart Association. Circulation 2020, 141, E705–E736. [Google Scholar] [CrossRef]
- Priori, S.G.; Remme, C.A. Inherited conditions of arrhythmia: Translating disease mechanisms to patient management. Cardiovasc. Res. 2020, 116, 1539–1541. [Google Scholar] [CrossRef]
- Zorzi, A.; Vio, R.; Bettella, N.; Corrado, D. Criteria for interpretation of the athlete’s ECG: A critical appraisal. PACE Pacing Clin. Electrophysiol. 2020, 43, 882–890. [Google Scholar] [CrossRef]
- Sarto, P.; Zorzi, A.; Merlo, L.; Vessella, T.; Pegoraro, C.; Giorgiano, F.; Patti, A.; Crosato, M.; Thiene, G.; Drezner, J.A.; et al. Serial Versus Single Cardiovascular Screening of Adolescent Athletes. Circulation 2021, 143, 1729–1731. [Google Scholar] [CrossRef] [PubMed]
- Pelliccia, A.; Sharma, S.; Gati, S.; Bäck, M.; Börjesson, M.; Caselli, S.; Collet, J.P.; Corrado, D.; Drezner, J.A.; Halle, M.; et al. 2020 ESC Guidelines on Sports Cardiology and Exercise in Patients with Cardiovascular Disease. Rev. Esp. Cardiol. 2021, 74, 545. [Google Scholar]
- Stadiotti, I.; Lippi, M.; Maione, A.S.; Compagnucci, P.; Andreini, D.; Casella, M.; Pompilio, G.; Sommariva, E. Cardiac biomarkers and autoantibodies in endurance athletes: Potential similarities with arrhythmogenic cardiomyopathy pathogenic mechanisms. Int. J. Mol. Sci. 2021, 22, 6500. [Google Scholar] [CrossRef] [PubMed]
- Rivaud, M.R.; Blok, M.; Jongbloed, M.R.M.; Boukens, B.J. How Cardiac Embryology Translates into Clinical Arrhythmias. J. Cardiovasc. Dev. Dis. 2021, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Trakmulkichkarn, T.; Ghadiry-Tavi, R.; Fruitman, D.; Niederhoffer, K.Y.; Caluseriu, O.; Lauzon, J.L.; Wewala, G.; Hornberger, L.K.; Urschel, S.; Conway, J.; et al. Clinical presentation, genetic etiologies and outcomes associated with fetal cardiomyopathies in a more recent era. Ultrasound Obstet. Gynecol. 2021. [Google Scholar] [CrossRef]
- Tschirhart, J.N.; Zhang, S. Fentanyl-induced block of hERG channels is exacerbated by hypoxia, hypokalemia, alkalosis, and the presence of HERG1b. Mol. Pharmacol. 2020, 98, 508–517. [Google Scholar] [CrossRef]
- Huang, Y.; Alsabbagh, M.W. Comparative risk of cardiac arrhythmias associated with acetylcholinesterase inhibitors used in treatment of dementias—A narrative review. Pharmacol. Res. Perspect. 2020, 8, e00622. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Cardiac fibrosis. Cardiovasc. Res. 2021, 117, 1450–1488. [Google Scholar] [CrossRef] [PubMed]
- Bamgboje, A.; Akintan, F.O.; Gupta, N.M.; Kaur, G.; Pekler, G.; Mushiyev, S. Lyme carditis: A reversible cause of acquired third-degree av block. Am. J. Case Rep. 2021, 22, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Bratincsak, A.; Liu, J.; Yalamanchili, R.; Purohit, P.J.; Xoinis, K.P.; Yamauchi, M.S.W. Junctional tachycardia as a diagnostic criterion in acute rheumatic fever. Pediatrics 2021, 147, e2020049361. [Google Scholar] [CrossRef] [PubMed]
- Lewars, J.; Wazir, H.; Gordon, B.; Faluyi, U.; Girgis, Y. Noninvasive Diagnostic Modalities in an Isolated Case of Cardiac Amyloidosis. Cureus 2021, 13, e14608. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, B.; Dave Javier, A.; Grewal, J.; Gabarin, N.; Colman, J.; Kiess, M.; Wald, R.M.; Sermer, M.; Siu, S.C.; Silversides, C.K. Risk Associated With Valvular Regurgitation During Pregnancy. J. Am. Coll. Cardiol. 2021, 77, 2656–2664. [Google Scholar] [CrossRef] [PubMed]
- Epstein, A.E.; DiMarco, J.P.; Ellenbogen, K.A.; Estes, N.A.M.; Freedman, R.A.; Gettes, L.S.; Gillinov, A.M.; Gregoratos, G.; Hammill, S.C.; Hayes, D.L.; et al. ACC/AHA/HRS 2008 Guidelines for Device-Based Therapy of Cardiac Rhythm Abnormalities. Circulation 2008, 117, e1–e62. [Google Scholar]
- Di Marco, A.; Brown, P.F.; Bradley, J.; Nucifora, G.; Claver, E.; de Frutos, F.; Dallaglio, P.D.; Comin-Colet, J.; Anguera, I.; Miller, C.A.; et al. Improved Risk Stratification for Ventricular Arrhythmias and Sudden Death in Patients with Nonischemic Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2021, 77, 2890–2905. [Google Scholar] [CrossRef]
- Shi, Y.P.; Pang, Z.; Venkateshappa, R.; Gunawan, M.; Kemp, J.; Truong, E.; Chang, C.; Lin, E.; Shafaattalab, S.; Faizi, S.; et al. The hERG channel activator, RPR260243, enhances protective IKr current early in the refractory period reducing arrhythmogenicity in zebrafish hearts. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H251–H261. [Google Scholar] [CrossRef] [PubMed]
- Kochi, A.N.; Tagliari, A.P.; Forleo, G.B.; Fassini, G.M.; Tondo, C. Cardiac and arrhythmic complications in patients with COVID-19. J. Cardiovasc. Electrophysiol. 2020, 31, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.I.; Postema, P.G.; Arbelo, E.; Behr, E.R.; Bezzina, C.R.; Napolitano, C.; Robyns, T.; Probst, V.; Schulze-Bahr, E.; Remme, C.A.; et al. SARS-CoV-2, COVID-19, and inherited arrhythmia syndromes. Heart Rhythm 2020, 17, 1456–1462. [Google Scholar] [CrossRef]
- Phyu Phyu Aung, T.; Buddhavarapu, S.; Park, W.J.; Ayala-Rodriguez, C.; Oo, Z.T.; Kyaw, H. A Visual Resolution of Cardiotoxicity: A Case Report of Digoxin-Induced Bidirectional Ventricular Tachycardia. Cureus 2021, 13, e15134. [Google Scholar] [CrossRef]
- Zhang, X. Atypical takotsubo cardiomyopathy with persistent left ventricular aneurysm after cardiac surgery. Heart Surg. Forum 2021, 24, E442–E444. [Google Scholar] [CrossRef]
- Martens, T.; François, K.; De Wilde, H.; Campens, L.; Demulier, L.; De Backer, J.; Dewolf, D.; Bove, T. QRS Duration During Follow-Up of Tetralogy of Fallot: How Valuable is it? Analysis of ECG Changes in Relation to Pulmonary Valve Implantation. Pediatr. Cardiol. 2021, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Muntané-Carol, G.; Nombela-Franco, L.; Serra, V.; Urena, M.; Amat-Santos, I.; Vilalta, V.; Chamandi, C.; Lhermusier, T.; Veiga-Fernandez, G.; Kleiman, N.; et al. Late arrhythmias in patients with new-onset persistent left bundle branch block after transcatheter aortic valve replacement using a balloon-expandable valve. Heart Rhythm 2021, 18, 1733–1740. [Google Scholar] [CrossRef]
- Ismail, I.; Wert, L.; Hanke, J.S.; Dogan, G.; Chatterjee, A.; Feldmann, C.; Cebotari, S.; Haverich, A.; Schmitto, J.D. Mid-term outcome of the edge-to-edge mitral valve repair via aortic outflow tract in high risk patients. Semin. Thorac. Cardiovasc. Surg. 2021. [Google Scholar] [CrossRef] [PubMed]
- Blomström-Lundqvist, C.; Traykov, V.; Erba, P.A.; Burri, H.; Nielsen, J.C.; Bongiorni, M.G.; Poole, J.; Boriani, G.; Costa, R.; Deharo, J.C.; et al. European Heart Rhythm Association (EHRA) international consensus document on how to prevent, diagnose, and treat cardiac implantable electronic device infections-endorsed by the Heart Rhythm Society (HRS), the Asia Pacific Heart Rhythm Society (APHRS). Eur. Heart J. 2020, 41, 2012–2032. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.; Scherr, D.; Lenarczyk, R.; Gandjbachkh, E.; Boulé, S.; Spartalis, M.D.; Behr, E.R.; Wilde, A.; Potpara, T. Diagnosis, family screening, and treatment of inherited arrhythmogenic diseases in Europe: Results of the European Heart Rhythm Association Survey. Europace 2020, 22, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Mullens, W.; Auricchio, A.; Martens, P.; Witte, K.; Cowie, M.R.; Delgado, V.; Dickstein, K.; Linde, C.; Vernooy, K.; Leyva, F.; et al. Optimized implementation of cardiac resynchronization therapy: A call for action for referral and optimization of care: A joint position statement from the Heart Failure Association (HFA), European Heart Rhythm Association (EHRA), and European Association. Eur. J. Heart Fail. 2020, 22, 2349–2369. [Google Scholar] [CrossRef] [PubMed]
- Collet, J.P.; Thiele, H.; Barbato, E.; Bauersachs, J.; Dendale, P.; Edvardsen, T.; Gale, C.P.; Jobs, A.; Lambrinou, E.; Mehilli, J.; et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur. Heart J. 2021, 42, 1289–1367. [Google Scholar] [CrossRef]
- Varma, N.; Cygankiewicz, I.; Turakhia, M.; Heidbuchel, H.; Hu, Y.; Chen, L.Y.; Couderc, J.; Cronin, E.M.; Estep, J.D.; Grieten, L.; et al. 2021 ISHNE/HRS/EHRA/APHRS collaborative statement on mHealth in Arrhythmia Management: Digital Medical Tools for Heart Rhythm Professionals. J. Arrhythmia 2021, 37, 271–319. [Google Scholar] [CrossRef]
- Iop, L. Toward the Effective Bioengineering of a Pathological Tissue for Cardiovascular Disease Modeling: Old Strategies and New Frontiers for Prevention, Diagnosis, and Therapy. Front. Cardiovasc. Med. 2020, 7, 591583. [Google Scholar] [CrossRef]
- Wander, G.S.; Babu, R.; Khattri, H.N.; Wahi, P.L.; Bidwai, P.S. Cardiac pacing. Indian Pediatr. 1988, 25, 141–147. [Google Scholar]
- Adán, V.; Crown, L.A. Diagnosis and treatment of sick sinus syndrome. Am. Fam. Phys. 2003, 67, 1725–1732. [Google Scholar]
- Dobrzynski, H.; Boyett, M.R.; Anderson, R.H. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation 2007, 115, 1921–1932. [Google Scholar] [CrossRef]
- Kusumoto, F.M.; Schoenfeld, M.H.; Barrett, C.; Edgerton, J.R.; Ellenbogen, K.A.; Gold, M.R.; Goldschlager, N.F.; Hamilton, R.M.; Joglar, J.A.; Kim, R.J.; et al. 2018 ACC/AHA/HRS Guideline on the Evaluation and Management of Patients With Bradycardia and Cardiac Conduction Delay: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2019, 140, e333–e381. [Google Scholar] [CrossRef] [PubMed]
- Montalescot, G.; Levy, Y.; Farge, D.; Brochard, L.; Fantin, B.; Arnoux, C.; Hatt, P.Y. Lithium causing a serious sinus-node dysfunction at therapeutic doses. Clin. Cardiol. 1984, 7, 617–620. [Google Scholar] [CrossRef]
- Lei, M.; Huang, C.L.H.; Zhang, Y. Genetic Na+ channelopathies and sinus node dysfunction. Prog. Biophys. Mol. Biol. 2008, 98, 171–178. [Google Scholar] [CrossRef]
- Snipes, G.M.; Hafeez, A.; Marek, G.; Winchester, D.E. Sinus bradycardia with haemodynamic compromise following lithium intoxication. BMJ Case Rep. 2021, 14, e242946. [Google Scholar] [CrossRef]
- Baulac, M.; Byrnes, W.; Williams, P.; Borghs, S.; Webster, E.; De Backer, M.; Dedeken, P. Lacosamide and sodium channel-blocking antiepileptic drug cross-titration against levetiracetam background therapy. Acta Neurol. Scand. 2017, 135, 434–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shibata, M.; Hoshino, R.; Shimizu, C.; Sato, M.; Furuta, N.; Ikeda, Y. Lacosamide-induced sinus node dysfunction followed by severe agranulocytosis. BMC Neurol. 2021, 21, 217. [Google Scholar] [CrossRef]
- Remme, C.A. Cardiac sodium channelopathy associated with SCN5A mutations: Electrophysiological, molecular and genetic aspects. J. Physiol. 2013, 591, 4099–4116. [Google Scholar] [CrossRef] [PubMed]
