Acid Sphingomyelinase Controls Early Phases of Skeletal Muscle Regeneration by Shaping the Macrophage Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Muscle Injury Model
2.2. Primary Cell Isolation and Culture
2.3. Histology and Immunofluorescence
2.4. Whole Body Tension
2.5. Flow Cytometry
2.6. ASMase Activity Assay
2.7. RNA Extraction and RT-qPCR
2.8. Statistical Analysis
3. Results
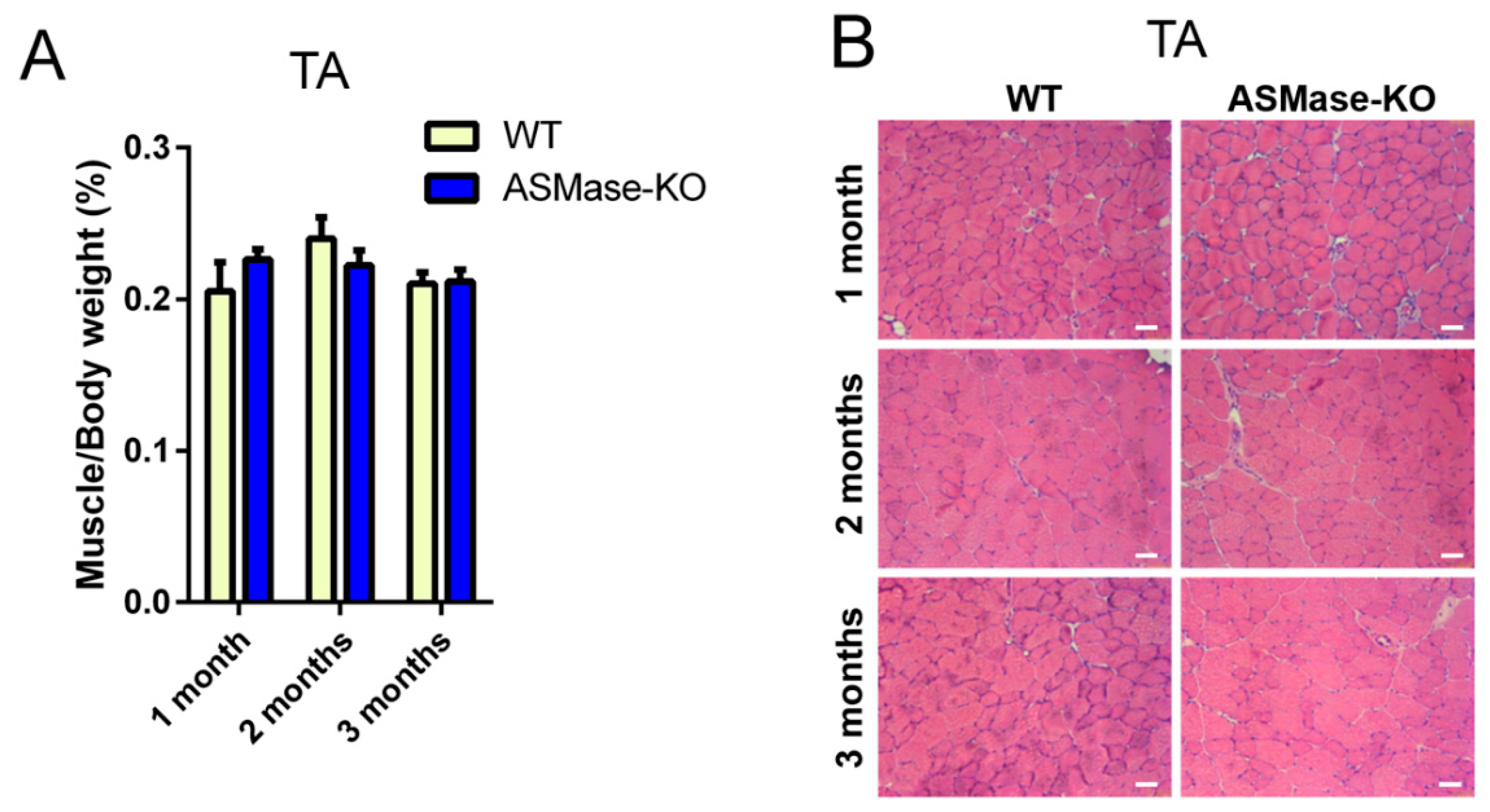
3.1. ASMase Ablation Does Not Affect the Phenotype/Function of Skeletal Muscle and the Content of Satellite Cells
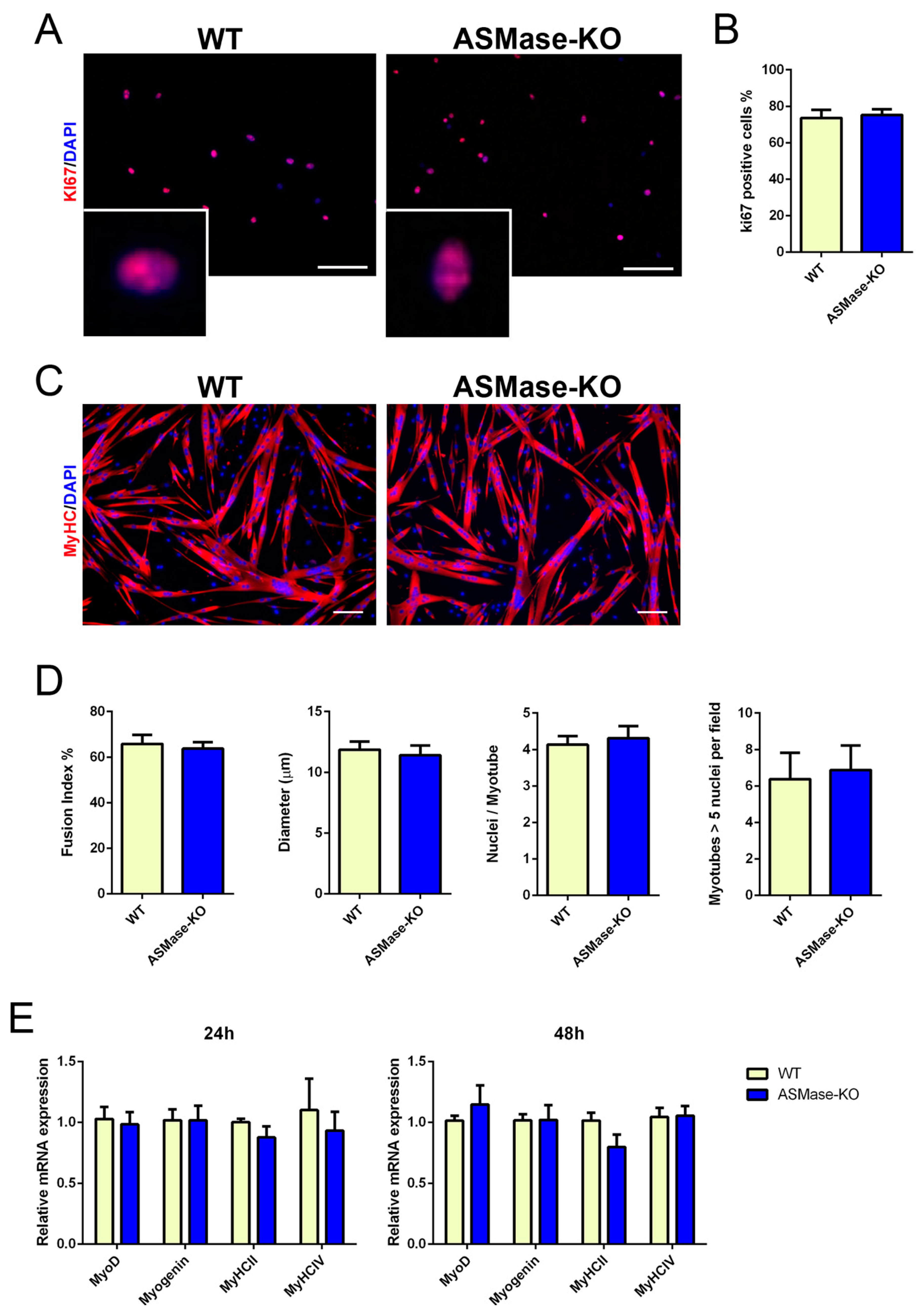
3.2. ASMase Ablation Does Not Affect the Proliferation and Differentiation of Satellite Cells
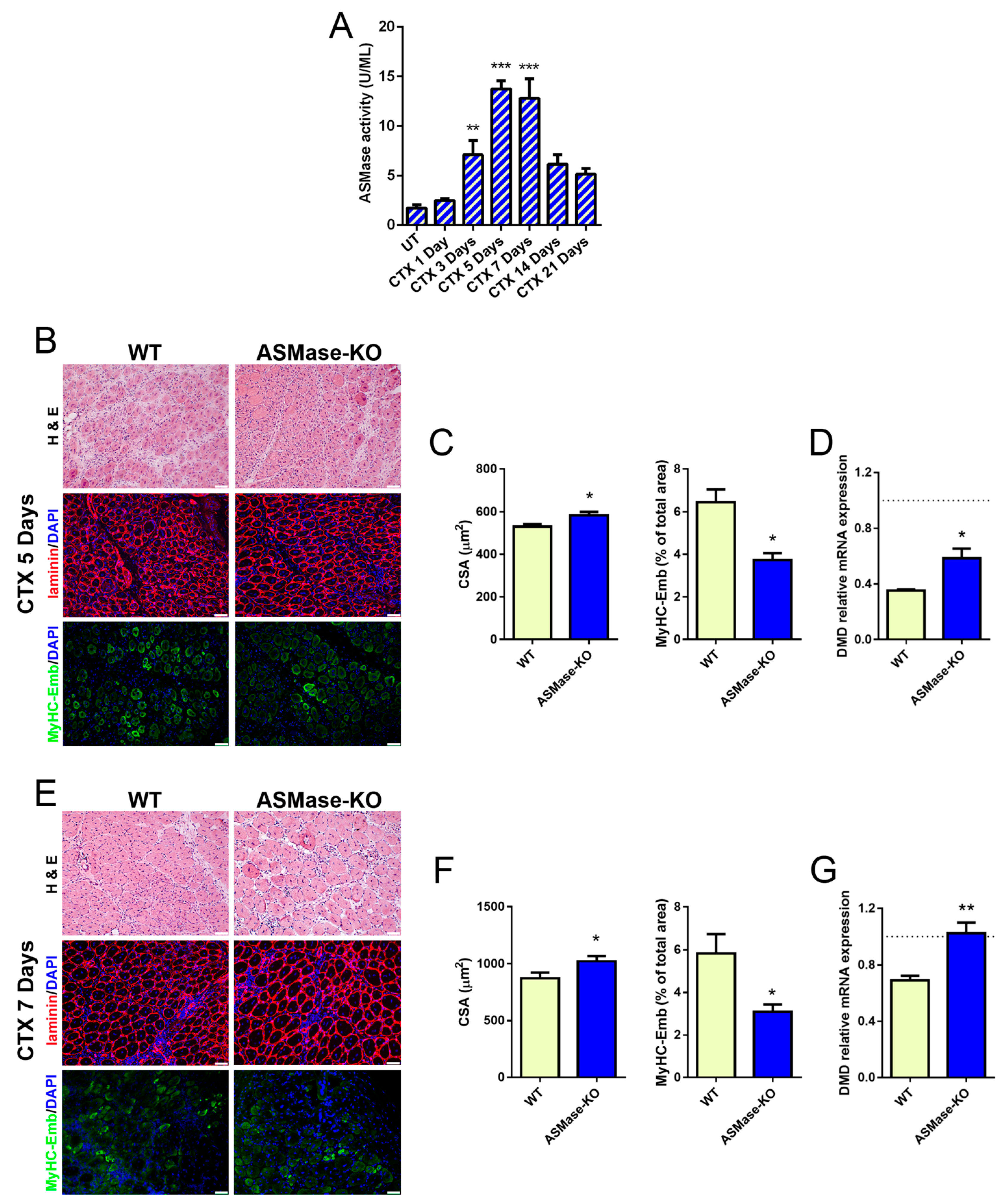
3.3. ASMase Is Activated in CTX-Induced Acute Damage and Its Loss Speeds Early Skeletal Muscle Regeneration
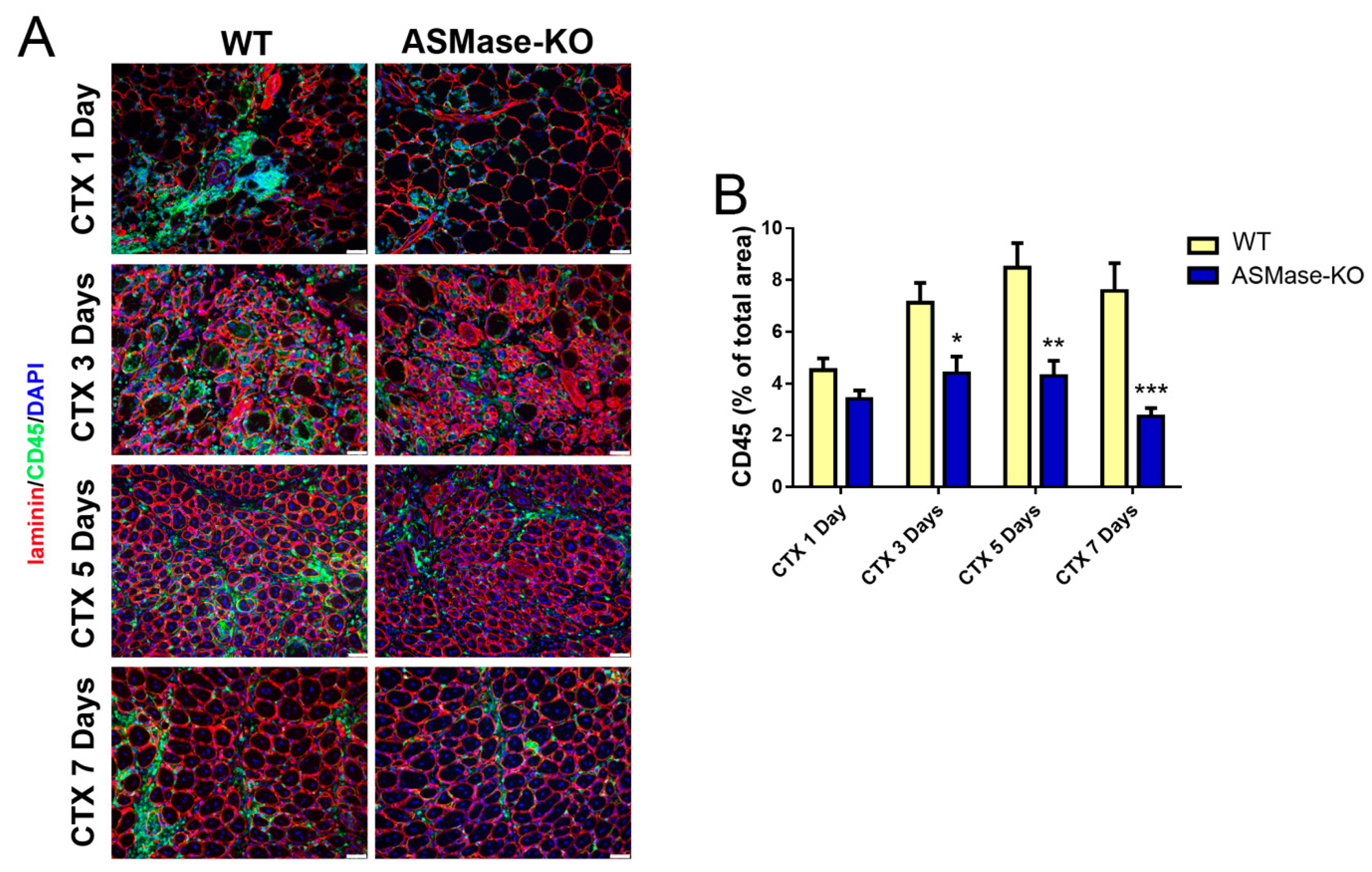
3.4. ASMase Modulates the Generation of Inflammatory Signals at the Injury Site
3.5. In the Absence of ASMase Macrophages Polarization Is Shifted towards an M2 Phenotype
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Palma, C.; Morisi, F.; Pambianco, S.; Assi, E.; Touvier, T.; Russo, S.; Perrotta, C.; Romanello, V.; Carnio, S.; Cappello, V.; et al. Deficient nitric oxide signalling impairs skeletal muscle growth and performance: Involvement of mitochondrial dysregulation. Skelet. Muscle 2014, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Forcina, L.; Cosentino, M.; Musarò, A. Mechanisms Regulating Muscle Regeneration: Insights into the Interrelated and Time-Dependent Phases of Tissue Healing. Cells 2020, 9, 1297. [Google Scholar] [CrossRef]
- Chargé, S.B.P.; Rudnicki, M.A. Cellular and Molecular Regulation of Muscle Regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Schmidt, M.; Schüler, S.C.; Hüttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell. Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef]
- Tidball, J.G. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 2017, 17, 165–178. [Google Scholar] [CrossRef]
- Pillon, N.J.; Bilan, P.J.; Fink, L.N.; Klip, A. Cross-talk between skeletal muscle and immune cells: Muscle-derived mediators and metabolic implications. Am. J. Physiol. Metab. 2013, 304, E453–E465. [Google Scholar] [CrossRef]
- Yang, W.; Hu, P. Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Transl. 2018, 13, 25–32. [Google Scholar] [CrossRef]
- Howard, E.E.; Pasiakos, S.M.; Blesso, C.N.; Fussell, M.A.; Rodriguez, N.R. Divergent Roles of Inflammation in Skeletal Muscle Recovery From Injury. Front. Physiol. 2020, 11, 87. [Google Scholar] [CrossRef]
- Muñoz-Cánoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef]
- Bosurgi, L.; Manfredi, A.A.; Rovere-Querini, P. Macrophages in injured skeletal muscle: A perpetuum mobile causing and limiting fibrosis, prompting or restricting resolution and regeneration. Front. Immunol. 2011, 2, 62. [Google Scholar] [CrossRef]
- Panci, G.; Chazaud, B. Inflammation during post-injury skeletal muscle regeneration. Semin. Cell Dev. Biol. 2021, 119, 32–38. [Google Scholar] [CrossRef]
