Identification of a Novel Mutation Exacerbated the PSI Photoinhibition in pgr5/pgrl1 Mutants; Caution for Overestimation of the Phenotypes in Arabidopsis pgr5-1 Mutant
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Transformation
2.3. Determination of Phenotypic Recovery in Complementation Lines
2.4. Simultaneous Measurement of Gas Exchange, Chlorophyll Fluorescence, and Absorbance Change Due to P700 Oxidation
2.5. Biochemical Analyses
2.6. Immunoblotting
2.7. Photoinhibition Experiment
2.8. Whole-genome Sequences Analysis
3. Results
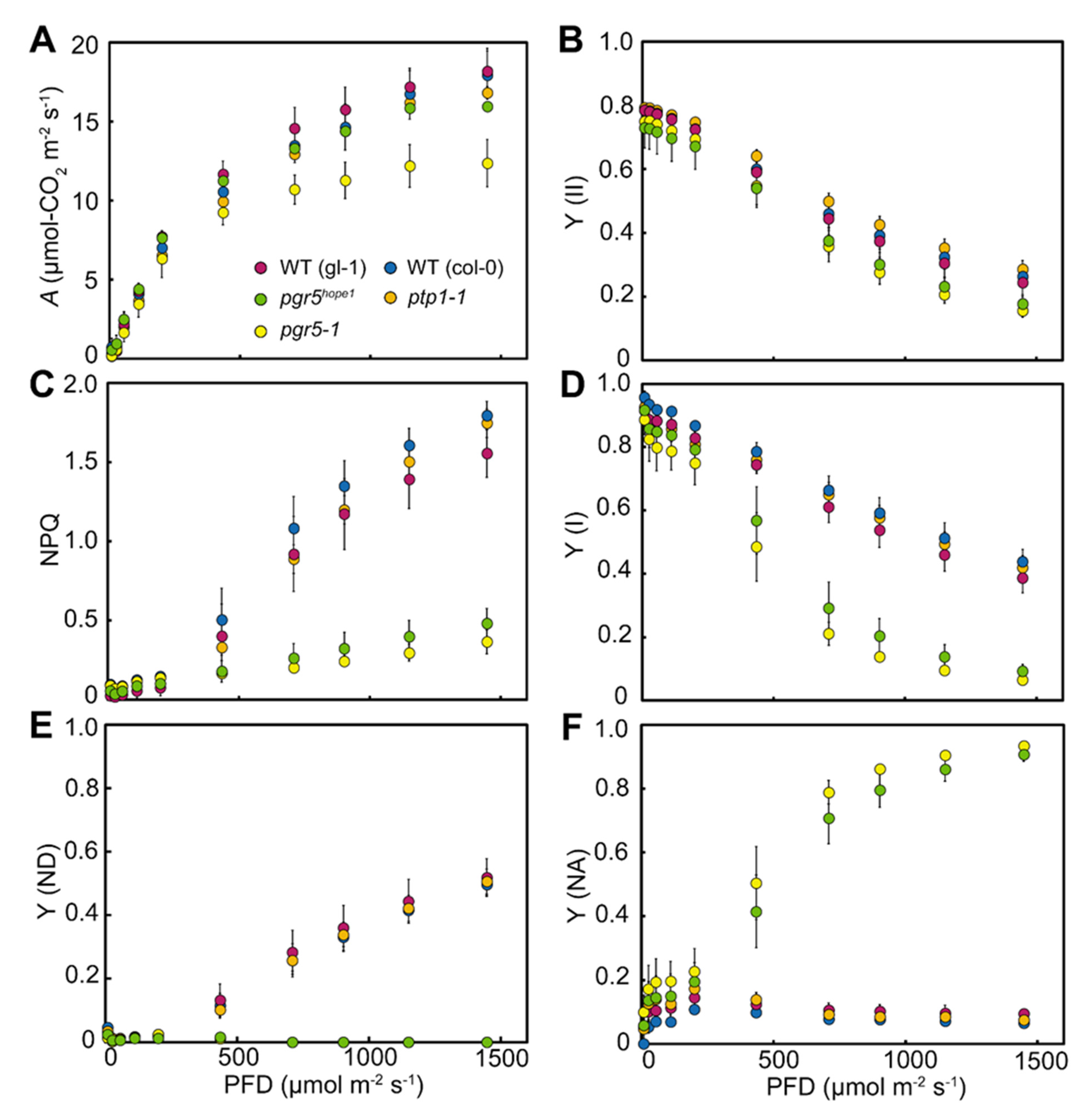
3.1. Identification of the PSI Photoprotection 1, PTP1; a Recessive Mutation in AT2G17240 Gene Enhanced the Growth Phenotype of pgr5-1, a Well-known PGR5 Deficient Mutant
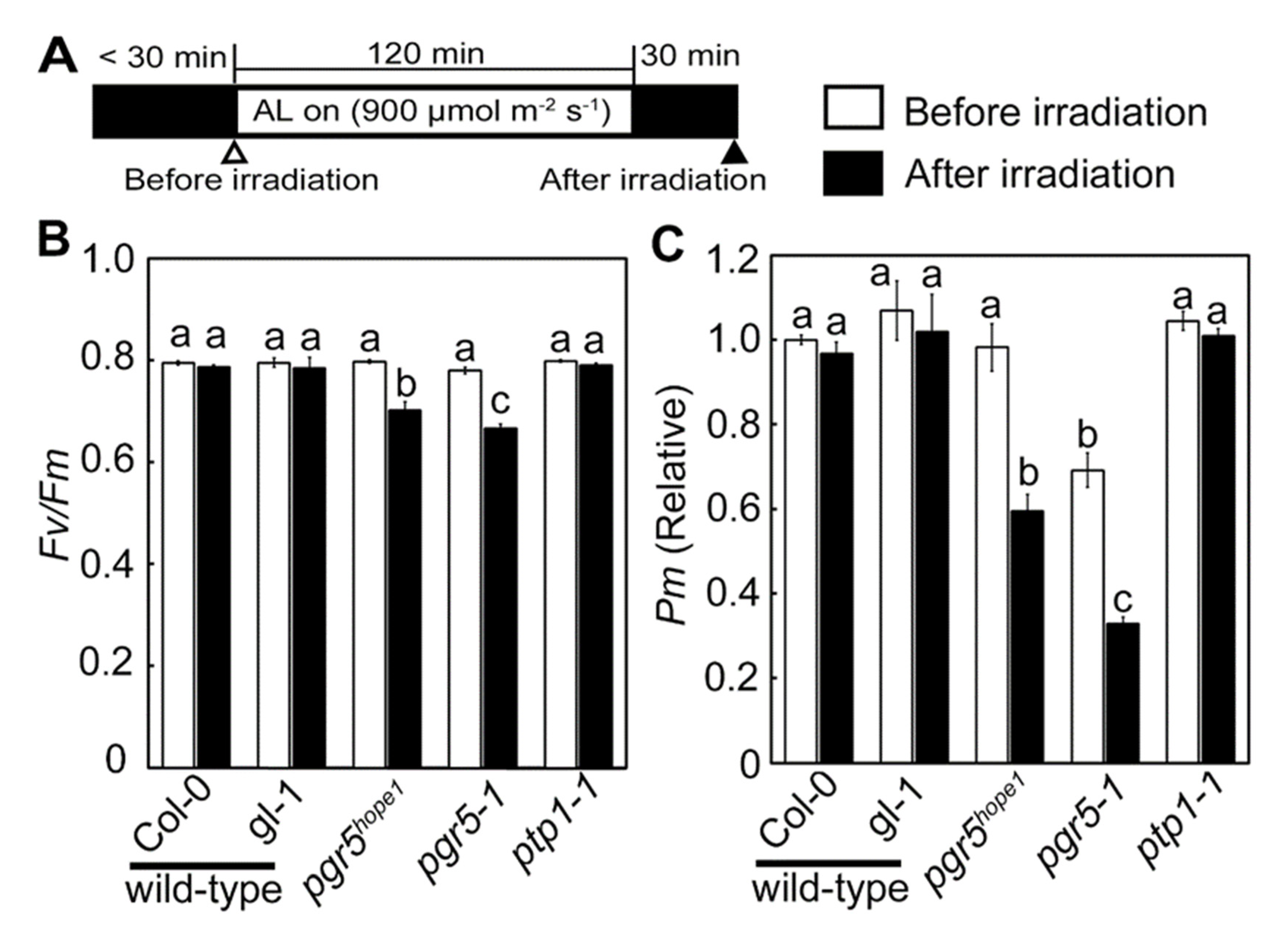
3.2. The Biochemical and Physiological Damages on Photosynthetic Apparatus Caused by the ptp1 Mutation
3.3. PSI Photoinhibition Was Triggered by PGR5/PGRL1 Deficiency
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Miyake, C. Molecular Mechanism of Oxidation of P700 and Suppression of ROS Production in Photosystem I in Response to Electron-Sink Limitations in C3 Plants. Antioxidants 2020, 9, 230. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. The water–water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. B Biol. Sci. 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [Green Version]
- Sejima, T.; Takagi, D.; Fukayama, H.; Makino, A.; Miyake, C. Repetitive Short-Pulse Light Mainly Inactivates Photosystem I in Sunflower Leaves. Plant Cell Physiol. 2014, 55, 1184–1193. [Google Scholar] [CrossRef]
- Shimakawa, G.; Miyake, C. Changing frequency of fluctuating light reveals the molecular mechanism for P700 oxidation in plant leaves. Plant Direct 2018, 2, e00073. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Kudoh, H.; Sonoike, K. Irreversible damage to photosystem I by chilling in the light: Cause of the degradation of chlorophyll after returning to normal growth temperature. Planta 2002, 215, 541–548. [Google Scholar] [CrossRef]