- Ziyadeh-Isleem, A.; Clatot, J.; Duchatelet, S.; Gandjbakhch, E.; Denjoy, I.; Hidden-Lucet, F.; Hatem, S.; Deschênes, I.; Coulombe, A.; Neyroud, N.; et al. A truncating SCN5A mutation combined with genetic variability causes sick sinus syndrome and early atrial fibrillation. Heart Rhythm 2014, 11, 1015–1023. [Google Scholar] [CrossRef]
- Abe, K.; Machida, T.; Sumitomo, N.; Yamamoto, H.; Ohkubo, K.; Watanabe, I.; Makiyama, T.; Fukae, S.; Kohno, M.; Harrell, D.T.; et al. Sodium channelopathy underlying familial sick sinus syndrome with early onset and predominantly male characteristics. Circ. Arrhythmia Electrophysiol. 2014, 7, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Abe, I.; Wang, P.; Takahashi, M.; Ohno, S.; Ono, K.; Takahashi, N. Familial sick sinus syndrome possibly associated with novel SCN5A mutation diagnosed in pregnancy. Heart Case Rep. 2021, 7, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Qin, J.; Shen, Y.; Huang, J.; Zhang, Z.; Zhu, Z.L.; Lu, H.; Huang, Y.; Yin, Y.; Wang, A.; et al. Identification of rare heterozygous linkage R965C-R1309H mutations in the pore-forming region of SCN5A gene associated with complex arrhythmia. Mol. Genet. Genomic Med. 2021, 9, e1613. [Google Scholar] [CrossRef]
- Ishikawa, T.; Ohno, S.; Murakami, T.; Yoshida, K.; Mishima, H.; Fukuoka, T.; Kimoto, H.; Sakamoto, R.; Ohkusa, T.; Aiba, T.; et al. Sick sinus syndrome with HCN4 mutations shows early onset and frequent association with atrial fibrillation and left ventricular noncompaction. Heart Rhythm 2017, 14, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Fernández-Gatta, M.; Gallego-Delgado, M.; Caballero, R.; Villacorta, E.; Díaz-Peláez, E.; García-BerrocaL, B.; Crespo-García, T.; Plata-Izquierdo, B.; Marcos-Vadillo, E.; García-Cuenllas, L.; et al. A rare HCN4 variant with combined sinus bradycardia, left atrial dilatation, and hypertrabeculation/left ventricular noncompaction phenotype. Rev. Española Cardiol. 2020, 74, 781–789. [Google Scholar] [CrossRef]
- Balla, C.; Conte, E.; Selvatici, R.; Marsano, R.M.; Gerbino, A.; Farnè, M.; Blunck, R.; Vitali, F.; Armaroli, A.; Brieda, A.; et al. Functional characterization of two novel mutations in scn5a associated with brugada syndrome identified in Italian patients. Int. J. Mol. Sci. 2021, 22, 6513. [Google Scholar] [CrossRef]
- Hong, L.; Zhang, M.; Ly, O.T.; Chen, H.; Sridhar, A.; Lambers, E.; Chalazan, B.; Youn, S.W.; Maienschein-Cline, M.; Feferman, L.; et al. Human induced pluripotent stem cell-derived atrial cardiomyocytes carrying an SCN5A mutation identify nitric oxide signaling as a mediator of atrial fibrillation. Stem Cell Rep. 2021, 16, 1542–1554. [Google Scholar] [CrossRef]
- Catterall, W.A.; Lenaeus, M.J.; Gamal El-Din, T.M. Structure and pharmacology of voltage-gated sodium and calcium channels. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Robaei, D.; Ford, T.; Ooi, S.Y. Ankyrin-B Syndrome: A Case of Sinus Node Dysfunction, Atrial Fibrillation and Prolonged QT in a Young Adult. Heart Lung Circ. 2015, 24, e31–e34. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Luo, J.W.; Jiang, F.; Liu, G. Genetic analysis of sick sinus syndrome in a family harboring compound CACNA1C and TTN mutations. Mol. Med. Rep. 2018, 17, 7073–7080. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, M.V.; Fung, L.; Jensen, E.; Oh, F.; Cung, K.; McCarthy, L.A.; Tran, C.K.; Hoang, V.; Hakim, S.A.; Grosberg, A. Exome sequencing identifies a novel LMNA splice-site mutation and multigenic heterozygosity of potential modifiers in a family with sick sinus syndrome, dilated cardiomyopathy, and sudden cardiac d3eath. PLoS ONE 2016, 11, e0155421. [Google Scholar] [CrossRef]
- Yokokawa, T.; Ichimura, S.; Hijioka, N.; Kaneshiro, T.; Yoshihisa, A.; Kunii, H.; Nakazato, K.; Ishida, T.; Suzuki, O.; Ohno, S.; et al. Case reports of a c.475G>T, p.E159*lamin A/C mutation with a family history of conduction disorder, dilated cardiomyopathy and sudden cardiac death. BMC Cardiovasc. Disord. 2019, 19, 298. [Google Scholar] [CrossRef] [PubMed]
- Thorolfsdottir, R.B.; Sveinbjornsson, G.; Aegisdottir, H.M.; Benonisdottir, S.; Stefansdottir, L.; Ivarsdottir, E.V.; Halldorsson, G.H.; Sigurdsson, J.K.; Torp-Pedersen, C.; Weeke, P.E.; et al. Genetic insight into sick sinus syndrome. Eur. Heart J. 2021, 42, 1959–1971. [Google Scholar] [CrossRef]
- Baruscotti, M.; DiFrancesco, D.; Robinson, R.B. A TTX-sensitive inward sodium current contributes to spontaneous activity in newborn rabbit sino-atrial node cells. J. Physiol. 1996, 492, 21–30. [Google Scholar] [CrossRef]
- Baruscotti, M.; DiFrancesco, D.; Robinson, R.B. Na+ current contribution to the diastolic depolarization in newborn rabbit SA node cells. Am. J. Physiol. Heart Circ. Physiol. 2000, 279, H2303–H2309. [Google Scholar] [CrossRef]
- Lei, M.; Goddard, C.; Liu, J.; Léoni, A.L.; Royer, A.; Fung, S.S.M.; Xiao, G.; Ma, A.; Zhang, H.; Charpentier, F.; et al. Sinus node dysfunction following targeted disruption of the murine cardiac sodium channel gene Scn5a. J. Physiol. 2005, 567, 387–400. [Google Scholar] [CrossRef]
- Jeevaratnam, K.; Zhang, Y.; Guzadhur, L.; Duehmke, R.M.; Lei, M.; Grace, A.A.; Huang, C.L.-H. Differences in sino-atrial and atrio-ventricular function with age and sex attributable to the Scn5a+/- mutation in a murine cardiac model. Acta Physiol. 2010, 200, 23–33. [Google Scholar]
- Hao, X.; Zhang, Y.; Zhang, X.; Nirmalan, M.; Davies, L.; Konstantinou, D.; Yin, F.; Dobrzynski, H.; Wang, X.; Grace, A.; et al. TGF-β1-mediated fibrosis and ion channel remodeling are key mechanisms in producing the sinus node dysfunction associated with SCN5A deficiency and aging. Circ. Arrhythmia Electrophysiol. 2011, 4, 397–406. [Google Scholar] [CrossRef]
- Butters, T.D.; Aslanidi, O.V.; Inada, S.; Boyett, M.R.; Hancox, J.C.; Lei, M.; Zhang, H. Mechanistic links between Na+ channel (SCN5A) mutations and impaired cardiac pacemaking in sick sinus syndrome. Circ. Res. 2010, 107, 126–137. [Google Scholar] [CrossRef]
- Difrancesco, D. Funny channel gene mutations associated with arrhythmias. J. Physiol. 2013, 591, 4117–4124. [Google Scholar] [CrossRef] [PubMed]
- Fenske, S.; Krause, S.C.; Hassan, S.I.H.; Becirovic, E.; Auer, F.; Bernard, R.; Kupatt, C.; Lange, P.; Ziegler, T.; Wotjak, C.T.; et al. Sick sinus syndrome in HCN1-Deficient mice. Circulation 2013, 128, 2585–2594. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Nakamura, K.; Hayashi, T.; Inagaki, N.; Takahashi, M.; Arimura, T.; Morita, H.; Higashiuesato, Y.; Hirano, Y.; Yasunami, M.; et al. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J. Biol. Chem. 2004, 279, 27194–27198. [Google Scholar] [CrossRef]
- Verkerk, A.O.; Wilders, R. Pacemaker activity of the human sinoatrial node: Effects of HCN4 mutations on the hyperpolarization-activated current. Europace 2014, 16, 384–395. [Google Scholar] [CrossRef]
- Möller, M.; Silbernagel, N.; Wrobel, E.; Stallmayer, B.; Amedonu, E.; Rinné, S.; Peischard, S.; Meuth, S.G.; Wünsch, B.; Strutz-Seebohm, N.; et al. In vitro analyses of novel HCN4 gene mutations. Cell. Physiol. Biochem. 2018, 49, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Torrente, A.G.; Zhang, R.; Wang, H.; Zaini, A.; Kim, B.; Yue, X.; Philipson, K.D.; Goldhaber, J.I. Contribution of small conductance K+ channels to sinoatrial node pacemaker activity: Insights from atrial-specific Na+/Ca2+ exchange knockout mice. J. Physiol. 2017, 595, 3847–3865. [Google Scholar] [CrossRef]
- Morotti, S.; Ni, H.; Peters, C.H.; Rickert, C.; Asgari-Targhi, A.; Sato, D.; Glukhov, A.V.; Proenza, C.; Grandi, E. Intracellular na+ modulates pacemaking activity in murine sinoatrial node myocytes: An in silico analysis. Int. J. Mol. Sci. 2021, 22, 5645. [Google Scholar] [CrossRef]
- Mezzano, V.; Liang, Y.; Wright, A.T.; Lyon, R.C.; Pfeiffer, E.; Song, M.Y.; Gu, Y.; Dalton, N.D.; Scheinman, M.; Peterson, K.L.; et al. Desmosomal junctions are necessary for adult sinus node function. Cardiovasc. Res. 2016, 111, 274–286. [Google Scholar] [CrossRef]
- Mavroidis, M.; Athanasiadis, N.C.; Rigas, P.; Kostavasili, I.; Kloukina, I.; Te Rijdt, W.P.; Kavantzas, N.; Chaniotis, D.; Peter van Tintelen, J.; Skaliora, I.; et al. Desmin is essential for the structure and function of the sinoatrial node: Implications for increased arrhythmogenesis. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H557–H570. [Google Scholar] [CrossRef]
- Battipaglia, I.; Scalone, G.; Macchione, A.; Pinnacchio, G.; Laurito, M.; Milo, M.; Pelargonio, G.; Bencardino, G.; Bellocci, F.; Pieroni, M.; et al. Association of heart rate variability with arrhythmic events in patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia. Circ. J. 2012, 76, 618–623. [Google Scholar] [CrossRef]
- Khudiakov, A.; Perepelina, K.; Klauzen, P.; Zlotina, A.; Gusev, K.; Kaznacheyeva, E.; Malashicheva, A.; Kostareva, A. Generation of two iPSC lines (FAMRCi004-A and FAMRCi004-B) from patient with familial progressive cardiac conduction disorder carrying genetic variant DSP p.His1684Arg. Stem Cell Res. 2020, 43, 101702. [Google Scholar] [CrossRef]
- Gusev, K.; Khudiakov, A.; Zaytseva, A.; Perepelina, K.; Makeenok, S.; Kaznacheyeva, E.; Kostareva, A. Impact of the DSP-H1684R genetic variant on ion channels activity in iPSC-derived cardiomyocytes. Cell. Physiol. Biochem. 2020, 54, 696–706. [Google Scholar] [PubMed]
- Qiao, Y.; Lipovsky, C.; Hicks, S.; Bhatnagar, S.; Li, G.; Khandekar, A.; Guzy, R.; Woo, K.V.; Nichols, C.G.; Efimov, I.R.; et al. Transient notch activation induces long-term gene expression changes leading to sick sinus syndrome in mice. Circ. Res. 2017, 121, 549–563. [Google Scholar] [CrossRef]
- Alghamdi, A.M.; Boyett, M.R.; Hancox, J.C.; Zhang, H. Cardiac Pacemaker Dysfunction Arising from Different Studies of Ion Channel Remodeling in the Aging Rat Heart. Front. Physiol. 2020, 11, 546508. [Google Scholar] [CrossRef] [PubMed]
- Paravicini, T.M.; Touyz, R.M. NADPH oxidases, reactive oxygen species, and hypertension: Clinical implications and therapeutic possibilities. Diabetes Care 2008, 31 (Suppl. S2), S170–S180. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Pesce, M. Cell-based mechanosensation, epigenetics, and non-coding RNAs in progression of cardiac fibrosis. Int. J. Mol. Sci. 2020, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.S.; Chiu, W.T.; Hsu, P.L.; Lin, S.C.; Peng, I.C.; Wang, C.Y.; Tsai, S.J. Pathophysiological implications of hypoxia in human diseases. J. Biomed. Sci. 2020, 27, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Fan, T.; Jin, X. Identification of Key Genes Involved in Acute Myocardial Infarction by Comparative Transcriptome Analysis. Biomed Res. Int. 2020, 2020, 1470867. [Google Scholar] [CrossRef] [PubMed]