- Rigamonti, E.; Zordan, P.; Sciorati, C.; Rovere-Querini, P.; Brunelli, S. Macrophage plasticity in skeletal muscle repair. Biomed Res. Int. 2014, 2014, 560629. [Google Scholar] [CrossRef]
- Lavin, Y.; Mortha, A.; Rahman, A.; Merad, M. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015, 15, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Saclier, M.; Cuvellier, S.; Magnan, M.; Mounier, R.; Chazaud, B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013, 280, 4118–4130. [Google Scholar] [CrossRef] [PubMed]
- Chazaud, B.; Sonnet, C.; Lafuste, P.; Bassez, G.; Rimaniol, A.C.; Poron, F.; Authier, F.J.; Dreyfus, P.A.; Gherardi, R.K. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J. Cell Biol. 2003, 163, 1133–1143. [Google Scholar] [CrossRef]
- Saclier, M.; Yacoub-Youssef, H.; Mackey, A.L.; Arnold, L.; Ardjoune, H.; Magnan, M.; Sailhan, F.; Chelly, J.; Pavlath, G.K.; Mounier, R.; et al. Differentially Activated Macrophages Orchestrate Myogenic Precursor Cell Fate During Human Skeletal Muscle Regeneration. Stem Cells 2013, 31, 384–396. [Google Scholar] [CrossRef]
- Juban, G.; Saclier, M.; Yacoub-Youssef, H.; Kernou, A.; Arnold, L.; Boisson, C.; Ben Larbi, S.; Magnan, M.; Cuvellier, S.; Théret, M.; et al. AMPK Activation Regulates LTBP4-Dependent TGF-β1 Secretion by Pro-inflammatory Macrophages and Controls Fibrosis in Duchenne Muscular Dystrophy. Cell Rep. 2018, 25, 2163–2176.e6. [Google Scholar] [CrossRef]
- Lea, S.R.; Metcalfe, H.J.; Plumb, J.; Beerli, C.; Poll, C.; Singh, D.; Abbott-Banner, K.H. Neutral sphingomyelinase-2, acid sphingomyelinase, and ceramide levels in COPD patients compared to controls. Int. J. COPD 2016, 11, 2139–2147. [Google Scholar] [CrossRef]
- Lee, J.K.; Jin, H.K.; Park, M.H.; Kim, B.R.; Lee, P.H.; Nakauchi, H.; Carter, J.E.; He, X.; Schuchman, E.H.; Bae, J.S. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. J. Exp. Med. 2014, 211, 1551–1570. [Google Scholar] [CrossRef]
- Boini, K.M.; Xia, M.; Koka, S.; Gehr, T.W.; Li, P.-L. Instigation of NLRP3 inflammasome activation and glomerular injury in mice on the high fat diet: Role of acid sphingomyelinase gene. Oncotarget 2016, 7, 19031–19044. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-Y.; Hupe, D.C.; Otto, G.P.; Sprenger, M.; Bunck, A.C.; Dorer, M.J.; Bockmeyer, C.L.; Deigner, H.-P.; Gräler, M.H.; Claus, R.A. Acid Sphingomyelinase Promotes Endothelial Stress Response in Systemic Inflammation and Sepsis. Mol. Med. 2016, 22, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yang, M.; Li, X.; Hou, L.; Liu, X.; Xiao, W. Acid Sphingomyelinase and Acid β-Glucosidase 1 Exert Opposite Effects on Interleukin-1β-Induced Interleukin 6 Production in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Inflammation 2021, 44, 1592–1606. [Google Scholar] [CrossRef]
- Assi, E.; Cervia, D.; Bizzozero, L.; Capobianco, A.; Pambianco, S.; Morisi, F.; De Palma, C.; Moscheni, C.; Pellegrino, P.; Clementi, E.; et al. Modulation of Acid Sphingomyelinase in Melanoma Reprogrammes the Tumour Immune Microenvironment. Mediators Inflamm. 2015, 2015, 370482. [Google Scholar] [CrossRef] [PubMed]