- Li, X.-G.; Wang, X.-M.; Meng, Q.-W.; Zou, Q. Factors Limiting Photosynthetic Recovery in Sweet Pepper Leaves After Short-Term Chilling Stress Under Low Irradiance. Photosynthetica 2004, 42, 257–262. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Yu, J.Q.; Huang, L.F.; Nogues, S. The relationship between CO2 assimilation, photosynthetic electron transport and water-water cycle in chill-exposed cucumber leaves under low light and subsequent recovery. Plant Cell Environ. 2004, 27, 1503–1514. [Google Scholar] [CrossRef]
- Zhang, S.; Scheller, H. Photoinhibition of Photosystem I at Chilling Temperature and Subsequent Recovery in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1595–1602. [Google Scholar] [CrossRef]
- Zivcak, M.; Brestic, M.; Kunderlikova, K.; Sytar, O.; Allakhverdiev, S.I. Repetitive light pulse-induced photoinhibition of photosystem I severely affects CO2 assimilation and photoprotection in wheat leaves. Photosynth. Res. 2015, 126, 449–463. [Google Scholar] [CrossRef]
- Shimakawa, G.; Miyake, C. What Quantity of Photosystem I Is Optimum for Safe Photosynthesis? Plant Physiol. 2019, 179, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Sonoike, K. Photoinhibition of photosystem I. Physiol. Plant 2011, 142, 56–64. [Google Scholar] [CrossRef]
- Foyer, C.; Furbank, R.; Harbinson, J.; Horton, P. The mechanisms contributing to photosynthetic control of electron transport by carbon assimilation in leaves. Photosynth. Res. 1990, 25, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Harbinson, J.; Hedley, C.L. Changes in P-700 Oxidation during the Early Stages of the Induction of Photosynthesis. Plant Physiol. 1993, 103, 649–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klughammer, C.; Schreiber, U. An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta 1994, 192, 261–268. [Google Scholar] [CrossRef]