- Schiano, C.; Benincasa, G.; Franzese, M.; Della Mura, N.; Pane, K.; Salvatore, M.; Napoli, C. Epigenetic-sensitive pathways in personalized therapy of major cardiovascular diseases. Pharmacol. Ther. 2020, 210, 107514. [Google Scholar] [CrossRef]
- Wang, X.; Shen, X.; Weil, B.R.; Young, R.F.; Canty, J.M.; Qu, J. Quantitative proteomic and phosphoproteomic profiling of ischemic myocardial stunning in swine. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H1256–H1271. [Google Scholar] [CrossRef]
- Simmonds, S.J.; Cuijpers, I.; Heymans, S.; Jones, E.A.V. Cellular and Molecular Differences between HFpEF and HFrEF: A Step Ahead in an Improved Pathological Understanding. Cells 2020, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Byrne, N.J.; Rajasekaran, N.S.; Abel, E.D.; Bugger, H. Therapeutic potential of targeting oxidative stress in diabetic cardiomyopathy. Free Radic. Biol. Med. 2021, 169, 317–342. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wu, J.; Zhou, W.; Liu, X.; Liu, Y.; Zhang, J.; Jia, S.; Li, J.; Wang, H. Identification and analysis of key genes associated with acute myocardial infarction by integrated bioinformatics methods. Medicine 2021, 100, e25553. [Google Scholar] [CrossRef]
- Sumi, M.P.; Mahajan, B.; Sattar, R.S.A.; Nimisha; Apurva; Kumar, A.; Sharma, A.K.; Ahmad, E.; Ali, A.; Saluja, S.S. Elucidation of Epigenetic Landscape in Coronary Artery Disease: A Review on Basic Concept to Personalized Medicine. Epigenet. Insights 2021, 14, 2516865720988567. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, E.A.; Rose, R.A.; Quinn, T.A. Neurohumoral Control of Sinoatrial Node Activity and Heart Rate: Insight From Experimental Models and Findings From Humans. Front. Physiol. 2020, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Adameova, A.; Shah, A.K.; Dhalla, N.S. Role of oxidative stress in the genesis of ventricular arrhythmias. Int. J. Mol. Sci. 2020, 21, 4200. [Google Scholar] [CrossRef]
- Fu, Y.; Zhong, H.; Nanamori, M.; Mortensen, R.M.; Huang, X.; Lan, K.; Neubig, R.R. RGS-insensitive G-protein mutations to study the role of endogenous RGS proteins. Methods Enzymol. 2004, 389, 229–243. [Google Scholar]
- Fu, Y.; Huang, X.; Zhong, H.; Mortensen, R.M.; D’Alecy, L.G.; Neubig, R.R. Endogenous RGS proteins and Gα subtypes differentially control muscarinic and adenosine-mediated chronotropic effects. Circ. Res. 2006, 98, 659–666. [Google Scholar] [CrossRef]
- Fu, Y.; Huang, X.; Piao, L.; Lopatin, A.N.; Neubig, R.R. Endogenous RGS proteins modulate SA and AV nodal functions in isolated heart: Implications for sick sinus syndrome and AV block. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H2532–H2539. [Google Scholar] [CrossRef]
- Yang, B.; Huang, Y.; Zhang, H.; Huang, Y.; Zhou, H.J.; Young, L.; Xiao, H.; Min, W. Mitochondrial thioredoxin-2 maintains HCN4 expression and prevents oxidative stress-mediated sick sinus syndrome. J. Mol. Cell. Cardiol. 2020, 138, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, B.; Ren, J.; Lu, F.; Qi, Y.; Weng, W.; Gao, R. Two methods for modeling of sick sinus syndrome in rats: Ischemia reperfusion and sodium hydroxide induced injury. Biomed. Pharmacother. 2019, 111, 778–784. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L.; Hao, M.; Chen, K.; Lu, Y.; Qi, J.; Chen, W.; Ren, L.; Cai, X.; Chen, C.; et al. Yixin-Fumai granules improve sick sinus syndrome in aging mice through Nrf-2/HO-1 pathway: A new target for sick sinus syndrome. J. Ethnopharmacol. 2021, 277, 114254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Zhao, H.Y.; Wang, Y.; Di, L.; Liu, X.Y.; Qian, F.; Liu, S.R. Zenglv fumai granule protects cardiomyocytes against hypoxia/reoxygenation-induced apoptosis via inhibiting TRIM28 expression. Mol. Med. Rep. 2020, 23, 1. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Liu, Y.; Peng, J.; Guan, X. The effect of astragaloside on pacemaker current and the cytoskeleton in rabbit sinoatrial node cells under the ischemia and reperfusion condition. Front. Pharmacol. 2018, 9, 551. [Google Scholar] [CrossRef]
- Bai, X.; Wang, K.; Yuan, Y.; Li, Q.; Dobrzynski, H.; Boyett, M.R.; Hancox, J.C.; Zhang, H. Mechanism underlying impaired cardiac pacemaking rhythm during ischemia: A simulation study. Chaos 2017, 27, 93934. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.P. Side effects of calcium channel blockers. Hypertension 1988, 11, II42. [Google Scholar] [CrossRef]
- Grant, A.O. Propafenone: An effective agent for the management of supraventricular arrhythmias. J. Cardiovasc. Electrophysiol. 1996, 7, 353–364. [Google Scholar] [CrossRef]
- Wasada, T.; Katsumori, K.; Hasumi, S.; Arii, H.; Saeki, A.; Kuroki, H.; Saito, S.; Omori, Y.; Kasanuki, H. Association of Sick Sinus Syndrome with Hyperinsulinemia and Insulin Resistance in Patients with Non-Insulin-Dependent Diabetes Mellitus: Report of Four Cases. Intern. Med. 1995, 34, 1174–1177. [Google Scholar] [CrossRef][Green Version]
- Kondo, H.; Kira, S.; Oniki, T.; Gotoh, K.; Fukui, A.; Abe, I.; Ikebe, Y.; Kawano, K.; Saito, S.; Aoki, K.; et al. Interleukin-10 treatment attenuates sinus node dysfunction caused by streptozotocin-induced hyperglycaemia in mice. Cardiovasc. Res. 2019, 115, 57–70. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, T.; Lian, G.; Xu, C.; Wang, H.; Xie, L. TRPM7 regulates angiotensin II-induced sinoatrial node fibrosis in sick sinus syndrome rats by mediating Smad signaling. Heart Vessels 2018, 33, 1094–1105. [Google Scholar] [CrossRef]
- Zhang, H.; Hao, M.; Li, L.; Chen, K.; Qi, J.; Chen, W.; Cai, X.; Chen, C.; Liu, Z.; Hou, P. Shenxian-Shengmai Oral Liquid Improves Sinoatrial Node Dysfunction through the PKC/NOX-2 Signaling Pathway. Evidence-Based Complement. Altern. Med. 2021, 2021, 5572140. [Google Scholar] [CrossRef]
- Roh, S.Y.; Kim, J.Y.; Cha, H.K.; Lim, H.Y.; Park, Y.; Lee, K.N.; Shim, J.; Choi, J., II; Kim, Y.H.; Son, G.H. Molecular signatures of sinus node dysfunction induce structural remodeling in the right atrial tissue. Mol. Cells 2020, 43, 408–418. [Google Scholar]
- Gourraud, J.B.; Barc, J.; Thollet, A.; Le Scouarnec, S.; Le Marec, H.; Schott, J.J.; Redon, R.; Probst, V. The Brugada Syndrome: A Rare Arrhythmia Disorder with Complex Inheritance. Front. Cardiovasc. Med. 2016, 3, 9. [Google Scholar] [CrossRef]
- Veltmann, C.; Barajas-Martinez, H.; Wolpert, C.; Borggrefe, M.; Schimpf, R.; Pfeiffer, R.; Cáceres, G.; Burashnikov, E.; Antzelevitch, C.; Hu, D. Further Insights in the Most Common SCN5A Mutation Causing Overlapping Phenotype of Long QT Syndrome, Brugada Syndrome, and Conduction Defect. J. Am. Heart Assoc. 2016, 5, e003379. [Google Scholar] [CrossRef] [PubMed]
- Wijeyeratne, Y.D.; Tanck, M.W.; Mizusawa, Y.; Batchvarov, V.; Barc, J.; Crotti, L.; Bos, J.M.; Tester, D.J.; Muir, A.; Veltmann, C.; et al. Scn5a mutation type and a genetic risk score associate variably with brugada syndrome phenotype in scn5a families. Circ. Genomic Precis. Med. 2020, 13, 599–608. [Google Scholar] [CrossRef]
- Monasky, M.M.; Micaglio, E.; Ciconte, G.; Pappone, C. Brugada syndrome: Oligogenic or mendelian disease? Int. J. Mol. Sci. 2020, 21, 1687. [Google Scholar] [CrossRef] [PubMed]
- Nademanee, K.; Veerakul, G.; Chandanamattha, P.; Chaothawee, L.; Ariyachaipanich, A.; Jirasirirojanakorn, K.; Likittanasombat, K.; Bhuripanyo, K.; Ngarmukos, T. Prevention of ventricular fibrillation episodes in brugada syndrome by catheter ablation over the anterior right ventricular outflow tract epicardium. Circulation 2011, 123, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Nademanee, K.; Raju, H.; De Noronha, S.V.; Papadakis, M.; Robinson, L.; Rothery, S.; Makita, N.; Kowase, S.; Boonmee, N.; Vitayakritsirikul, V.; et al. Fibrosis, connexin-43, and conduction abnormalities in the Brugada syndrome. J. Am. Coll. Cardiol. 2015, 66, 1976–1986. [Google Scholar] [CrossRef] [PubMed]
- Pappone, C.; Monasky, M.M.; Micaglio, E.; Ciconte, G. Right ventricular electromechanical abnormalities in Brugada syndrome: Is this a cardiomyopathy? Eur. Hear. J. Suppl. 2020, 22, E101–E104. [Google Scholar] [CrossRef]
- Papadatos, G.A.; Wallerstein, P.M.R.; Head, C.E.G.; Ratcliff, R.; Brady, P.A.; Benndorf, K.; Saumarez, R.C.; Trezise, A.E.O.; Huang, C.L.H.; Vandenberg, J.I.; et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl. Acad. Sci. USA 2002, 99, 6210–6215. [Google Scholar] [CrossRef] [PubMed]
- Leoni, A.L.; Gavillet, B.; Rougier, J.S.; Marionneau, C.; Probst, V.; Le Scouarnec, S.; Schott, J.J.; Demolombe, S.; Bruneval, P.; Huang, C.L.H.; et al. Variable Nav1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a +/- mouse model. PLoS ONE 2010, 5, e9298. [Google Scholar] [CrossRef]
- Matthews, G.D.K.; Guzadhur, L.; Sabir, I.N.; Grace, A.A.; Huang, C.L.H. Action potential wavelength restitution predicts alternans and arrhythmia in murine Scn5a+/- hearts. J. Physiol. 2013, 591, 4167–4188. [Google Scholar] [CrossRef]
- Kelly, A.; Salerno, S.; Connolly, A.; Bishop, M.; Charpentier, F.; Stølen, T.; Smith, G.L. Normal interventricular differences in tissue architecture underlie right ventricular susceptibility to conduction abnormalities in a mouse model of Brugada syndrome. Cardiovasc. Res. 2018, 114, 724–736. [Google Scholar] [CrossRef]
- Finlay, M.; Bhar-Amato, J.; Ng, K.E.; Santos, D.; Orini, M.; Vyas, V.; Taggart, P.; Grace, A.A.; Huang, C.L.H.; Lambiase, P.D.; et al. Autonomic modulation of the electrical substrate in mice haploinsufficient for cardiac sodium channels: A model of the Brugada syndrome. Am. J. Physiol. Cell Physiol. 2019, 317, C576–C583. [Google Scholar] [CrossRef]
- Wichter, T.; Matheja, P.; Eckardt, L.; Kies, P.; Schäfers, K.; Schulze-Bahr, E.; Haverkamp, W.; Borggrefe, M.; Schober, O.; Breithardt, G.; et al. Cardiac autonomic dysfunction in Brugada syndrome. Circulation 2002, 105, 702–706. [Google Scholar] [CrossRef]
- Pérez-Hernández, M.; Matamoros, M.; Alfayate, S.; Nieto-Marín, P.; Utrilla, R.G.; Tinaquero, D.; de Andrés, R.; Crespo, T.; Ponce-Balbuena, D.; Willis, B.C.; et al. Brugada syndrome trafficking-defective Nav1.5 channels can trap cardiac Kir2.1/2.2 channels. JCI Insight 2018, 3, 18. [Google Scholar] [CrossRef]