- Perrotta, C.; Cervia, D.; Di Renzo, I.; Moscheni, C.; Bassi, M.T.; Campana, L.; Martelli, C.; Catalani, E.; Giovarelli, M.; Zecchini, S.; et al. Nitric oxide generated by tumor-associated macrophages is responsible for cancer resistance to cisplatin and correlated with syntaxin 4 and acid sphingomyelinase inhibition. Front. Immunol. 2018, 9, 1186. [Google Scholar] [CrossRef]
- Assi, E.; Cazzato, D.; De Palma, C.; Perrotta, C.; Clementi, E.; Cervia, D. Sphingolipids and brain resident macrophages in neuroinflammation: An emerging aspect of nervous system pathology. Clin. Dev. Immunol. 2013, 2013, 309302. [Google Scholar] [CrossRef]
- Perrotta, C.; Cervia, D.; De Palma, C.; Assi, E.; Pellegrino, P.; Bassi, M.T.; Clementi, E. The emerging role of Acid Sphingomyelinase in autophagy. Apoptosis 2015, 20, 635–644. [Google Scholar] [CrossRef]
- Cervia, D.; Assi, E.; De Palma, C.; Giovarelli, M.; Bizzozero, L.; Pambianco, S.; Di Renzo, I.; Zecchini, S.; Moscheni, C.; Vantaggiato, C.; et al. Essential role for acid sphingomyelinase-inhibited autophagy in melanoma response to cisplatin. Oncotarget 2016, 7, 24995–25009. [Google Scholar] [CrossRef]
- Perrotta, C.; Clementi, E. Biological roles of acid and neutral sphingomyelinases and their regulation by nitric oxide. Physiology 2010, 25, 64–71. [Google Scholar] [CrossRef]
- Perrotta, C.; Bizzozero, L.; Cazzato, D.; Morlacchi, S.; Assi, E.; Simbari, F.; Zhang, Y.; Gulbins, E.; Bassi, M.T.; Rosa, P.; et al. Syntaxin 4 is required for acid sphingomyelinase activity and apoptotic function. J. Biol. Chem. 2010, 285, 40240–40251. [Google Scholar] [CrossRef] [PubMed]
- Bizzozero, L.; Cazzato, D.; Cervia, D.; Assi, E.; Simbari, F.; Pagni, F.; De Palma, C.; Monno, A.; Verdelli, C.; Querini, P.R.; et al. Acid sphingomyelinase determines melanoma progression and metastatic behaviour via the microphtalmia-associated transcription factor signalling pathway. Cell Death Differ. 2014, 21, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, K.A.; Krupenko, N.I. Ceramide Signaling and p53 Pathways. Adv. Cancer Res. 2018, 140, 191–215. [Google Scholar] [CrossRef]
- Arana, L.; Gangoiti, P.; Ouro, A.; Trueba, M.; Gómez-Muñoz, A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010, 9, 15. [Google Scholar] [CrossRef]
- Barsacchi, R.; Perrotta, C.; Sestili, P.; Cantoni, O.; Moncada, S.; Clementi, E. Cyclic GMP-dependent inhibition of acid sphingomyelinase by nitric oxide: An early step in protection against apoptosis. Cell Death Differ. 2002, 9, 1248–1255. [Google Scholar] [CrossRef]
- Falcone, S.; Perrotta, C.; De Palma, C.; Pisconti, A.; Sciorati, C.; Capobianco, A.; Rovere-Querini, P.; Manfredi, A.A.; Clementi, E. Activation of acid sphingomyelinase and its inhibition by the nitric oxide/cyclic guanosine 3′,5′-monophosphate pathway: Key events in Escherichia coli-elicited apoptosis of dendritic cells. J. Immunol. 2004, 173, 4452–4463. [Google Scholar] [CrossRef]
- Chung, H.Y.; Wickel, J.; Stuhlmüller, B.; Kinne, R.W.; Claus, R.A. Acid sphingomyelinase activity is elevated in the serum of rheumatoid arthritis patients, suppressed by anti-TNF-α treatment. Clin. Exp. Rheumatol. 2019, 37, 18–19. [Google Scholar]
