Assessment of the Hematopoietic Differentiation Potential of Human Pluripotent Stem Cells in 2D and 3D Culture Systems
Abstract
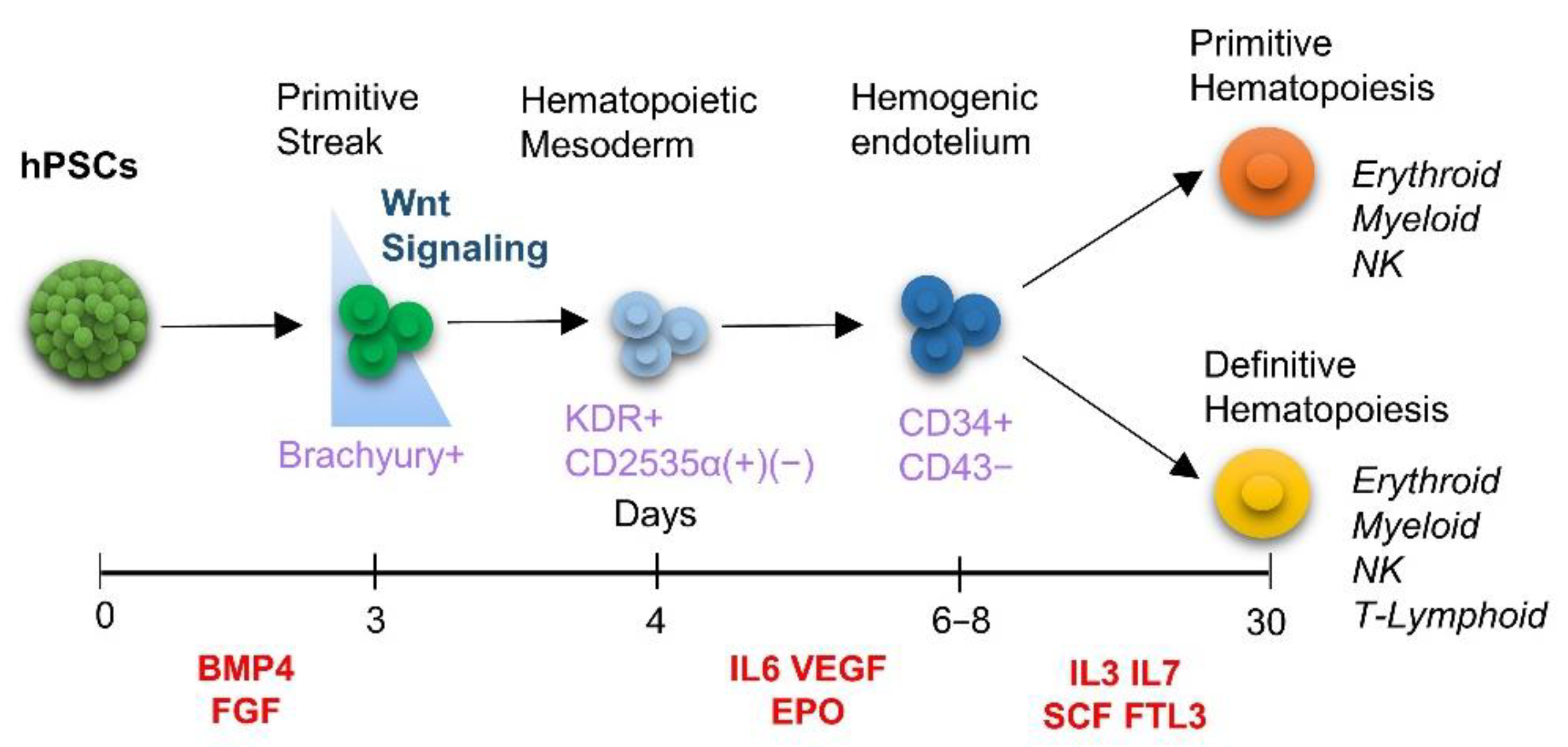
1. Introduction
2. Materials and Methods
2.1. Embryonic Stem Cell and iPS Cell Lines
2.2. Cell Culture under 2D/Monolayer and 3D/Embryoid Body Conditions
2.3. Mesodermal, Hematoendothelial and Hematopoietic Differentiation
2.4. Hematopoietic Colony Assay
2.5. AFT024 Co-Culture for Further Hematopoietic Differentiation
2.6. Flow Cytometry Analysis
2.7. Immunofluorescence
2.8. Gene Expression Analysis by RT-qPCR
2.9. Statistical Analysis
3. Results
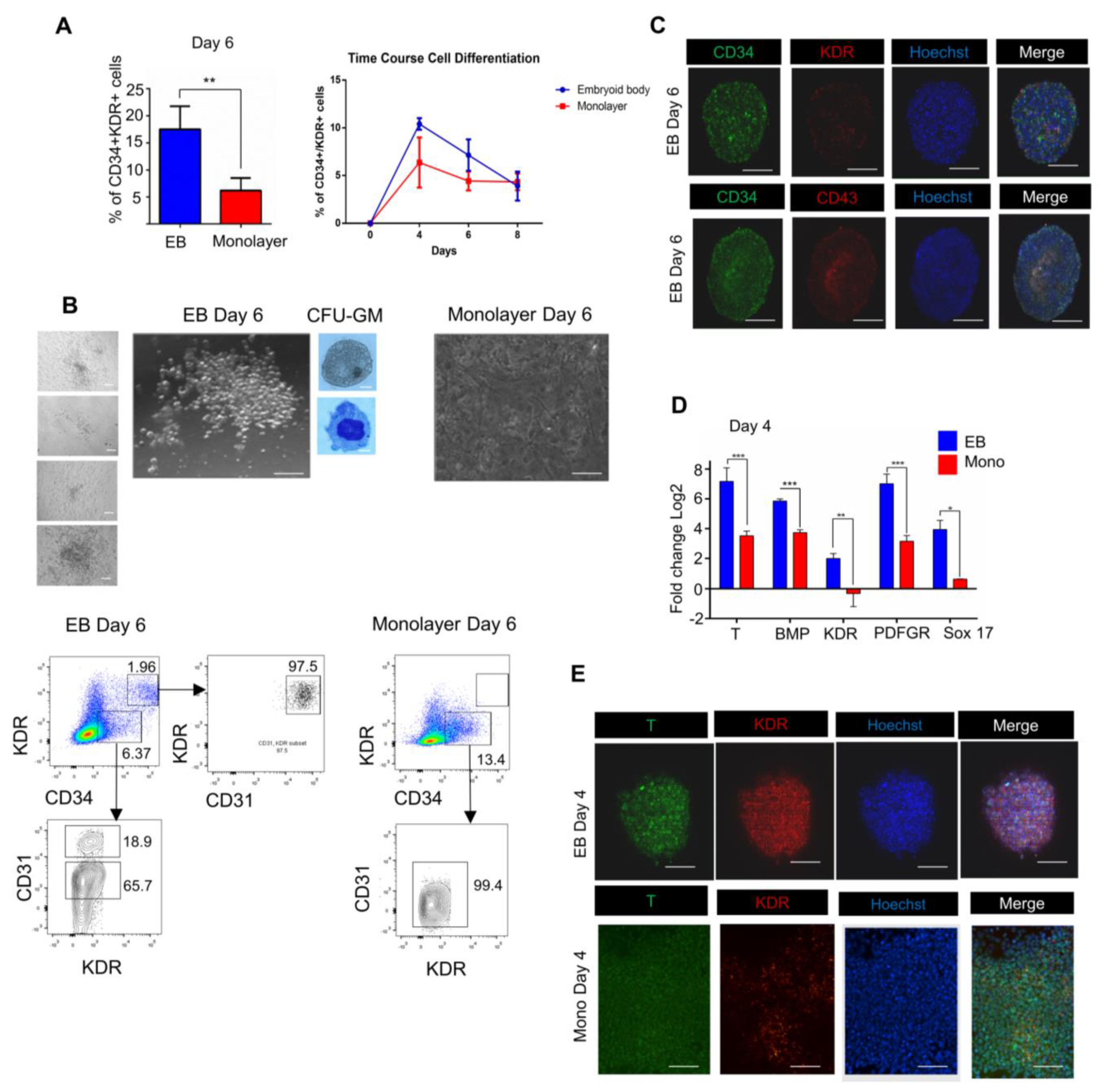
3.1. 3D/EB and 2D/Monolayer Culture Conditions Differ in Their Capability to Produce Hematopoietic Cells
3.2. Endogenous Wnt/Catenin Signaling Is Active in the 3D/EB System but Not in 2D/Monolayer System
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murry, C.E.; Keller, G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell 2008, 132, 661–680. [Google Scholar] [CrossRef]
- Ditadi, A.; Sturgeon, C.M. Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods 2016, 101, 65–72. [Google Scholar] [CrossRef]
- Sadlon, T.J.; Lewis, I.D.; D’Andrea, R.J. BMP4: Its role in development of the hematopoietic system and potential as a hematopoietic growth factor. Stem Cells 2004, 22, 457–474. [Google Scholar] [CrossRef]
- Keller, G.; Kennedy, M.; Papayannopoulou, T.; Wiles, M.V. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell Biol. 1993, 13, 473–486. [Google Scholar] [CrossRef]
- Kennedy, M.; D’Souza, S.L.; Lynch-Kattman, M.; Schwantz, S.; Keller, G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood 2007, 109, 2679–2687. [Google Scholar] [CrossRef]