- Wilde, A.A.M.; Brugada, R. Phenotypical manifestations of mutations in the genes encoding subunits of the cardiac sodium channel. Circ. Res. 2011, 108, 884–897. [Google Scholar] [CrossRef]
- Remme, C.A.; Verkerk, A.O.; Nuyens, D.; Van Ginneken, A.C.G.; Van Brunschot, S.; Belterman, C.N.W.; Wilders, R.; Van Roon, M.A.; Tan, H.L.; Wilde, A.A.M.; et al. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation 2006, 114, 2584–2594. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.I.; Rougier, J.S.; Kucera, J.P.; Benammar, N.; Fressart, V.; Guicheney, P.; Madle, A.; Fromer, M.; Schläpfer, J.; Abriel, H. Brugada syndrome and fever: Genetic and molecular characterization of patients carrying SCN5A mutations. Cardiovasc. Res. 2005, 67, 510–519. [Google Scholar] [CrossRef]
- Clatot, J.; Ziyadeh-Isleem, A.; Maugenre, S.; Denjoy, I.; Liu, H.; Dilanian, G.; Hatem, S.N.; Deschênes, I.; Coulombe, A.; Guicheney, P.; et al. Dominant-negative effect of SCN5A N-terminal mutations through the interaction of Nav1.5 α-subunits. Cardiovasc. Res. 2012, 96, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Doisne, N.; Grauso, M.; Mougenot, N.; Clergue, M.; Souil, C.; Coulombe, A.; Guicheney, P.; Neyroud, N. In vivo Dominant-Negative Effect of an SCN5A Brugada Syndrome Variant. Front. Physiol. 2021, 12, 687. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Vermij, S.H.; Sottas, V.; Shestak, A.; Ross-Kaschitza, D.; Zaklyazminskaya, E.V.; Hudmon, A.; Pitt, G.S.; Rougier, J.S.; Abriel, H. Calmodulin binds to the N-terminal domain of the cardiac sodium channel Nav1.5. Channels 2020, 14, 268–286. [Google Scholar] [CrossRef]
- Park, D.S.; Cerrone, M.; Morley, G.; Vasquez, C.; Fowler, S.; Liu, N.; Bernstein, S.A.; Liu, F.Y.; Zhang, J.; Rogers, C.S.; et al. Genetically engineered SCN5A mutant pig hearts exhibit conduction defects and arrhythmias. J. Clin. Investig. 2015, 125, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.P.; Casini, S.; Van Den Berg, C.W.; Hoekstra, M.; Remme, C.A.; Dambrot, C.; Salvatori, D.; Van Oostwaard, D.W.; Wilde, A.A.M.; Bezzina, C.R.; et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 2012, 125, 3079–3091. [Google Scholar] [CrossRef] [PubMed]
- Portero, V.; Casini, S.; Hoekstra, M.; Verkerk, A.O.; Mengarelli, I.; Belardinelli, L.; Rajamani, S.; Wilde, A.A.M.; Bezzina, C.R.; Veldkamp, M.W.; et al. Anti-arrhythmic potential of the late sodium current inhibitor GS-458967 in murine Scn5a-1798insD+/- and human SCN5A-1795insD+/- iPSC-derived cardiomyocytes. Cardiovasc. Res. 2017, 113, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Sallam, K.; Wu, H.; Li, Y.; Itzhaki, I.; Garg, P.; Zhang, Y.; Vermglinchan, V.; Lan, F.; Gu, M.; et al. Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell–Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome. J. Am. Coll. Cardiol. 2016, 68, 2086–2096. [Google Scholar] [CrossRef]
- Selga, E.; Sendfeld, F.; Martinez-Moreno, R.; Medine, C.N.; Tura-Ceide, O.; Wilmut, S.I.; Pérez, G.J.; Scornik, F.S.; Brugada, R.; Mills, N.L. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J. Mol. Cell. Cardiol. 2018, 114, 10–19. [Google Scholar] [CrossRef]
- Aiba, T.; Shimizu, W.; Hidaka, I.; Uemura, K.; Noda, T.; Zheng, C.; Kamiya, A.; Inagaki, M.; Sugimachi, M.; Sunagawa, K. Cellular Basis for Trigger and Maintenance of Ventricular Fibrillation in the Brugada Syndrome Model: High-Resolution Optical Mapping Study. J. Am. Coll. Cardiol. 2006, 47, 2074–2085. [Google Scholar] [CrossRef]
- Ma, D.; Liu, Z.; Loh, L.J.; Zhao, Y.; Li, G.; Liew, R.; Islam, O.; Wu, J.; Chung, Y.Y.; Teo, W.S.; et al. Identification of an I Na-dependent and I to-mediated proarrhythmic mechanism in cardiomyocytes derived from pluripotent stem cells of a Brugada syndrome patient. Sci. Rep. 2018, 8, 1–11. [Google Scholar]
- Angsutararux, P.; Luanpitpong, S.; Chingsuwanrote, P.; Supraditaporn, K.; Waeteekul, S.; Terbto, P.; Lorthongpanich, C.; Laowtammathron, C.; U.-Pratya, Y.; Issaragrisil, S. Generation of human induced pluripotent stem cell line carrying SCN5AC2204>T Brugada mutation (MUSli009-A-1) introduced by CRISPR/Cas9-mediated genome editing. Stem Cell Res. 2019, 41, 101618. [Google Scholar] [CrossRef]
- De la Roche, J.; Angsutararux, P.; Kempf, H.; Janan, M.; Bolesani, E.; Thiemann, S.; Wojciechowski, D.; Coffee, M.; Franke, A.; Schwanke, K.; et al. Comparing human iPSC-cardiomyocytes versus HEK293T cells unveils disease-causing effects of Brugada mutation A735V of NaV1.5 sodium channels. Sci. Rep. 2019, 9, 11173. [Google Scholar] [CrossRef] [PubMed]
- El-Battrawy, I.; Albers, S.; Cyganek, L.; Zhao, Z.; Lan, H.; Li, X.; Xu, Q.; Kleinsorge, M.; Huang, M.; Liao, Z.; et al. A cellular model of Brugada syndrome with SCN10A variants using human-induced pluripotent stem cell-derived cardiomyocytes. Europace 2019, 21, 1410–1421. [Google Scholar] [CrossRef]
- Ozhathil, L.C.; Rougier, J.S.; Arullampalam, P.; Essers, M.C.; Ross-Kaschitza, D.; Abriel, H. Deletion of trpm4 alters the function of the nav1.5 channel in murine cardiac myocytes. Int. J. Mol. Sci. 2021, 22, 3401. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Wu, X.; Yang, Z.; Wu, L.; Yong, S.L.; Zhang, C.; Hu, K.; Wang, Q.K.; Chen, Q. MOG1 rescues defective trafficking of nav1.5 mutations in brugada syndrome and sick sinus syndrome. Circ. Arrhythmia Electrophysiol. 2013, 6, 392–401. [Google Scholar] [CrossRef]
- Yu, G.; Liu, Y.; Qin, J.; Wang, Z.; Hu, Y.; Wang, F.; Li, Y.; Chakrabarti, S.; Chen, Q.; Wang, Q.K. Mechanistic insights into the interaction of the MOG1 protein with the cardiac sodium channel Nav1.5 clarify the molecular basis of Brugada syndrome. J. Biol. Chem. 2018, 293, 18207–18217. [Google Scholar] [CrossRef]
- Veerman, C.C.; Mengarelli, I.; Guan, K.; Stauske, M.; Barc, J.; Tan, H.L.; Wilde, A.A.M.; Verkerk, A.O.; Bezzina, C.R. HiPSC-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Morita, H.; Zipes, D.P.; Morita, S.T.; Wu, J. Differences in arrhythmogenicity between the canine right ventricular outflow tract and anteroinferior right ventricle in a model of Brugada syndrome. Heart Rhythm 2007, 4, 66–74. [Google Scholar] [CrossRef]
- Tsumoto, K.; Ashihara, T.; Naito, N.; Shimamoto, T.; Amano, A.; Kurata, Y.; Kurachi, Y. Specific decreasing of Na + channel expression on the lateral membrane of cardiomyocytes causes fatal arrhythmias in Brugada syndrome. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Calvo, M.; Le Rolle, V.; Perez, D.R.; Behar, N.; Gomis, P.; Mabo, P.; Hernandez, A.I. Analysis of a cardiovascular model for the study of the autonomic response of Brugada syndrome patients. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2016, 2016, 5591–5594. [Google Scholar]
- Crea, P.; Picciolo, G.; Luzza, F.; Oreto, G. A three-dimensional computed model of ST segment abnormality in type 1 Brugada Pattern: A key role of right ventricular outflow tract orientation? J. Electrocardiol. 2019, 53, 31–35. [Google Scholar] [CrossRef]
- Kornej, J.; Börschel, C.S.; Börschel, C.S.; Benjamin, E.J.; Benjamin, E.J.; Schnabel, R.B.; Schnabel, R.B. Epidemiology of Atrial Fibrillation in the 21st Century: Novel Methods and New Insights. Circ. Res. 2020, 127, 4–20. [Google Scholar] [CrossRef]
- Hindricks, G.; Potpara, T.; Dagres, N.; Bax, J.J.; Boriani, G.; Dan, G.A.; Fauchier, L.; Kalman, J.M.; Lane, D.A.; Lettino, M.; et al. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS). Eur. Heart J. 2021, 42, 373–498. [Google Scholar] [CrossRef]
- Wilhelm, M. Atrial fibrillation in endurance athletes. Eur. J. Prev. Cardiol. 2014, 21, 1040–1048. [Google Scholar] [CrossRef]
- Newman, W.; Parry-Williams, G.; Wiles, J.; Edwards, J.; Hulbert, S.; Kipourou, K.; Papadakis, M.; Sharma, R.; O’Driscoll, J. Risk of atrial fibrillation in athletes: A systematic review and meta-analysis. Br. J. Sports Med. 2021, 55, 1233–1238. [Google Scholar] [CrossRef]
- Greenberg, J.W.; Lancaster, T.S.; Schuessler, R.B.; Melby, S.J. Postoperative atrial fibrillation following cardiac surgery: A persistent complication. Eur. J. Cardio-Thorac. Surg. 2017, 52, 665–672. [Google Scholar] [CrossRef]
- Qureshi, M.; Ahmed, A.; Massie, V.; Marshall, E.; Harky, A. Determinants of atrial fibrillation after cardiac surgery. Rev. Cardiovasc. Med. 2021, 22, 329–341. [Google Scholar] [CrossRef]
- Soto-Pedre, E.; Siddiqui, M.K.; Maroteau, C.; Dawed, A.Y.; Doney, A.S.; Palmer, C.N.A.; Pearson, E.R.; Leese, G.P. Polymorphism in INSR Locus Modifies Risk of Atrial Fibrillation in Patients on Thyroid Hormone Replacement Therapy. Front. Genet. 2021, 12, 652878. [Google Scholar] [CrossRef]
- Haruta, D.; Landes, R.D.; Hida, A.; Imaizumi, M.; Ohishi, W.; Akahoshi, M.; Maemura, K. Relationship Between Radiation Exposure and Incident Atrial Fibrillation Among Atomic Bomb Survivors. Circ. Rep. 2021, 3, 21. [Google Scholar] [CrossRef]
- Roselli, C.; Rienstra, M.; Ellinor, P.T. Genetics of Atrial Fibrillation in 2020—GWAS, genome sequencing, polygenic risk and beyond. Circ. Res. 2020, 127, 21. [Google Scholar] [CrossRef]
- Hong, K.; Piper, D.R.; Diaz-Valdecantos, A.; Brugada, J.; Oliva, A.; Burashnikov, E.; Santos-De-Soto, J.; Grueso-Montero, J.; Diaz-Enfante, E.; Brugada, P.; et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc. Res. 2005, 68, 433–440. [Google Scholar] [CrossRef]
- Das, S.; Makino, S.; Melman, Y.F.; Shea, M.A.; Goyal, S.B.; Rosenzweig, A.; MacRae, C.A.; Ellinor, P.T. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm 2009, 6, 1146–1153. [Google Scholar] [CrossRef]
- Bartos, D.C.; Duchatelet, S.; Burgess, D.E.; Klug, D.; Denjoy, I.; Peat, R.; Lupoglazoff, J.M.; Fressart, V.; Berthet, M.; Ackerman, M.J.; et al. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Hear. Rhythm 2011, 8, 48–55. [Google Scholar] [CrossRef]
- Bartos, D.C.; Anderson, J.B.; Bastiaenen, R.; Johnson, J.N.; Gollob, M.H.; Tester, D.J.; Burgess, D.E.; Homfray, T.; Behr, E.R.; Ackerman, M.J.; et al. A KCNQ1 mutation causes a high penetrance for familial atrial fibrillation. J. Cardiovasc. Electrophysiol. 2013, 24, 562–569. [Google Scholar] [CrossRef]
- Hasegawa, K.; Ohno, S.; Ashihara, T.; Itoh, H.; Ding, W.G.; Toyoda, F.; Makiyama, T.; Aoki, H.; Nakamura, Y.; Delisle, B.P.; et al. A novel KCNQ1 missense mutation identified in a patient with juvenile-onset atrial fibrillation causes constitutively open IKs channels. Heart Rhythm. 2014, 11, 67–75. [Google Scholar] [CrossRef]
- Chen, Y.H.; Xu, S.J.; Bendahhou, S.; Wang, X.L.; Wang, Y.; Xu, W.Y.; Jin, H.W.; Sun, H.; Su, X.Y.; Zhuang, Q.N.; et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 2003, 299, 251–254. [Google Scholar] [CrossRef]