- Golding, A.J.; Johnson, G.N. Down-regulation of linear and activation of cyclic electron transport during drought. Planta 2003, 218, 107–114. [Google Scholar] [CrossRef]
- Miyake, C.; Miyata, M.; Shinzaki, Y.; Tomizawa, K.-I. CO2 Response of Cyclic Electron Flow around PSI (CEF-PSI) in Tobacco Leaves—Relative Electron fluxes through PSI and PSII Determine the Magnitude of Non-photochemical Quenching (NPQ) of Chl Fluorescence. Plant Cell Physiol. 2005, 46, 629–637. [Google Scholar] [CrossRef]
- Eberhard, S.; Finazzi, G.; Wollman, F.-A. The Dynamics of Photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef] [Green Version]
- Shaku, K.; Shimakawa, G.; Hashiguchi, M.; Miyake, C. Reduction-induced suppression of electron flow (RISE) in the photosynthetic electron transport system of Synechococcus elongates PCC 7942. Plant Cell Physiol. 2015, 57, 1443–1453. [Google Scholar]
- Furutani, R.; Ifuku, K.; Suzuki, Y.; Noguchi, K.; Shimakawa, G.; Wada, S.; Makino, A.; Sohtome, T.; Miyake, C. P700 oxidation suppresses the production of reactive oxygen species in photosystem I. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 151–176. [Google Scholar]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 Is Involved in Cyclic Electron Flow around Photosystem I and Is Essential for Photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef] [Green Version]
- DalCorso, G.; Pesaresi, P.; Masiero, S.; Aseeva, E.; Schünemann, D.; Finazzi, G.; Joliot, P.; Barbato, R.; Leister, D. A Complex Containing PGRL1 and PGR5 Is Involved in the Switch between Linear and Cyclic Electron Flow in Arabidopsis. Cell 2008, 132, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Nuijs, A.M.; Shuvalov, V.A.; van Gorkom, H.J.; Plijter, J.J.; Duysens, L.N. Picosecond absorbance difference spectroscopy on the primary reactions and the antenna-excited states in Photosystem I particles. Biochim. Biophys. Acta (BBA)-Bioenerg. 1986, 850, 310–318. [Google Scholar] [CrossRef]