- Crisan, M.; Kartalaei, P.S.; Vink, C.S.; Yamada-Inagawa, T.; Bollerot, K.; van IJcken, W.; van der Linden, R.; de Sousa Lopes, S.M.; Monteiro, R.; Mummery, C.; et al. BMP signalling differentially regulates distinct haematopoietic stem cell types. Nat. Commun. 2015, 6, 8040. [Google Scholar] [CrossRef]
- D’Souza, S.S.; Maufort, J.; Kumar, A.; Zhang, J.; Smuga-Otto, K.; Thomson, J.A.; Slukvin, I.I. GSK3β Inhibition Promotes Efficient Myeloid and Lymphoid Hematopoiesis from Non-human Primate-Induced Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 243–256. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, M. The hemangioblast revisited. Blood 2014, 124, 2472–2473. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lacaud, G.; Kouskoff, V. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp. Hematol. 2017, 49, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, J.; Liu, S.; Li, Y.; Hu, J.; Kou, Z.; Zhang, Y.; Sun, X.; Gao, S. Differentiation of reprogrammed somatic cells into functional hematopoietic cells. Differentiation 2009, 78, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kennedy, M.; Kazarov, A.; Papadimitriou, J.C.; Keller, G. A common precursor for hematopoietic and endothelial cells. Development 1998, 125, 725–732. [Google Scholar] [CrossRef]
- Huber, T.L.; Kouskoff, V.; Fehling, H.J.; Palis, J.; Keller, G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 2004, 432, 625–630. [Google Scholar] [CrossRef]
- Knorr, D.A.; Ni, Z.; Hermanson, D.; Hexum, M.K.; Bendzick, L.; Cooper, L.J.; Lee, D.A.; Kaufman, D.S. Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl. Med. 2013, 2, 274–283. [Google Scholar] [CrossRef]