- Hofmeister, R.; Wiegmann, K.; Korherr, C.; Bernardo, K.; Krönke, M.; Falk, W. Activation of Acid Sphingomyelinase by Interleukin-1 (IL-1) Requires the IL-1 Receptor Accessory Protein. J. Biol. Chem. 1997, 272, 27730–27736. [Google Scholar] [CrossRef]
- Bauer, J.; Huy, C.; Brenmoehl, J.; Obermeier, F.; Bock, J. Matrix metalloproteinase-1 expression induced by IL-1β requires acid sphingomyelinase. FEBS Lett. 2009, 583, 915–920. [Google Scholar] [CrossRef]
- Becker, K.A.; Riethmüller, J.; Lüth, A.; Döring, G.; Kleuser, B.; Gulbins, E. Acid Sphingomyelinase Inhibitors Normalize Pulmonary Ceramide and Inflammation in Cystic Fibrosis. Am. J. Respir. Cell Mol. Biol. 2012, 42, 716–724. [Google Scholar] [CrossRef]
- Perry, D.M.; Newcomb, B.; Adada, M.; Wu, B.X.; Roddy, P.; Kitatani, K.; Siskind, L.; Obeid, L.M.; Hannun, Y.A. Defining a role for acid sphingomyelinase in the p38/interleukin-6 pathway. J. Biol. Chem. 2014, 289, 22401–22412. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, X.; Lu, Z.; Perry, D.M.; Li, Y.; Russo, S.B.; Cowart, L.A.; Hannun, Y.A.; Huang, Y. Acid sphingomyelinase plays a key role in palmitic acid-amplified inflammatory signaling triggered by lipopolysaccharide at low concentrations in macrophages. Am. J. Physiol. Metab. 2013, 305, E853–E867. [Google Scholar] [CrossRef]
- Li, C.; Guo, S.; Pang, W.; Zhao, Z. Crosstalk between Acid Sphingomyelinase and Inflammasome Signaling and Their Emerging Roles in Tissue Injury and Fibrosis. Front. Cell Dev. Biol. 2020, 7, 378. [Google Scholar] [CrossRef]
- Rigamonti, E.; Touvier, T.; Clementi, E.; Manfredi, A.A.; Brunelli, S.; Rovere-Querini, P. Requirement of Inducible Nitric Oxide Synthase for Skeletal Muscle Regeneration after Acute Damage. J. Immunol. 2013, 190, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Touvier, T.; De Palma, C.; Rigamonti, E.; Scagliola, A.; Incerti, E.; Mazelin, L.; Thomas, J.-L.; D’Antonio, M.; Politi, L.; Schaeffer, L.; et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015, 6, e1663. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, K.; Erlich, S.; Perl, D.P.; Ferlinz, K.; Bisgaier, C.L.; Sandhoff, K.; Desnick, R.J.; Stewart, C.L.; Schuchman, E.H. Acid sphingomyelinase deficient mice: A model of types A and B Niemann–Pick disease. Nat. Genet. 1995, 10, 288–293. [Google Scholar] [CrossRef]
- Zecchini, S.; Giovarelli, M.; Perrotta, C.; Morisi, F.; Touvier, T.; Di Renzo, I.; Moscheni, C.; Bassi, M.T.; Cervia, D.; Sandri, M.; et al. Autophagy controls neonatal myogenesis by regulating the GH-IGF1 system through a NFE2L2- and DDIT3-mediated mechanism. Autophagy 2019, 15, 58–77. [Google Scholar] [CrossRef]
- Perrotta, C.; Buldorini, M.; Assi, E.; Cazzato, D.; De Palma, C.; Clementi, E.; Cervia, D. The thyroid hormone triiodothyronine controls macrophage maturation and functions: Protective role during inflammation. Am. J. Pathol. 2014, 184, 230–247. [Google Scholar] [CrossRef] [PubMed]