- Trissl, H.-W. Determination of the quenching efficiency of the oxidized primary donor of Photosystem I, P700+: Implications for the trapping mechanism. Photosynth. Res. 1997, 54, 237–240. [Google Scholar] [CrossRef]
- Bukhov, N.G.; Carpentier, R. Measurement of photochemical quenching of absorbed quanta in photosystem I of intact leaves using simultaneous measurements of absorbance changes at 830 nm and thermal dissipation. Planta 2003, 216, 630–638. [Google Scholar] [CrossRef]
- Yamamoto, H.; Shikanai, T. PGR5-Dependent Cyclic Electron Flow Protects Photosystem I under Fluctuating Light at Donor and Acceptor Sides. Plant Physiol. 2019, 179, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Rantala, S.; Lempiäinen, T.; Gerotto, C.; Tiwari, A.; Aro, E.-M.; Tikkanen, M. PGR5 and NDH-1 systems do not function as protective electron acceptors but mitigate the consequences of PSI inhibition. Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148154. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T. Physiological Functions of Cyclic Electron Transport Around Photosystem I in Sustaining Photosynthesis and Plant Growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef]
- Kono, M.; Noguchi, K.; Terashima, I. Roles of the Cyclic Electron Flow Around PSI (CEF-PSI) and O2-Dependent Alternative Pathways in Regulation of the Photosynthetic Electron Flow in Short-Term Fluctuating Light in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 990–1004. [Google Scholar] [CrossRef] [Green Version]
- Kono, M.; Terashima, I. Elucidation of Photoprotective Mechanisms of PSI Against Fluctuating Light photoinhibition. Plant Cell Physiol. 2016, 57, 1405–1414. [Google Scholar] [CrossRef]