- Campbell, C.M.; Campbell, J.D.; Thompson, C.H.; Galimberti, E.S.; Darbar, D.; Vanoye, C.G.; George, A.L. Selective targeting of gain-of-function KCNQ1 mutations predisposing to atrial fibrillation. Circ. Arrhythmia Electrophysiol. 2013, 6, 960–966. [Google Scholar] [CrossRef]
- Schultz, H.D.; Gardner, D.G.; Deschepper, C.F.; Coleridge, H.M.; Coleridge, J.C.G. Vagal C-fiber blockade abolishes sympathetic inhibition by atrial natriuretic factor. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1988, 255, R6–R13. [Google Scholar] [CrossRef]
- CROZIER, I.; RICHARDS, A.M.; FOY, S.G.; IKRAM, H. Electrophysiological Effects of atrial Natriuretic Peptide on the Cardiac Conduction System in Man. Pacing Clin. Electrophysiol. 1993, 16, 738–742. [Google Scholar] [CrossRef]
- Lonardo, G.; Cerbai, E.; Casini, S.; Giunti, G.; Bonacchi, M.; Battaglia, F.; Fiorani, B.; Stefano, P.L.; Sani, G.; Mugelli, A. Atrial natriuretic peptide modulates the hyperpolarization-activated current (If) in human atrial myocytes. Cardiovasc. Res. 2004, 63, 528–536. [Google Scholar] [CrossRef]
- Stambler, B.S.; Guo, G.B. Atrial natriuretic peptide has dose-dependent, autonomically mediated effects on atrial refractoriness and repolarization in anesthetized dogs. J. Cardiovasc. Electrophysiol. 2005, 16, 1341–1347. [Google Scholar] [CrossRef]
- Hodgson-Zingman, D.M.; Karst, M.L.; Zingman, L.V.; Heublein, D.M.; Darbar, D.; Herron, K.J.; Ballew, J.D.; de Andrade, M.; Burnett, J.C.; Olson, T.M. Atrial Natriuretic Peptide Frameshift Mutation in Familial Atrial Fibrillation. N. Engl. J. Med. 2008, 359, 158–165. [Google Scholar] [CrossRef]
- Li, Q.Y.; Newbury-Ecob, R.A.; Terrett, J.A.; Wilson, D.I.; Curtis, A.R.J.; Yi, C.H.; Gebühr, T.; Bullen, P.J.; Robson, S.C.; Strachan, T.; et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 1997, 15, 21–29. [Google Scholar] [CrossRef]
- Bruneau, B.G.; Logan, M.; Davis, N.; Levi, T.; Tabin, C.J.; Seidman, J.G.; Seidman, C.E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt- Oram syndrome. Dev. Biol. 1999, 211, 100–108. [Google Scholar] [CrossRef]
- Hatcher, C.J.; Goldstein, M.M.; Mah, C.S.; Susan Delia, C.; Basson, C.T. Identification and localization of TBX5 transcription factor during human cardiac morphogenesis. Dev. Dyn. 2000, 219, 90–95. [Google Scholar] [CrossRef]
- Hatcher, C.J.; Kim, M.S.; Mah, C.S.; Goldstein, M.M.; Wong, B.; Mikawa, T.; Basson, C.T. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev. Biol. 2001, 230, 177–188. [Google Scholar] [CrossRef]
- Zhou, Y.Q.; Zhu, Y.; Bishop, J.; Davidson, L.; Henkelman, R.M.; Bruneau, B.G.; Foster, F.S. Abnormal cardiac inflow patterns during postnatal development in a mouse model of Holt-Oram syndrome. Am. J. Physiol. Heart Circ. Physiol. 2005, 289, H992–H1001. [Google Scholar] [CrossRef]
- Postma, A.V.; Van De Meerakker, J.B.A.; Mathijssen, I.B.; Barnett, P.; Christoffels, V.M.; Ilgun, A.; Lam, J.; Wilde, A.A.M.; Deprez, R.H.L.; Moorman, A.F.M. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ. Res. 2008, 102, 1433–1442. [Google Scholar] [CrossRef]
- Guzzolino, E.; Pellegrino, M.; Ahuja, N.; Garrity, D.; D’Aurizio, R.; Groth, M.; Baumgart, M.; Hatcher, C.J.; Mercatanti, A.; Evangelista, M.; et al. miR-182-5p is an evolutionarily conserved Tbx5 effector that impacts cardiac development and electrical activity in zebrafish. Cell. Mol. Life Sci. 2020, 77, 3215–3229. [Google Scholar] [CrossRef]
- Hall, A.W.; Chaffin, M.; Roselli, C.; Lin, H.; Lubitz, S.A.; Bianchi, V.; Geeven, G.; Bedi, K.; Margulies, K.B.; De Laat, W.; et al. Epigenetic Analyses of Human Left Atrial Tissue Identifies Gene Networks Underlying Atrial Fibrillation. Circ. Genomic Precis. Med. 2020, 13, 588–598. [Google Scholar] [CrossRef]
- Yuan, F.; Qiu, X.B.; Li, R.G.; Qu, X.K.; Wang, J.; Xu, Y.J.; Liu, X.; Fang, W.Y.; Yang, Y.Q.; Liao, D.N. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int. J. Mol. Med. 2015, 35, 478–486. [Google Scholar] [CrossRef]
- Kasahara, H.; Wakimoto, H.; Liu, M.; Maguire, C.T.; Converso, K.L.; Shioi, T.; Huang, W.Y.; Manning, W.J.; Paul, D.; Lawitts, J.; et al. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J. Clin. investig. 2001, 108, 189–201. [Google Scholar] [CrossRef]
- Jay, P.Y.; Harris, B.S.; Buerger, A.; Rozhitskaya, O.; Maguire, C.T.; Barbosky, L.A.; Mccusty, E.; Berul, C.I.; O’Brien, T.X.; Gourdie, R.G.; et al. Function follows form: Cardiac conduction system defects in Nkx2-5 mutation. In Anatomical Record—Part A Discoveries in Molecular, Cellular, and Evolutionary Biology; American Association for Anatomy: Rockville, MD, USA, 2004; Volume 280, pp. 966–972. [Google Scholar]
- Chen, J.; Xu, S.; Li, W.; Wu, L.; Wang, L.; Li, Y.; Zhou, W. Nkx2.5 insufficiency leads to atrial electrical remodeling through Wnt signaling in HL-1 cells. Exp. Ther. Med. 2019, 18, 4631–4636. [Google Scholar] [CrossRef]
- Karakikes, I.; Termglinchan, V.; Cepeda, D.A.; Lee, J.; Diecke, S.; Hendel, A.; Itzhaki, I.; Ameen, M.; Shrestha, R.; Wu, H.; et al. A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell-based models for cardiovascular diseases. Circ. Res. 2017, 120, 1561–1571. [Google Scholar] [CrossRef]
- Laforest, B.; Dai, W.; Tyan, L.; Lazarevic, S.; Shen, K.M.; Gadek, M.; Broman, M.T.; Weber, C.R.; Moskowitz, I.P. Atrial fibrillation risk loci interact to modulate Ca2+-dependent atrial rhythm homeostasis. J. Clin. Investig. 2019, 129, 4937–4950. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Lin, Y.; Liu, H.; Chen, H.; Ju, W.; Cui, C.; Chen, M. Establishment of an iPSC line (JSPHi001-A) from a patient with familial dilated cardiomyopathy and atrial fibrillation caused by LMNA missense mutation (c.1003C > T). Stem Cell Res. 2021, 53, 102349. [Google Scholar] [CrossRef]
- Benzoni, P.; Campostrini, G.; Landi, S.; Bertini, V.; Marchina, E.; Iascone, M.; Ahlberg, G.; Olesen, M.S.; Crescini, E.; Mora, C.; et al. Human iPSC modelling of a familial form of atrial fibrillation reveals a gain of function of If and ICaL in patient-derived cardiomyocytes. Cardiovasc. Res. 2020, 116, 1147–1160. [Google Scholar] [CrossRef]
- Chiu, Y.T.; Tsai, M.H.; Chan, D.Z.H.; Ko, H.W.; Lu, H.E.; Huang, C.Y.F.; Lin, Y.J.; Chang, S.L.; Lo, L.W.; Huang, C.Y.; et al. Generation of IBMS-iPSC-015, -016, -017 human induced pluripotent stem cells (IBMSi013-A, IBMSi014-A, and IBMSi015-A) derived from patients with atrial fibrillation. Stem Cell Res. 2021, 54, 102419. [Google Scholar] [CrossRef]
- Fareh, S.; Bénardeau, A.; Thibault, B.; Nattel, S. The T-type Ca2+ channel blocker mibefradil prevents the development of a substrate for atrial fibrillation by tachycardia-induced atrial remodeling in dogs. Circulation 1999, 100, 2191–2197. [Google Scholar] [CrossRef]
- Denham, N.C.; Pearman, C.M.; Madders, G.W.P.; Smith, C.E.R.; Trafford, A.W.; Dibb, K.M. Optimising Large Animal Models of Sustained Atrial Fibrillation: Relevance of the Critical Mass Hypothesis. Front. Physiol. 2021, 12, 887. [Google Scholar] [CrossRef]
- Nofi, C.; Zhang, K.; Tang, Y.-D.; Li, Y.; Migirov, A.; Ojamaa, K.; Gerdes, A.M.; Zhang, Y. Chronic dantrolene treatment attenuates cardiac dysfunction and reduces atrial fibrillation inducibility in a rat myocardial infarction heart failure model. Hear. Rhythm O2 2020, 1, 126–135. [Google Scholar] [CrossRef]
- Jung, C.B.; Moretti, A.; Mederos y Schnitzler, M.; Iop, L.; Storch, U.; Bellin, M.; Dorn, T.; Ruppenthal, S.; Pfeiffer, S.; Goedel, A.; et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol. Med. 2012, 4, 180–191. [Google Scholar] [CrossRef]
- Penttinen, K.; Swan, H.; Vanninen, S.; Paavola, J.; Lahtinen, A.M.; Kontula, K.; Aalto-Setälä, K. Antiarrhythmic effects of dantrolene in patients with catecholaminergic polymorphic ventricular tachycardia and replication of the responses using iPSC models. PLoS ONE 2015, 10, e0125366. [Google Scholar]
- Heijman, J.; Sutanto, H.; Crijns, H.J.G.M.; Nattel, S.; Trayanova, N.A. Computational models of atrial fibrillation: Achievements, challenges, and perspectives for improving clinical care. Cardiovasc. Res. 2021, 117, 1682–1699. [Google Scholar] [CrossRef]
- Belletti, R.; Romero, L.; Martinez-Mateu, L.; Cherry, E.M.; Fenton, F.H.; Saiz, J. Arrhythmogenic Effects of Genetic Mutations Affecting Potassium Channels in Human Atrial Fibrillation: A Simulation Study. Front. Physiol. 2021, 12, 681943. [Google Scholar] [CrossRef]
- Abellana, R.; Gonzalez-Loyola, F.; Verdu-Rotellar, J.; Bustamante, A.; Palà, E.; Clua-Espuny, J.L.; Montaner, J.; Pedrote, A.; Val-Garcia, J.L.; Ribas Segui, D.; et al. Predictive model for atrial fibrillation in hypertensive diabetic patients. Eur. J. Clin. Investig. 2021, e13633. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Lu, D.-Y.; Ventoulis, I.; Greenland, G.V.; Yalcin, H.; Guan, Y.; Marine, J.E.; Olgin, J.E.; Zimmerman, S.L.; Abraham, T.P.; et al. Machine Learning Methods for Identifying Atrial Fibrillation Cases and Their Predictors in Patients With Hypertrophic Cardiomyopathy: The HCM-AF-Risk Model. CJC Open 2021, 3, 801–813. [Google Scholar] [CrossRef]
- Hunt, B.; Kwan, E.; McMillan, M.; Dosdall, D.; MacLeod, R.; Ranjan, R. Deep Learning Based Prediction of Atrial Fibrillation Disease Progression with Endocardial Electrograms in a Canine Model. In Proceedings of the 2020 Computing in Cardiology, Rimini, Italy, 13–16 September 2020. [Google Scholar]
- Dueñas-Pamplona, J.; García, J.G.; Sierra-Pallares, J.; Ferrera, C.; Agujetas, R.; López-Mínguez, J.R. A comprehensive comparison of various patient-specific CFD models of the left atrium for atrial fibrillation patients. Comput. Biol. Med. 2021, 133, 104423. [Google Scholar] [CrossRef]
- Bai, J.; Lu, Y.; Zhang, H. In silico study of the effects of anti-arrhythmic drug treatment on sinoatrial node function for patients with atrial fibrillation. Sci. Rep. 2020, 10, 1–14. [Google Scholar]
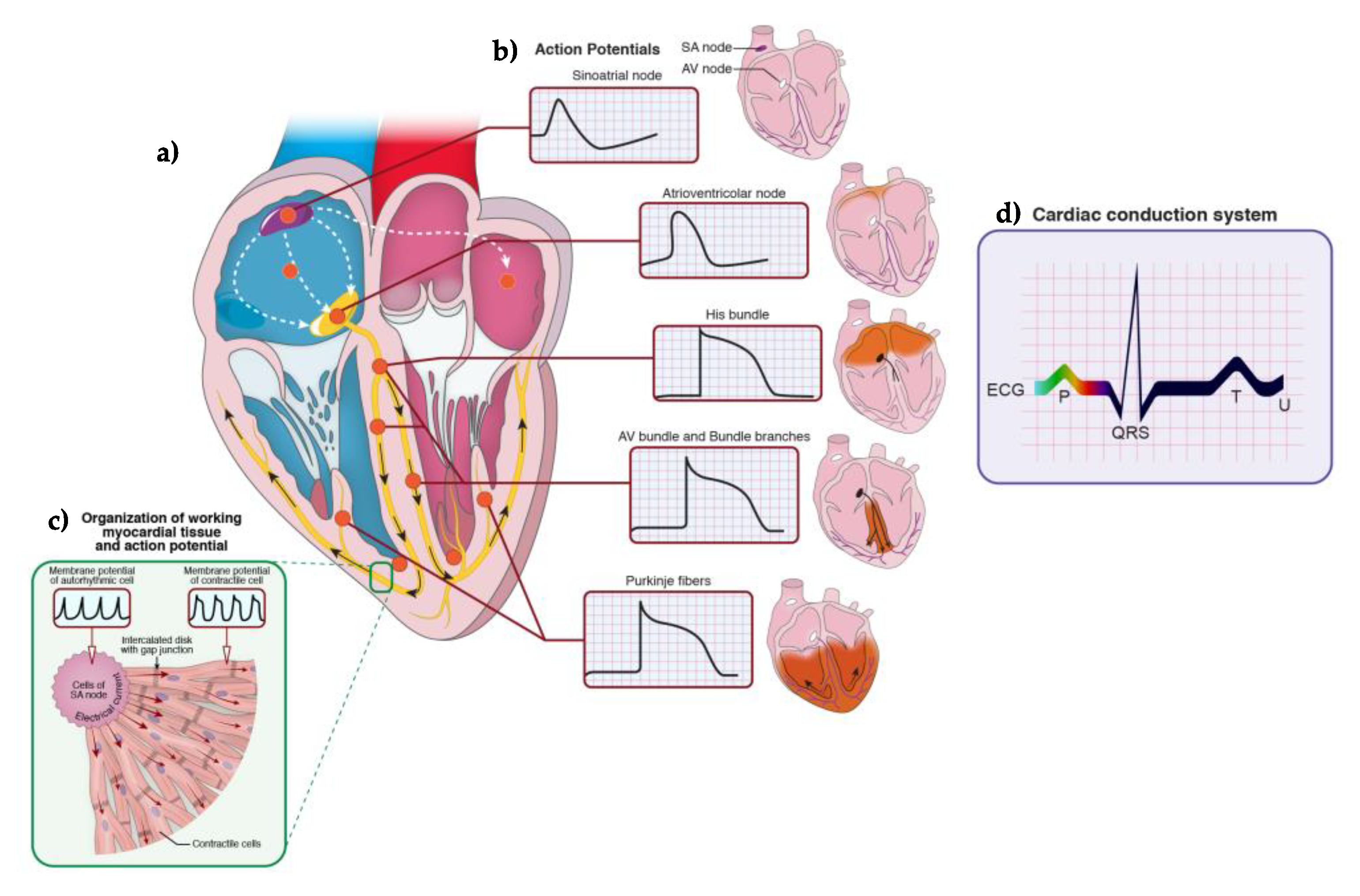
| Disease Causes | Modeling Strategies | Disease Recapitulation Pattern | Novel Mechanistic Insights | Clinical Translation Potential | Ref. |
|---|---|---|---|---|---|
| Genetic mutations | |||||
| SCN5A | In vivo model with Scn5a+/− mice | SAN bradycardia, conduction, and exit block, typical of SSS, are recapitulated. | Role of SAN INa current on atrial activation | [161] | |
| Prolonged RR intervals (ventricular repolarization) | Sex- and age-dependent effects caused by the genetic variant | [162] | |||