- Sciorati, C.; Touvier, T.; Buono, R.; Pessina, P.; François, S.; Perrotta, C.; Meneveri, R.; Clementi, E.; Brunelli, S. Necdin is expressed in cachectic skeletal muscle to protect fibers from tumor-induced wasting. J. Cell Sci. 2009, 122, 1119–1125. [Google Scholar] [CrossRef][Green Version]
- Amato, R.; Catalani, E.; Dal Monte, M.; Cammalleri, M.; Di Renzo, I.; Perrotta, C.; Cervia, D.; Casini, G. Autophagy-mediated neuroprotection induced by octreotide in an ex vivo model of early diabetic retinopathy. Pharmacol. Res. 2018, 128, 167–178. [Google Scholar] [CrossRef]
- Catalani, E.; Proietti Serafini, F.; Zecchini, S.; Picchietti, S.; Fausto, A.M.; Marcantoni, E.; Buonanno, F.; Ortenzi, C.; Perrotta, C.; Cervia, D. Natural products from aquatic eukaryotic microorganisms for cancer therapy: Perspectives on anti-tumour properties of ciliate bioactive molecules. Pharmacol. Res. 2016, 113, 409–420. [Google Scholar] [CrossRef]
- Guardiola, O.; Lafuste, P.; Brunelli, S.; Iaconis, S.; Touvier, T.; Mourikis, P.; De Bock, K.; Lonardo, E.; Andolfi, G.; Bouché, A.; et al. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc. Natl. Acad. Sci. USA 2012, 109, E3231–E3240. [Google Scholar] [CrossRef]
- Cazzato, D.; Assi, E.; Moscheni, C.; Brunelli, S.; De Palma, C.; Cervia, D.; Perrotta, C.; Clementi, E. Nitric oxide drives embryonic myogenesis in chicken through the upregulation of myogenic differentiation factors. Exp. Cell Res. 2014, 320, 269–280. [Google Scholar] [CrossRef]
- Giovarelli, M.; Zecchini, S.; Catarinella, G.; Moscheni, C.; Sartori, P.; Barbieri, C.; Roux-Biejat, P.; Napoli, A.; Vantaggiato, C.; Cervia, D.; et al. Givinostat as metabolic enhancer reverting mitochondrial biogenesis deficit in Duchenne Muscular Dystrophy. Pharmacol. Res. 2021, 170, 105751. [Google Scholar] [CrossRef]
- Pasut, A.; Jones, A.E.; Rudnicki, M.A. Isolation and Culture of Individual Myofibers and their Satellite Cells from Adult Skeletal Muscle. J. Vis. Exp. 2013, 73, e50074. [Google Scholar] [CrossRef]
- Lauria, S.; Perrotta, C.; Casati, S.; Di Renzo, I.; Ottria, R.; Eberini, I.; Palazzolo, L.; Parravicini, C.; Ciuffreda, P. Design, synthesis, molecular modelling and in vitro cytotoxicity analysis of novel carbamate derivatives as inhibitors of Monoacylglycerol lipase. Bioorganic Med. Chem. 2018, 26, 2561–2572. [Google Scholar] [CrossRef]
- Perrotta, C.; Buonanno, F.; Zecchini, S.; Giavazzi, A.; Proietti Serafini, F.; Catalani, E.; Guerra, L.; Belardinelli, M.C.; Picchietti, S.; Fausto, A.M.; et al. Climacostol reduces tumour progression in a mouse model of melanoma via the p53-dependent intrinsic apoptotic programme. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Zecchini, S.; Proietti Serafini, F.; Catalani, E.; Giovarelli, M.; Coazzoli, M.; Di Renzo, I.; De Palma, C.; Perrotta, C.; Clementi, E.; Buonanno, F.; et al. Dysfunctional autophagy induced by the pro-apoptotic natural compound climacostol in tumour cells. Cell Death Dis. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Yoshimoto, Y.; Ikemoto-Uezumi, M.; Hitachi, K.; Fukada, S.; Uezumi, A. Methods for Accurate Assessment of Myofiber Maturity During Skeletal Muscle Regeneration. Front. Cell Dev. Biol. 2020, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, L.; Giacinti, C.; Nardis, C.; Borsellino, G.; Rizzuto, E.; Nicoletti, C.; Wannenes, F.; Battistini, L.; Rosenthal, N.; Molinaro, M.; et al. Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 2007, 21, 1393–1402. [Google Scholar] [CrossRef]