- Tiwari, A.; Mamedov, F.; Grieco, M.; Suorsa, M.; Jajoo, A.; Styring, S.; Tikkanen, M.; Aro, E.-M. Photodamage of iron–sulphur clusters in photosystem I induces non-photochemical energy dissipation. Nat. Plants 2016, 2, 16035. [Google Scholar] [CrossRef]
- Suorsa, M.; Järvi, S.; Grieco, M.; Nurmi, M.; Pietrzykowska, M.; Rantala, M.; Kangasjärvi, S.; Paakkarinen, V.; Tikkanen, M.; Jansson, S.; et al. Proton gradient regulation5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 2012, 24, 2934–2948. [Google Scholar] [CrossRef] [Green Version]
- Gollan, P.; Lima-Melo, Y.; Tiwari, A.; Tikkanen, M.; Aro, E.-M. Interaction between photosynthetic electron transport and chloroplast sinks triggers protection and signalling important for plant productivity. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160390. [Google Scholar] [CrossRef] [Green Version]
- Reiter, B.; Vamvaka, E.; Marino, G.; Kleine, T.; Jahns, P.; Bolle, C.; Leister, D.; Rühle, T. The Arabidopsis Protein CGL20 Is Required for Plastid 50S Ribosome Biogenesis. Plant Physiol. 2020, 182, 1222–1238. [Google Scholar] [CrossRef] [Green Version]
- Takagi, D.; Amako, K.; Hashiguchi, M.; Fukaki, H.; Ishizaki, K.; Goh, T.; Fukao, Y.; Sano, R.; Kurata, T.; Demura, T.; et al. Chloroplastic ATP synthase builds up a proton motive force preventing production of reactive oxygen species in photosystem I. Plant J. 2017, 91, 306–324. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-P.; Björkman, O.; Shih, C.; Grossman, A.R.; Rosenquist, M.; Jansson, S.; Niyogi, K.K. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nat. Cell Biol. 2000, 403, 391–395. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Von Caemmerer, S.; Farquhar, G.D. Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 1981, 153, 376–387. [Google Scholar] [CrossRef]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New fluorescence parameters for the determination of Q A redox state and excitation energy fluxes. Photosynt. Res. 2004, 79, 209. [Google Scholar] [CrossRef]