| Electrophysiological alterations worsened in tandem with fibrosis | Up-regulation of TGF-beta1 induced colagen synthesis increase | [163] | |||
| In vivo, in vitro and in silico model based on Scn5a+/− rabbit | Characterization of post-natal SAN INa current in vivo; SAN cells displayed in vitro different pacing abilities based on their localization, with a dramatic decrease in the periphery | In silico modeling reconstructed INa current modifications related to cell localization, sex and aging. | [164] | ||
| HCN1 | In vivo and in vitro HCN1−/− mouse model | In vivo: prolonged SAN recovery time, RR interval, high beat-to-beat dispersion, and sinus pauses. In vitro: no alteration in the expression of other ion channels and proteins, but decreased beating rate, abnormal beat-to-beat interval, and delayed impulse formation | No established compensation revealed the HCN1 relevance in the generation of a basal depolarizing current. | HCN1 mutations have never been identified in humans, thus these findings are not completely translatable. | [166] |
| HCN4 | In vitro HCN4mut heterologous system | A rabbit HCN4 mutated gene was transfected in COS-7 cells. The D553N mutated channel displayed a modification in a region connecting two domains. Faster activation and slower deactivation were observed in transfected COS-7 cells when compared to their control. | The mutation did not affect channel voltage-dependence. | This mutation is similar to the one reported for a SSS patient, responsible for the generation of sinus bradycardia, cardiac arrest, polymorphic ventricular tachycardia, and torsade de pointes), thus personalized treatments could be developed targeting the connecting region of the mutated channel. | [167] |
| In vitro HCN4mut heterologous system and in silico modeling | Human HCN4 missense mutations of two young SSS patients were expressed in Xenopus laevis oocytes and COS-7. A complete loss of function was induced in homo- or heteromeric combinations due to a reduced HCN4 membrane expression. Collected data from control and transfected cells were used to develop a computational model. | A new hypothesis was formulated on the possible structural channel alteration interfering with trafficking and voltage sensing. | Personalized pharmacology approaches for SSS patients bearing these HCN4 mutations could be designed on this new mechanistic hypothesis. | [169] | |
| NCX | In vivo modeling through NCX−/− mice | Intracellular Na+ alterations due to Na+/Ca2+ exchanger variant protein were studied and revealed to be typical of SSS phenotype. | These pathological modifications were correlated to alterations in the feedback between Ca2+ handling and membrane potential clock. | [170] | |
| In silico modeling through NCX−/− mice | [171] | ||||
| DSP | In vitro modeling with iPS-cardiomyocytes of a DSPmut patient | Impaired function of Nav1.5 and L-type Ca2+ channels was revealed in these cells bearing a H1684R-inducing mutation. | Further studies with other DSPmut iPS are needed to confirm these alterations of the electrical phenotype, as typical manifestations of SAN dysfunction in the mutation context. | [176] | |
| Aging | |||||
| In silico modeling of rat SAN aging | The action potential of a SAN cell derived from an aging rat was reconstructed computationally through literature studies. | SAN dysfunction was demonstrated to be originated by concomitant, age-dependent alterations in several currents and electrical functions. | [178] | ||
| Inflammation | |||||
| G-protein coupled receptors in the context of catecholamines and angiotensin II storm during cardiac injury | In vitro and in vivo modeling with ES- derived cardiomycytes and knock-in mice | Suppression of binding and inhibition of G-protein coupled receptors with their negative modulators, i.e., regulators of G protein signaling (RGS) was studied in vitro and in vivo in the settings of isoproterenol, adenosine, and muscarinic M2 receptor agonist administration to simulate SAN automaticity during inflammation. | Catecholaminergic stimulation did not have effect, while adenosine and muscarinic M2 receptor agonist induced SAN bradycardia in RGS-insensitive mutant ES-cardiomyocytes and in knocked-in mice. These findings revealed the ability of RGS to control SAN automaticity independently from neurohormonal stimulation. | [191,192,193] | |
| Mitochondrial oxidative stress | In vivo modeling with thioredoxin 2−/− mice | Thioredoxin 2 deletion in whole mouse hearts induced dilated cardiomyopathy, AV block, and sinus bradycardia, following HCN4 downregulation. | HDAC4-MEF2C signaling pathway modifications were reported as the mechanistic link between mitochondrial oxidative stress and reduced HCN4 expression in SAN cells. | [194] | |
| Myocardial ischemia/reperfusion injury | In vivo modeling by experimental clamping of SAN region in rats | The clamping of rat SAN region with hemostatic forceps induced injury in terms of anatomical cell distribution. A reduction in HCN4 and SCN5A expression and Ca2+ handling concurred to generate decreased heart rate and long RR intervals. | The herbal pharmacological compound Zengl Fumai granule was administered by acting on pathways related to myocardial ischemia/reperfusion injury, as TRIM genes. | Zengl Fumai granules were shown to rescue SSS phenotype in vivo. Its action was confirmed in vitro with human AC16 cardiomyocyte line. Further experiments are needed to demonstrate whether it could have a similar effect in the clinical pharmacology treatment of SSS. | [195,196,197] |
| In vitro ischemia/reperfusion injury model with rabbit SAN cells | A remarkable reduction of the If current was observed in rabbit SAN cells after ischemia/reperfusion injury. | The herbal medicine astragaloside increased HCN4 expression and protected cells from stress responses. | Astragaloside ability to suppress SSS signs needs to be verified in other experimental settings and in the clinic. | [198] | |
| In silico multiscale simulation of SAN-atrium dysfunction | Experimental observations on ion current modifications in rabbit SAN cells after ischemia/reperfusion injury were used to develop a computational model. INaCa and IK were impaired in ischemic settings. Additionally, SAN heart rate and atrial conduction velocity were decreased. | Simulated acetylcholine vagal stimulation worsened both ischemic consequences and SAN dysfunction, leading to arrest and exit block. | [199] | ||
| Drug or chemical cardiotoxicity | |||||
| Streptozotocin | In vivo rodent model | Both hyperglycemia and streptozotocin (used to induce diabetes) induced SSS in rats. Streprozotocin provoked systemic inflammation, high ROS levels, cardiac fibroblast apoptosis, decreased HCN4 expression, increased TGF-beta1 expression, fibrosis, macrophage infiltration and reduced baroflex sensitivity. | The complex pathological phenotype of diabetes and SSS could be reversed in vivo by administering IL-10. Further studies are needed to confirm the ability of IL-10 in suppressing SSS signs related to diabetes. | [203] | |
| Angiotensin II | In vivo and/or in vitro cardiac overload | Angiotensin II/Smad pathway activation induced SAN fibrotic lesions. | The transient receptor potential subfamily M member 7 (TRPM7) was found implicated in cardiac fibrosis. | TRPM7 could be investigated as a target to prevent SAN fibrosis in SSS. | [204] |
| Angiotensin II continuous administration in rats provoked typical SSS signs. | In HL-1 atrial cardiomyocytes (used as SAN cells surrogate) in vitro, angiotensin II mediated PKC/NOX-2 signaling upregulation, as well as reduced HCN4 and HDAC4 levels. | Shenxian-Shengmai herbal medicine generated protection from oxidative stress both in vitro and in vitro. The ability of this herbal medicine to contrast SSS signs due to angiotensin II overload needs to be confirmed. | [205] | ||
| Pinpoint press permeation | In vivo models | Pinpoint permeation with 10% sodium hydroxide reduced SAN automaticity less than surgical clamping, but equal, prolonged times of sinoatrial recovery and conduction. | Useful artificial models to reproduce SSS phenotype, although not physiological. | [195] | |
| Local administration of 20% formaldehyde provoked atrial remodeling with inflammation and alteration of extracellular matrix homeostasis. | [206] | ||||
| Disease Causes | Modeling Strategies | Disease Recapitulation Pattern | Novel Mechanistic Insights | Clinical Translation Potential | Ref. |
|---|---|---|---|---|---|
| Genetic mutations | |||||
| SCN5A haploinsufficiency by targeted gene disruption | In vivo, in vitro, and/or ex vivo models based on Scn5a+/− mice | Scn5a+/− mice showed normal QT interval duration in respect to wild-type animals, but conduction block, re-entrant arrhythmias, and ventricular tachycardia were observed. Their ventricular cardiomyocytes display a 50% reduction of INa current without gating properties changes, slowed conduction velocity. | Scn5a−/− mice could survive only in uterus until gestational day E10.5 and manifested ventricular morphogenetic defects. | Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [214] |
| QRS interval prolongation degree was used to classify severity, which was more important in old Scn5a+/− mice observed in vivo and ex vivo. | A correlation between severity and INa current reduction was observed, as associated to normal mRNA levels but decreased protein product. | QRS interval prolongation degree is more effective than aging as severity marker. Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [215] | ||
| In Langendorff ex vivo system, maximum conduction velocity was higher in the endocardial surface than in the epicardial one of Scn5a+/− right ventricles. Fleicanamide and quinidine worsened this conduction block. | Wavelength restitution was found to be a better prognostic marker of action potential duration. More effective prediction could be obtained by alternans magnitude, instead of considering the sole diastolic interval. Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [216] | |||
| Scn5a+/− mice showed a cleared transmural gradient in action potential duration between the right and left ventricles differently from wild-type animals. By increasing pacing, pacing delays were observed only in the right ventricles of mutants. | The physiological anatomical dissimilarities between the right and left ventricles contribute to create a structural, electrophysiological substrate for BrS manifestations if Nav1.5 channels are mutant. Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [217] | |||