- Tonkin, J.; Temmerman, L.; Sampson, R.D.; Gallego-Colon, E.; Barberi, L.; Bilbao, D.; Schneider, M.D.; Musarò, A.; Rosenthal, N. Monocyte/Macrophage-derived IGF-1 Orchestrates Murine Skeletal Muscle Regeneration and Modulates Autocrine Polarization. Mol. Ther. 2015, 23, 1189–1200. [Google Scholar] [CrossRef]
- Audousset, C.; McGovern, T.; Martin, J.G. Role of Nrf2 in Disease: Novel Molecular Mechanisms and Therapeutic Approaches—Pulmonary Disease/Asthma. Front. Physiol. 2021, 12, 1637. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-A.; Kwak, M.-K. The Nrf2 System as a Potential Target for the Development of Indirect Antioxidants. Molecules 2010, 15, 7266. [Google Scholar] [CrossRef]
- Petrillo, S.; Pelosi, L.; Piemonte, F.; Travaglini, L.; Forcina, L.; Catteruccia, M.; Petrini, S.; Verardo, M.; D’Amico, A.; Musaró, A.; et al. Oxidative stress in Duchenne muscular dystrophy: Focus on the NRF2 redox pathway. Hum. Mol. Genet. 2017, 26, 2781–2790. [Google Scholar] [CrossRef]
- Ma, Q. Role of Nrf2 in Oxidative Stress and Toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401. [Google Scholar] [CrossRef]
- Laumonier, T.; Menetrey, J. Muscle injuries and strategies for improving their repair. J. Exp. Orthop. 2016, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Donati, C.; Meacci, E.; Nuti, F.; Becciolini, L.; Farnararo, M.; Bruni, P. Sphingosine 1-phosphate regulates myogenic differentiation: A major role for S1P 2 receptor. FASEB J. 2004, 19, 1–22. [Google Scholar] [CrossRef]
- Fortier, M.; Figeac, N.; White, R.B.; Knopp, P.; Zammit, P.S. Sphingosine-1-phosphate receptor 3 influences cell cycle progression in muscle satellite cells. Dev. Biol. 2013, 382, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Mebarek, S.; Komati, H.; Naro, F.; Zeiller, C.; Alvisi, M.; Lagarde, M.; Prigent, A.F.; Némoz, G. Inhibition of de novo ceramide synthesis upregulates phospholipase D and enhances myogenic differentation. J. Cell Sci. 2007, 120, 407–416. [Google Scholar] [CrossRef]
- Henry, B.; Ziobro, R.; Becker, K.A.; Kolesnick, R.; Gulbins, E. Acid sphingomyelinase. Handb. Exp. Pharmacol. 2013, 215, 77–88. [Google Scholar] [CrossRef]
- Shela, S.B.; Narasimhan, M.; Shanmugam, G.; Litovsky, S.H.; Gounder, S.S.; Karan, G.; Arulvasu, C.; Kensler, T.W.; Hoidal, J.R.; Darley-Usmar, V.M.; et al. Disruption of nuclear factor (erythroid-derived-2)-like 2 antioxidant signaling: A mechanism for impaired activation of stem cells and delayed regeneration of skeletal muscle. FASEB J. 2016, 30, 1865–1879. [Google Scholar] [CrossRef]