- Makino, A.; Sato, T.; Nakano, H.; Mae, T. Leaf photosynthesis, plant growth and nitrogen allocation in rice under different irradiances. Planta 1997, 203, 390–398. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenol-oxidase in B vulgaris. Plant Physiol. 1969, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Munekage, Y.N.; Genty, B.; Peltier, G. Effect of PGR5 Impairment on Photosynthesis and Growth in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1688–1698. [Google Scholar] [CrossRef] [Green Version]
- Teicher, H.B.; Møller, B.L.; Scheller, H.V. Photoinhibition of Photosystem I in field-grown barley (Hordeum vulgare L.): Induction, recovery and acclimation. Photosynth. Res. 2000, 64, 53–61. [Google Scholar] [CrossRef]
- Lima-Melo, Y.; Gollan, P.J.; Tikkanen, M.; Silveira, J.A.G.; Aro, E.-M. Consequences of photosystem-I damage and repair on photosynthesis and carbon use in Arabidopsis thaliana. Plant J. 2018, 97, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.-I.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nat. Cell Biol. 2004, 429, 579–582. [Google Scholar] [CrossRef]
- Kadota, K.; Furutani, R.; Makino, A.; Suzuki, Y.; Wada, S.; Miyake, C. Oxidation of P700 Induces Alternative Electron Flow in Photosystem I in Wheat Leaves. Plants 2019, 8, 152. [Google Scholar] [CrossRef] [Green Version]
- Wada, S.; Yamamoto, H.; Suzuki, Y.; Yamori, W.; Shikanai, T.; Makino, A. Flavodiiron Protein Substitutes for Cyclic Electron Flow without Competing CO2 Assimilation in Rice. Plant Physiol. 2018, 176, 1509–1518. [Google Scholar] [CrossRef] [Green Version]
- Shikanai, T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim. Biophys. Acta (BBA)-Bioenerg. 2016, 1857, 1015–1022. [Google Scholar] [CrossRef]