| Ex vivo, hearts from old Scn5a+/− mice and cardiac tissue preparations from young animals were stimulated with carbachol and isoprenaline. Longer effective refractory periods were observed for Scn5a+/− cardiac organs and tissues. | These results suggest a loss of the ability to reply to autonomic stimulation in case of Scn5a haploinsufficiency. Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [218] | |||
| In the context of Scn5a haploinsufficiency, a reduction of INa and IK1 was observed, which were due to changes in Nav1.5 and Kir 2.X mutants | Kir2.1 overexpression in Scn5a+/− mice increased IK1 and INa, too, with rescuing effect on BrS pathologic phenotype. Further studies need to be performed to evaluate whether a Kir2.1 overexpression gene therapy could be effective in the treatment of human BrS. Scn5a+/− mice generated by gene targeted disruption have BrS signs similar to humans but do not bear the same genetic mutation. | [220] | |||
| SCN5A point mutations | In vivo and in vitro mouse model based on human 1798insD mutation | Mice bearing 1798insD mutation in homozygosis did not survive. Heterozygous mice showed prolonged PQ, QRS, and QTc intervals. Flecainide exacerbated these alterations, causing sinus bradycardia and/or arrest. In addition, total activation time and effective refractory periods were found different in right and left ventricles. In vitro, cardiomyocytes isolated from these animals displayed longer action potential and reduced upstroke velocity. This model reproduced mixed SSS-BrS phenotype (predominant RV affection, SAN dysfunction, and absent ventricular arrhythmias), already observed in the Dutch family harboring 1798insD mutation. | This mouse model could provide further insights on the altered molecular pathways and identify targets for a personalized pharmacological approach for this cardiopatic patients. | [222] | |
| In vitro heterologous system to express L325R variant-inducing mutation and in silico model | 1:1 overexpression of wild-type and L325R variant-inducing SCN5A mutation in HEK-293 cells evidenced a decrease in Nav1.5 channels and derived current. Computational simulations demonstrated premature repolarization and dome loss in the action potential of cardiomyocytes bearing this protein variant. | The computational model allowed to observe a correlation between INa current reduction and fever, clinically observed in patients bearing this SCN5A mutation. For the first time, the concept of dominant-negative effect was introduced in BrS diagnosis. | [223] | ||
| In vitro heterologous system, cardiomyocyte overexpression and in vivo mouse modeling of R104W, R121W, and/or Y87C mutants | The unique overexpression of R104W variant in HEK-293 and rat neonatal cardiomyocytes led to a complete INa suppression. 1:1 overexpression with wild-type gene induced a less critical INa reduction. Mice bearing this mutation manifested lower heart rate, INa reduction, prolonged RR intervals, longer P-wave. | R104W mutant was not able to reach the cell membrane, due to trafficking impairment. Through these different models, it was possible to confirm the dominant-negative effect of this mutation, related to a defective N-terminal. | [224,225] | ||
| Several N-terminal variants, including R104W and R121W, and the mutated protein Y87C were overexpressed in human kidney TsA-201 and COS-7 cell lines. Mutant proteins showed binding to calmodulin (weak for R121W variant) and wild-type Nav1.5 channels. | These experiments allowed to confirm a dominant-negative effect for both R104W and Y87C variants (not for R121W). Moreover, they identified the existence of a binding site for calmodulin in the N-terminal of Nav1.5 protein. These novel mechanistic insights need to be confirmed in a more physiological model. | [226] | |||
| In vivo modeling through E558X-expressing pigs | This human mutation was overexpressed in Yucatan minipigs. No events of sudden death were reported in two-year follow up, but animals had slow conduction, prolonged P wave, PR, and QRS intervals, sustained atrial-His and His-ventricular conduction periods. SAN recovery time was prolonged and conduction velocity decreased. Nav1.5 protein was decreased confirming electrophysiological observation of reduced INa current. Aging and temperature rise aggravated these signs, however without any difference in between right and left ventricles. | This model did not recapitulate other human BrS signs, as myocyte hypertrophy or fibrosis. | [227] | ||
| In vitro modeling with iPS-cardiomyocytes of a 1795insD SCN5A patient | Reduced INa current, decreased upstroke velocity and prolonged action potential were observed in patient-specific iPS-cardiomyocytes and were worsened by increasing pacing. | These findings were similar to the ones revealed in mice carrying the same mutation and their derived iPS. | The late sodium current inhibitor GS-458967 was able to rescue the BrS phenotype in both mouse and human Brs iPS cells. Electrophysiological immaturity of iPS-cardiomyocytes (absent IK1 current) renders unfeasible to generate phase-1 repolarization and hence to effectively simulate all BrS signs. | [229,230] | |
| In vitro modeling with patient iPS-cardiomyocytes expressing SCN5A variants (R620H/R811H and prematurely truncated channel) | For all variants, the action potential was abnormally prolonged, with enlarged variability of the peak-to-peak interval and slower depolarization. INa current was decreased, due to a reduced number of Nav1.5 channels. Ca2+ handling was altered with reduced transient amplitude and maximal intracellular concentration. | Downregulation of Ca2+ handling paralleled cardiac hypertrophy and beta-adrenergic stimulation. | Targeted genome editing by CRISPR-CAS9 technology could be used to rescue the phenotype by mutation correction. Electrophysiological immaturity of iPS-cardiomyocytes (absent IK1 current) renders unfeasible to generate phase-1 repolarization and hence to effectively simulate all BrS signs. | [231] | |
| In vitro modeling with patient iPS-cardiomyocytes expressing R367H-inducing SCN5A variant and comparison with heterologous system | BrS iPS-cardiomyocytes did not show morphological alterations, but electrophysiological changes, as INa current reduction (30%), increased activation, decreased steady-state deactivation, and accelerated recovery from inactivation. | Heterologous system based on tsa201 cells revealed the causal role in defective functionality of the mutant channels. iPS-cardiomyocytes possess however a stronger potential to mimic BrS. | Electrophysiological immaturity of iPS-cardiomyocytes (absent IK1 current) renders unfeasible to generate phase-1 repolarization and hence to effectively simulate all BrS signs. | [232] | |
| In vitro modeling with patient iPS-cardiomyocytes expressing compound A226V and R1629X-inducing SCN5A variant and comparison with heterologous system | R1629X variant expressed in tsA201 cells induced complete INa abolishment due to trafficking impairment, while A226V variant reduced by half INa current due to halved SCN5A transcript. iPS-cardiomyocytes displaying A226V variant did show current reduction but not so importantly and differ from the controls in terms of recovery rate from inactivation. The missing IK1 current was introduced, so that a dome-like pattern could be visible during phase-1 repolarization. By decreasing pacing and increasing temperature, the electrophysiological pattern was aggravated. | The introduction of IK1 current in iPS-cardiomyocytes allows to phenocopy the typical BrS action potential and thus, represents a more physiological disease model, in which to test possible pharmacological treatments. | [234] | ||
| In vitro modeling with healthy iPS-cardiomyocytes induced to express A735V SCN5A variant through CRISPR-CAS9 and comparison with heterologous system | Although Nav1.5 transcripts were equally expressed with respect to controls, a decreased upstroke velocity and a positively shifted curve of voltage-dependent activation could be appreciated. The use of the heterologous system confirmed the results observed in engineered iPS-cardiomyocytes. | Trafficking impairment was found not to be the reason for observed pattern. | Hybrid modeling demonstrated to be a valid experimental choice to gain more information on the disease. | [236] | |
| Other mutated genes | In vitro modeling with patient iPS-cardiomyocytes expressing compound c.3803G>A and c.3749G<A SCN10A variant | In iPS-cardiomyocytes, SCN5A and SCN10A transcripts resulted to be increasingly expressed. Peak INa current, ICa,L, and INaCa were reduced, as well as the action potential amplitude and maximum depolarization velocity, while delayed afterdepolarizations and ectopic beats became more frequent events. | [237] | ||
| In vitro heterologous MOG1 expression systems | E83D-inducing MOG1 mutation was shown to affect Nav1.5 trafficking in HEK293 and tsA201. | E83D-inducing mutation was demonstrated to compromise the binding of MOG1 protein with Nav1.5 channels. | MOG1 overexpression could be to induce a rescue, even in case of SCN5A mutation | [239,240] | |
| Unknown gene mutations | In vivo modeling with patient iPS-cardiomyocytes without any known genetic mutations | iPS-cardiomyocytes derived from three patients with unknown mutations after genomic screening did not display INa or other current impairment. | These observations led to formulate a new mechanistic hypothesis, for which Nav1.5 channel dysfunction is not a prerequisite for BrS onset. | [241] | |
| Drug cardiotoxicity | |||||
| Terfenadine, pinacidil, and pilsicainide | Ex vivo model of drug cardiotoxicity | Ex vivo perfusion of canine right ventricular tissue with a cocktail of terfenadine, pinacidil, and pilsicainide induced spontaneous phase-2 reentrant extrasystole, polymorphic ventricular tachycardia, and abnormal epicardial repolarization gradient. | [233] | ||
| In vivo model of drug cardiotoxicity | When the cocktail was administered to dogs, ventricular tachycardia manifested predominantly in right ventricles by interesting the epicardium, without alterations in the endocardium. | [242] | |||
| Ajmaline and flecainide | In vitro iPS-cardiomyocyte model of drug cardiotoxicity | Ajmaline and flecainide are used to unmask the ECG BrS signs. When administered to iPS-cardiomyocytes from a healthy donor, they provoked the classical BrS pattern. | [234] | ||