- Bronisz-Budzyńska, I.; Kozakowska, M.; Podkalicka, P.; Kachamakova-Trojanowska, N.; Łoboda, A.; Dulak, J. The role of Nrf2 in acute and chronic muscle injury. Skelet. Muscle 2020, 10, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, Y.; Inaba, S.; Zhang, L.; Tsujikawa, K.; Uezumi, A.; Fukada, S. Implication of basal lamina dependency in survival of Nrf2-null muscle stem cells via an antioxidative-independent mechanism. J. Cell. Physiol. 2019, 234, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Strle, K.; Broussard, S.R.; Mccusker, R.H.; Shen, W.-H.; Johnson, R.W.; Freund, G.G.; Dantzer, R.; Kelley, K.W. Proinflammatory Cytokine Impairment of Insulin-Like Growth Factor I-Induced Protein Synthesis in Skeletal Muscle Myoblasts Requires Ceramide. Endocrinology 2004, 145, 4592–4602. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. ARTICLE Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Mounier, R.; Théret, M.; Arnold, L.; Cuvellier, S.; Bultot, L.; Göransson, O.; Sanz, N.; Ferry, A.; Sakamoto, K.; Foretz, M.; et al. AMPKα1 Regulates Macrophage Skewing at the Time of Resolution of Inflammation during Skeletal Muscle Regeneration. Cell Metab. 2013, 18, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, E.; Sousa-Victor, P.; Ruiz-Bonilla, V.; Jardí, M.; Caelles, C.; Serrano, A.L.; Muñoz-Cánoves, P. p38/MKP-1–regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J. Cell Biol. 2011, 195, 307–322. [Google Scholar] [CrossRef]
- Saclier, M.; Lapi, M.; Bonfanti, C.; Rossi, G.; Antonini, S.; Messina, G. The Transcription Factor Nfix Requires RhoA-ROCK1 Dependent Phagocytosis to Mediate Macrophage Skewing during Skeletal Muscle Regeneration. Cells 2020, 9, 708. [Google Scholar] [CrossRef]
- Ruffell, D.; Mourkioti, F.; Gambardella, A.; Kirstetter, P.; Lopez, R.G.; Rosenthal, N.; Nerlov, C. A CREB-C/EBPβ cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar] [CrossRef]
- Peterson, J.M.; Bakkar, N.; Guttridge, D.C. NF-κB Signaling in Skeletal Muscle Health and Disease. Curr. Top. Dev. Biol. 2011, 96, 85–119. [Google Scholar]
- Chen, S.E.; Gerken, E.; Zhang, Y.; Zhan, M.; Mohan, R.K.; Li, A.S.; Reid, M.B.; Li, Y.P. Role of TNF-α signaling in regeneration of cardiotoxin-injured muscle. Am. J. Physiol.—Cell Physiol. 2005, 289, 1179–1187. [Google Scholar] [CrossRef]
- Warren, G.L.; Hulderman, T.; Jensen, N.; McKinstry, M.; Mishra, M.; Luster, M.I.; Simeonova, P.P. Physiological role of tumor necrosis factor α in traumatic muscle injury. FASEB J. 2002, 16, 1630–1632. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Hershko, D.D.; Robb, B.W.; Wray, C.J.; Hasselgren, P.-O. IL-1β stimulates IL-6 production in cultured skeletal muscle cells through activation of MAP kinase signaling pathway and NF-κB. Am. J. Physiol. Integr. Comp. Physiol. 2003, 284, R1249–R1254. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, N.; Sharma, D.; Gulbins, E.; Becker, K.A.; Edelmann, B. Inhibition of acid sphingomyelinase by tricyclic antidepressants and analogons. Front. Physiol. 2014, 5, 331. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.T.; Beckmann, N.; Winer, L.K.; Pugh, A.M.; Pritts, T.A.; Nomellini, V.; Gulbins, E.; Caldwell, C.C. Amitriptyline reduces inflammation and mortality in a murine model of sepsis. Cell. Physiol. Biochem. 2019, 52, 565–579. [Google Scholar] [CrossRef] [PubMed]