- Hashimoto, M.; Endo, T.; Peltier, G.; Tasaka, M.; Shikanai, T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003, 36, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, S.-B.; Cao, K.-F. Stimulation of Cyclic Electron Flow During Recovery After Chilling-Induced Photoinhibition of PSII. Plant Cell Physiol. 2010, 51, 1922–1928. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Nelson, C.; Trösch, J.; Castleden, I.; Huang, S.; Millar, A.H. Protein Degradation Rate in Arabidopsis thaliana Leaf Growth and Development. Plant Cell 2017, 29, 207–228. [Google Scholar] [CrossRef] [Green Version]
- Sonoike, K. Photoinhibition of photosystemI: Its physiological significance in the chilling sensitivity of plants. Plant Cell Physiol. 1996, 37, 239–247. [Google Scholar] [CrossRef]
- Sonoike, K.; Kamo, M.; Hihara, Y.; Hiyama, T.; Enami, I. The mechanism of the degradation of psaB gene product, one of the photosynthetic reaction center subunits of Photosystem I, upon photoinhibition. Photosynth. Res. 1997, 53, 55–63. [Google Scholar] [CrossRef]
- Myouga, F.; Hosoda, C.; Umezawa, T.; Iizumi, H.; Kuromori, T.; Motohashi, R.; Shono, Y.; Nagata, N.; Ikeuchi, M.; Shinozaki, K. A Heterocomplex of Iron Superoxide Dismutases Defends Chloroplast Nucleoids against Oxidative Stress and Is Essential for Chloroplast Development in Arabidopsis. Plant Cell 2008, 20, 3148–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallie, D.R.; Chen, Z. Chloroplast-localized iron superoxide dismutases FSD2 and FSD3 are functionally distinct in Arabidopsis. PLoS ONE 2019, 14, e0220078. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.J.; Avenson, T.J.; Thomas, J.B.; Herbert, S.K. A Cyanobacterium Lacking Iron Superoxide Dismutase Is Sensitized to Oxidative Stress Induced with Methyl Viologen but Is Not Sensitized to Oxidative Stress Induced with Norflurazon1. Plant Physiol. 1998, 116, 1593–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