| In silico modeling | |||||
| In silico modeling of BrS human ventricle | INa alterations were reproduced in silico. | A reduced Nav1.5 channel expression was found to impair INa current and induce the loss-of-dome and phase-2 reentry. | [243] | ||
| In silico modeling of ST-elevated segment | The typical ST-elevated segment registered at the precordial leads was reproduced. | This model allowed to confirm the hypothesized relationship between horizontal right ventricular outflow tract and BrS pattern. In silico models can be useful in stratifying the clinical risk. | [245] | ||
| Disease Causes | Modeling Strategies | Disease Recapitulation Pattern | Novel Mechanistic Insights | Clinical Translation Potential | Ref. |
|---|---|---|---|---|---|
| Genetic mutations | |||||
| KCNQ1 | In vitro heterologous systems and/or computational models | S140G-inducing KCNQ1 mutation, discovered in a 4-generation Chinese family with AF, was expressed in COS7 cells. No IKs current and altered gating and kinetic properties were observed. | Co-expression of S140G variant and several KCNE genes (normally not expressed in COS7 cells) revealed a gain of potassium channel function and increased outward current at depolarized potential, thus evidencing the electrical substrate leading to AF. | [260] | |
| The V141M-inducing missense mutation was expressed in Xenopus laevis oocytes. A computational model simulating human heart behavior in the context of V141M variant. | This mutation was found to induce a gain of function. Through the computational model, alterations of the action potential duration in human ventricular cardiomyocytes and sinoatrial node cells could be predicted, by excluding modifications of the resting membrane potential. | [255] | |||
| Compound S140G- and V141M-inducing KCNQ1 mutation, observed in Chinese AF patients, was expressed in CHO cells and adult rabbit left atrial myocytes leading to several electrophysiological signs typical of AF. | The drug HRM-1556 was able to revert AF signs when tested both on CHO cells and rabbit atrial cardiomyocytes. Further tests need to be performed in a more physiological model. | [261] | |||
| NPPA | Ex vivo rat model | A mutant ANP resulting from a two-base-pair deletion observed in AF patients, was circulated in an ex vivo rat heart perfusion model. Treated hearts manifested a significant reduction in action potential duration at 90% and effective refractory period. | The strong implications in the shortening of atrial action potentials in AF patients with this NPPA mutation were revealed. | [266] | |
| Transcription factor genes | In vivo modeling with Tbx5+/del mice | Holt Oram syndrome, related to TBX5 mutation expressed in heterozygosis in European and Asiatic AF patients, was modeled in mice by allelic deletion of Tbx5, by recapitulating typical inflow tract alterations, consequent hemodynamic alterations in A and E waves, and left ventricle diastolic dysfunction. | No investigation on electrical function was performed, thus no novel insights on the relationship between altered hemodynamics and electrophysiological impairment were achieved. | [271] | |
| In vitro heterologous system of TBX5 mutation | G125-inducing TBX5 mutation was expressed in HEK293 cells. The variant bound normally to NKX2.5 but increased to other DNA fragments. | The mutation was revealed to exert a positive effect on the transcription of several genes, as NPPA and GJA. | [272] | ||
| In vivo overexpression of TBX5-modulating miRNA in Zebrafish model | miR-182-5p, found increased in Tbx5+/del mice, was overexpressed in Zebrafish, leading to profound effects in heart morphology and calcium handling, as well as heart arrhythmic behavior. | These findings related to miR-183 family overexpression could open up to new pharmacological treatments targeting miR-182-5p. | [273] | ||
| In vivo and in vitro modeling simulating NKX2.5 mutations | A myocardial infarction-heart failure rat model was developed to reproduce AF in vivo. Nkx2.5 was found to be reduced in its transcription, particularly in the left atrium. In HL-1 cardiomyocytes, RNA interference decreased both NKX2.5 transcripts and protein. | The in vitro model revealed that NKX2.5 downregulation induced increased HCN4 expression, while importantly reduced the expression of proteins involved in Ca2+ handling. | [278] | ||
| In vitro modeling with iPS-cardiomyocytes by knocking out of transcription factor genes | TALEN technology was used to induce knock out of transcription factor genes in iPS and differentiated cardiomyocytes were studied, revealing a fundamental role of TBX5 on extracellular matrix synthesis. | Novel targets of TBX5 were identified: VCAN, FN1, and HSPG2 could be targeted for a pharmacological treatment of Holt Oran syndrome. | [279] | ||
| In vivo modeling with Nkx2.5, Tbx5, and Gata4 haploinsufficient mice | Several combinatorial haploinsufficient mice were generated. | Only the combinatorial haploinsufficiency for Tbx5 and Gata4 was able to rescue AF phenotype by normalizing Ca2+ handling. Further tests should be performed to confirm these findings in a more physiological system. | [280] | ||
| Other genes or unknown genetic causes | In vitro modeling with LMNA-mutated iPS-cardiomyocytes | Peripheral blood of a Chinese AF patient carrying a LMNA mutation was used to generate an iPS line. | This iPS line could be employed in an in vitro model shedding more light on the AF pattern in the context of LMNA mutations. | [281] | |
| In vitro modeling with patient iPS-cardiomyocytes expressing compound E428K- and N470K-inducing SCN5A variant and comparison with heterologous system | These SCN5S mutations were responsible for the AF pattern of an American patient. Patient-specific iPS-cardiomyocytes evidenced heart rate alteration, action potential prolongation, and triggered arrhythmias. Variants studied in HEK293 cells did not reveal INa current modifications, which were instead significant in iPS-cardiomyocytes. | Gene expression analyses and RNA sequencing of iPS-cardiomyocytes revealed dysregulation of several genes and alteration of nitric oxide signaling. | The administration of the nitric oxide blocker N-ethylmaleimide reduced the late INa current. In addition, ranolazine rescue the arrhythmic behavior of AF iPS-cardiomyocytes. | [152] | |
| In vitro modeling with patient iPS-cardiomyocytes with unknown genetic etiology | No unique gene mutation could be identified in three AF patients unresponsive to pharmacological treatment. Derived iPS were differentiated to cardiomyocytes and used to study the AF pattern in vitro. Higher beating rate, longer action potential, and increased ICa,L were observed. Isoproterenol administration provoked delayed after-depolarizations and ectopic beats. | This model could be employed to develop effective pharmacological treatments so far unavailable for these three AF patients. | [282] | ||
| Experimental induction | |||||
| Neurostimulation | In vivo modeling with dogs | A 400 bpm-neurostimulation of dog hearts for 7 days induced AF signs. | The administration of the drug mibefradil rescued the diseased phenotype. | [284] | |
| In vivo modeling with sheep | A 50 Hz pacing of sheep hearts for 8 days induced AF signs. | Statistical analyses performed on lectrophysiological observations led to identify the fibrillation number as a predictor of sustained AF. | [285] | ||
| Myocardial infarction-heart failure- induced AF | In vivo modeling by coronary artery ligation and neurostimulation in rats | Rat hearts were submitted to coronary artery ligation and 200 bpm-neurostimulation, by successfully inducing AF onset. | Dantrolene suppressed triggered arrhythmias. | [286] | |
| In silico modeling | |||||
| In silico modeling of V17M-KCNE3, T895M-KCNH2, and T436M-KCNH2 | Computational simulations revealed that all these variants induce a gain-of-function of the K channels. | This model was able to predict that V17M-KCNE3 variant was responsible for the worse AF scenario, and hence might be relevant for risk stratification. | [290] | ||
| Deep learning from an experimentally paced canine model | Observations collected from a neurostimulated canine heart model were submitted to deep learning. | Based on conduction heterogenicity, action potential characteristics, other rhythm modifications, and concomitant cardiovascular diseases, AF progression can be predicted. | [293] | ||
| Computational model of AF left atrium fluid dynamics | Modifications of fluid dynamics, also induced by surgery, were modeled in silico. | Such a modeling system allows to identify predicting indexes for AF progression and severity, as thrombosis-mediated stasis. | [294] | ||
| Population-based computational simulations | Observations collected from a large cohort of patients with the same transcription factor gene mutation were loaded in a computational model able to reproduce the electrical remodeling and where several drugs, as disopyramide, quinidine, and propafenone, could be assessed to predict their potential efficacy. | This in silico model allowed to establish that only disopyramide could be effective in the treatment of this AF genetic form. Thus, similar modeling approaches have a strong potential for the advancement of personalized medicine treatments. | [295] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iop, L.; Iliceto, S.; Civieri, G.; Tona, F. Inherited and Acquired Rhythm Disturbances in Sick Sinus Syndrome, Brugada Syndrome, and Atrial Fibrillation: Lessons from Preclinical Modeling. Cells 2021, 10, 3175. https://doi.org/10.3390/cells10113175
Iop L, Iliceto S, Civieri G, Tona F. Inherited and Acquired Rhythm Disturbances in Sick Sinus Syndrome, Brugada Syndrome, and Atrial Fibrillation: Lessons from Preclinical Modeling. Cells. 2021; 10(11):3175. https://doi.org/10.3390/cells10113175
Chicago/Turabian StyleIop, Laura, Sabino Iliceto, Giovanni Civieri, and Francesco Tona. 2021. "Inherited and Acquired Rhythm Disturbances in Sick Sinus Syndrome, Brugada Syndrome, and Atrial Fibrillation: Lessons from Preclinical Modeling" Cells 10, no. 11: 3175. https://doi.org/10.3390/cells10113175
APA StyleIop, L., Iliceto, S., Civieri, G., & Tona, F. (2021). Inherited and Acquired Rhythm Disturbances in Sick Sinus Syndrome, Brugada Syndrome, and Atrial Fibrillation: Lessons from Preclinical Modeling. Cells, 10(11), 3175. https://doi.org/10.3390/cells10113175