| Name | Primers Sequences |
|---|---|
| CD80 | F: 5′-AGTTTCTCTTTTTCAGGTTGTGAA-3′ R: 5′-CACCCGGCAGATGCTAAAGA-3′ |
| CD163 | F: 5′-CTCCTGTGGACTCTGAAGCG-3′ R: 5′-CTCTGAATGACCCCCGAGGA-3′ |
| CD206 | F: 5′-ATGGATTGCCCTGAACAGCA-3′ R: 5′-TGTACCGCACCCTCCATCTA-3′ |
| IL1β | F: 5′-CCCTGCAGCTGGAGAGTGTGGA-3′ R: 5′-TGTGCTCTGCTTGTGAGGTGCTG-3′ |
| NOS2 | F: 5′-GTTCTCAGCCCAACAATACAAGA-3′ R: 5′-GTGGACGGGTCGATGTCAC-3′ |
| F4/80 | F: 5′-TGACTCACCTTGTGGTCCTAA-3′ R: 5′-CTTCCCAGAATCCAGTCTTTCC-3′ |
| MyoD | F: 5-CTGGCGCCGCTGCCTTCTAC-3′ R: 5′-GGCCGCTGTAATCCATCATGCCA-3′ |
| Myogenin | F: 5′-GACCCTACAGACGCCCACAATC-3′ R: 5′-ACACCCAGCCTGACAGACAATC-3′ |
| MyHCII | F: 5′-AAGCGAAGAGTAAG CTGTC-3′ R: 5′-GTGATTGCTTGCAAAGGAAC-3′ |
| MyHCIV | F: 5′-ACAAGCTGCGGGTGAAGAGC-3′ R: 5′-CAGGACAGTGACAAAGAACG-3′ |
| DMD | F: 5′-GGAAGAAGTAGAGGACTGTTATG-3′ R: 5′-AGGTCTAGGAGGCGTTTTCC-3′ |
| Myoz1 | F: 5′-GGAACTTGGCATTGACCTACTG-3′ R: 5′-AAACTTGGGCATCTGGAAGG-3′ |
| TNF-α | F: 5′-CCCACGTCGTAGCAAACCACC-3′ R: 5′-TCGGGGCAGCCTTGTCCCTT-3′ |
| Nrf2 | F: 5′-CATTCCCGAATTACAGTGTC-3′ R: 5′-GGAGATCGATGAGTAAAAATGG-3′ |
| HO-1 | F: 5′-CATGAAGAACTTTCAGAAGGG-3′ R: 5′-TAGATATGGTACAAGGAAGCC-3′ |
| GPX1 | F: 5′-GGAGAATGGCAAGAATGAAG-3′ R: 5′-TTCGCACTTCTCAAACAATG-3′ |
| SOD3 | F: 5′-AGAGAGAGTATTTGGGAACC-3′ R: 5′-AAACTAAGCTGCAAAGTCTC-3′ |
| RPL32 | F: 5′-TTAAGCGAAACTGGCGGAAAC-3′ R: 5′-TTGTTGCTCCCATAACCGATG-3′ |
| RPL38 | F: 5′-GAAGGATGCCAAGTCTGTCAA-3′ R: 5′-GAGGGCTGGTTCATTTCAGA-3′ |
| GAPDH | F: 5′-ACCACAGTCCATGCCATCAC-3′ R:5′- TCCACCACCCTGTTGCTGTA-3′ |
| 36B4 | F: 5′-AGGATATGGGATTCGGTCTCTTC-3′ R: 5′-TCATCCTGCTTAAGTGAACAAACT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roux-Biejat, P.; Coazzoli, M.; Marrazzo, P.; Zecchini, S.; Di Renzo, I.; Prata, C.; Napoli, A.; Moscheni, C.; Giovarelli, M.; Barbalace, M.C.; et al. Acid Sphingomyelinase Controls Early Phases of Skeletal Muscle Regeneration by Shaping the Macrophage Phenotype. Cells 2021, 10, 3028. https://doi.org/10.3390/cells10113028
Roux-Biejat P, Coazzoli M, Marrazzo P, Zecchini S, Di Renzo I, Prata C, Napoli A, Moscheni C, Giovarelli M, Barbalace MC, et al. Acid Sphingomyelinase Controls Early Phases of Skeletal Muscle Regeneration by Shaping the Macrophage Phenotype. Cells. 2021; 10(11):3028. https://doi.org/10.3390/cells10113028
Chicago/Turabian StyleRoux-Biejat, Paulina, Marco Coazzoli, Pasquale Marrazzo, Silvia Zecchini, Ilaria Di Renzo, Cecilia Prata, Alessandra Napoli, Claudia Moscheni, Matteo Giovarelli, Maria Cristina Barbalace, and et al. 2021. "Acid Sphingomyelinase Controls Early Phases of Skeletal Muscle Regeneration by Shaping the Macrophage Phenotype" Cells 10, no. 11: 3028. https://doi.org/10.3390/cells10113028
APA StyleRoux-Biejat, P., Coazzoli, M., Marrazzo, P., Zecchini, S., Di Renzo, I., Prata, C., Napoli, A., Moscheni, C., Giovarelli, M., Barbalace, M. C., Catalani, E., Bassi, M. T., De Palma, C., Cervia, D., Malaguti, M., Hrelia, S., Clementi, E., & Perrotta, C. (2021). Acid Sphingomyelinase Controls Early Phases of Skeletal Muscle Regeneration by Shaping the Macrophage Phenotype. Cells, 10(11), 3028. https://doi.org/10.3390/cells10113028