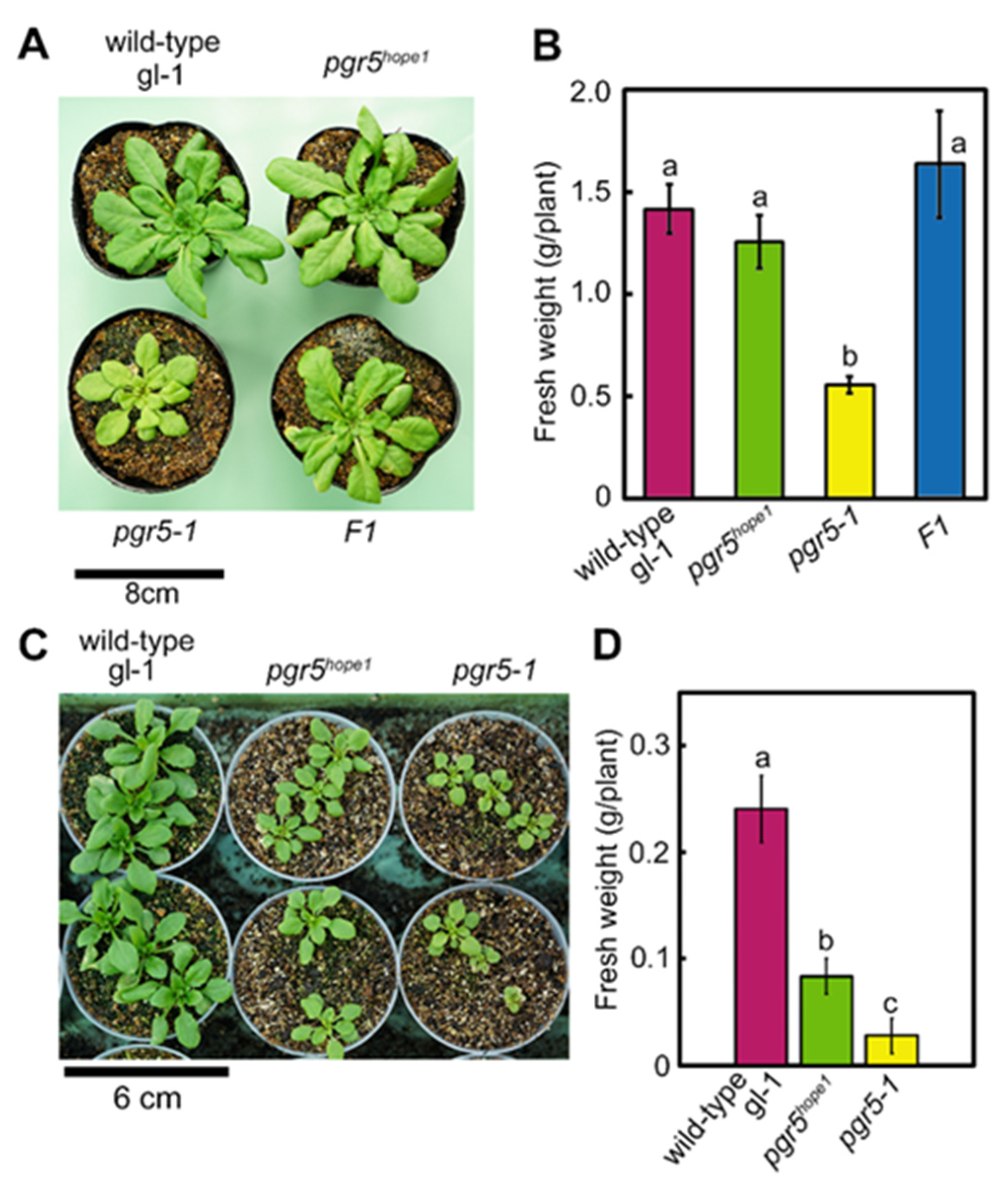
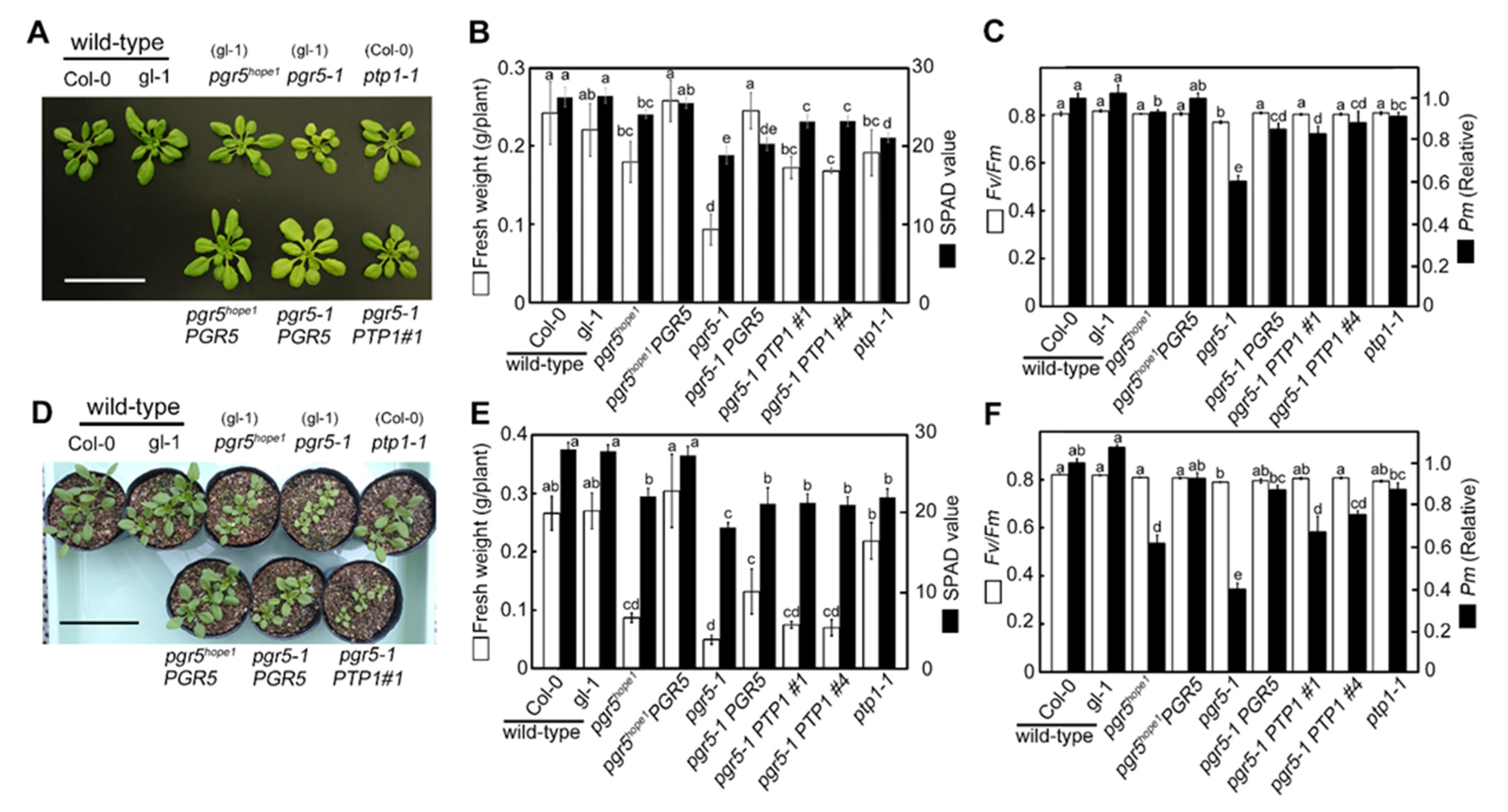
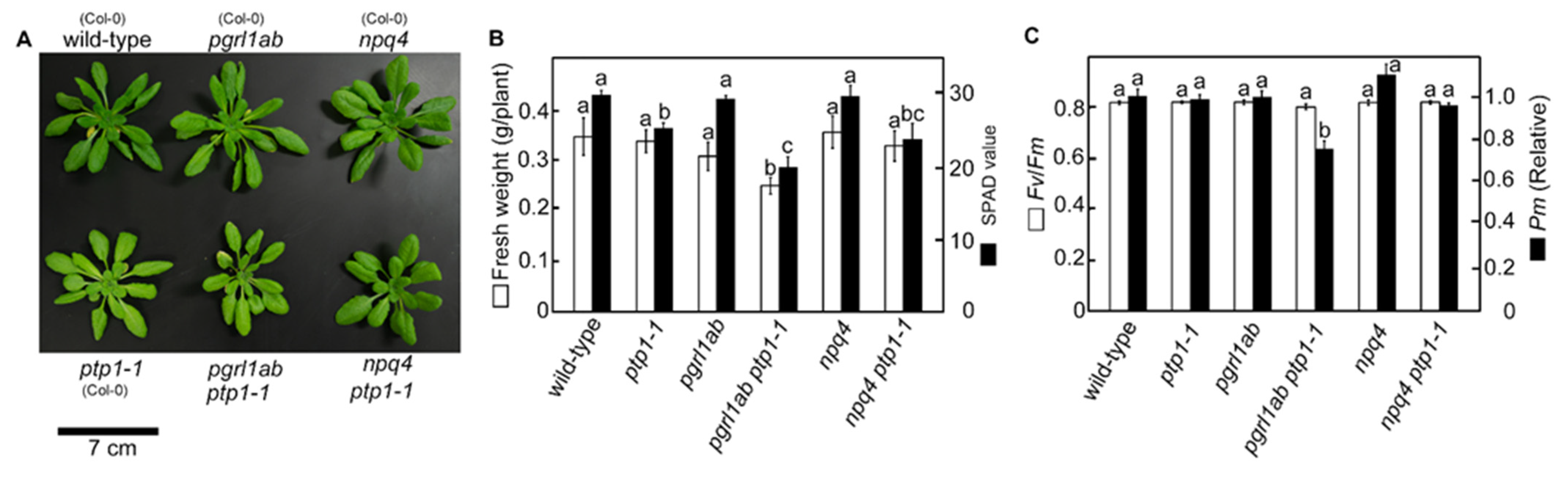
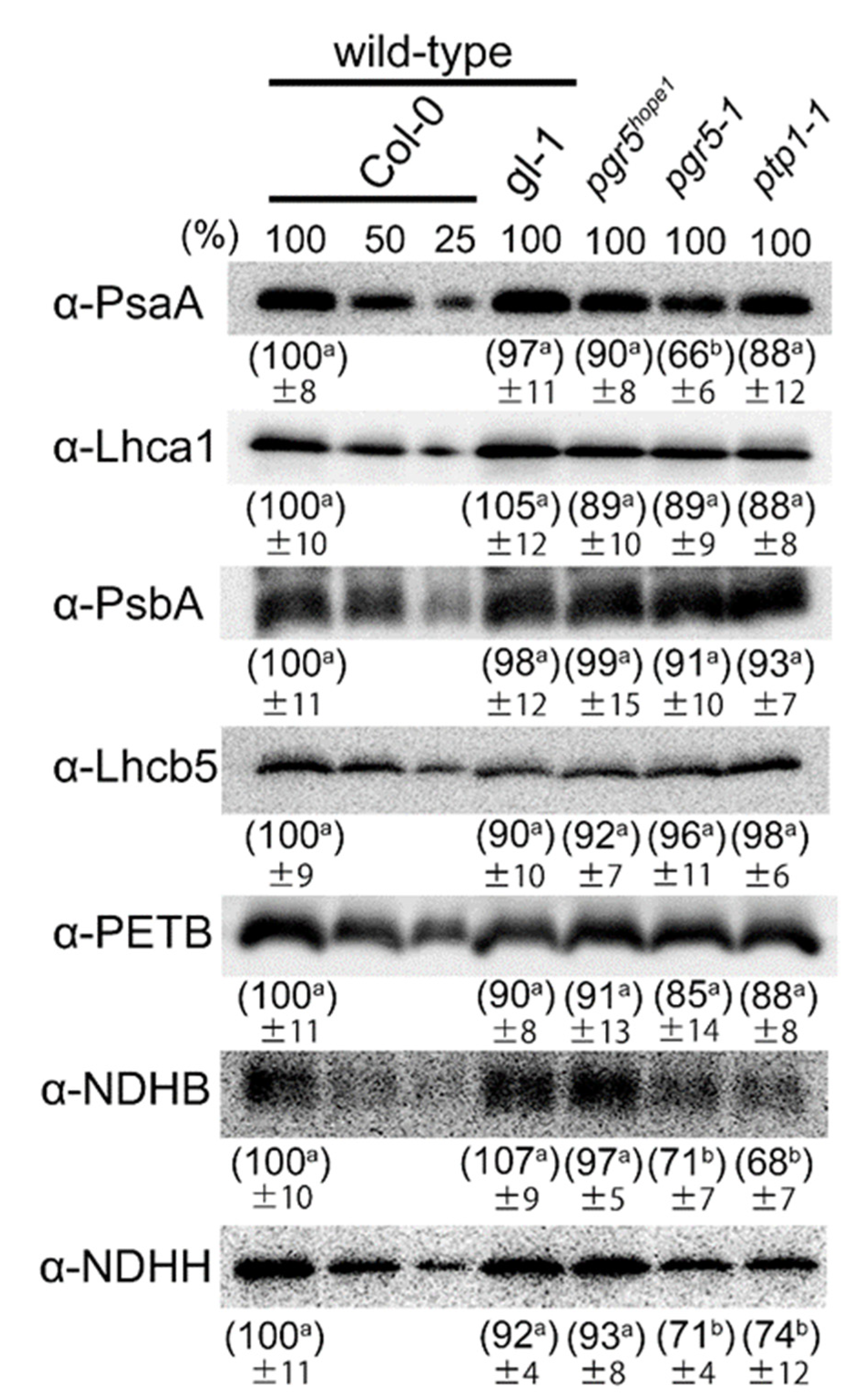
| Genotypes (Background) | Chlorophyll mmol m−2 | Chlorophyll a/b | Leaf N mmol m−2 | Rubisco g m−2 |
|---|---|---|---|---|
| wild-type (Col-0) | 0.377 ± 0.041 a | 3.08 ± 0.20 a | 80.6 ± 6.9 a | 1.45 ± 0.17 a |
| wild-type (gl-1) | 0.366 ± 0.024 a | 3.09 ± 0.40 a | 85.8 ± 4.6 a | 1.42 ± 0.13 a |
| pgr5hope1 (gl-1) | 0.385 ± 0.009 a | 2.96 ± 0.30 a | 82.4 ± 5.6 a | 1.48 ± 0.15 a |
| pgr5-1 (gl-1) | 0.287 ± 0.030 b | 3.30 ± 0.20 a | 78.4 ± 4.0 a | 1.27 ± 0.09 a |
| ptp1-1 (Col-0) | 0.293 ± 0.019 b | 3.29 ± 0.19 a | 80.2 ± 5.6 a | 1.51 ± 0.11 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wada, S.; Amako, K.; Miyake, C. Identification of a Novel Mutation Exacerbated the PSI Photoinhibition in pgr5/pgrl1 Mutants; Caution for Overestimation of the Phenotypes in Arabidopsis pgr5-1 Mutant. Cells 2021, 10, 2884. https://doi.org/10.3390/cells10112884
Wada S, Amako K, Miyake C. Identification of a Novel Mutation Exacerbated the PSI Photoinhibition in pgr5/pgrl1 Mutants; Caution for Overestimation of the Phenotypes in Arabidopsis pgr5-1 Mutant. Cells. 2021; 10(11):2884. https://doi.org/10.3390/cells10112884
Chicago/Turabian StyleWada, Shinya, Katsumi Amako, and Chikahiro Miyake. 2021. "Identification of a Novel Mutation Exacerbated the PSI Photoinhibition in pgr5/pgrl1 Mutants; Caution for Overestimation of the Phenotypes in Arabidopsis pgr5-1 Mutant" Cells 10, no. 11: 2884. https://doi.org/10.3390/cells10112884
APA StyleWada, S., Amako, K., & Miyake, C. (2021). Identification of a Novel Mutation Exacerbated the PSI Photoinhibition in pgr5/pgrl1 Mutants; Caution for Overestimation of the Phenotypes in Arabidopsis pgr5-1 Mutant. Cells, 10(11), 2884. https://doi.org/10.3390/cells10112884