- Niwa, A.; Heike, T.; Umeda, K.; Oshima, K.; Kato, I.; Sakai, H.; Suemori, H.; Nakahata, T.; Saito, M.K. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS ONE 2011, 6, e22261. [Google Scholar] [CrossRef]
- Niwa, A.; Umeda, K.; Chang, H.; Saito, M.; Okita, K.; Takahashi, K.; Nakagawa, M.; Yamanaka, S.; Nakahata, T.; Heike, T. Orderly hematopoietic development of induced pluripotent stem cells via Flk-1(+) hemoangiogenic progenitors. J. Cell. Physiol. 2009, 221, 367–377. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mills, J.A.; Paluru, P.; Weiss, M.J.; Gadue, P.; French, D.L. Hematopoietic differentiation of pluripotent stem cells in culture. Methods Mol. Biol. 2014, 1185, 181–194. [Google Scholar] [PubMed]
- Galat, Y.; Dambaeva, S.; Elcheva, I.; Khanolkar, A.; Beaman, K.; Iannaccone, P.M.; Galat, V. Cytokine-free directed differentiation of human pluripotent stem cells efficiently produces hemogenic endothelium with lymphoid potential. Stem Cell Res. Ther. 2017, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.P.; Chen, G.; Haro Mora, J.J.; Keyvanfar, K.; Liu, C.; Zou, J.; Beers, J.; Bloomer, H.; Qanash, H.; Uchida, N.; et al. Robust generation of erythroid and multilineage hematopoietic progenitors from human iPSCs using a scalable monolayer culture system. Stem Cell Res. 2019, 41, 101600. [Google Scholar] [CrossRef] [PubMed]
- Demirci, S.; Haro-Mora, J.J.; Leonard, A.; Drysdale, C.; Malide, D.; Keyvanfar, K.; Essawi, K.; Vizcardo, R.; Tamaoki, N.; Restifo, N.P.; et al. Definitive hematopoietic stem/progenitor cells from human embryonic stem cells through serum/feeder-free organoid-induced differentiation. Stem Cell Res. Ther. 2020, 11, 493. [Google Scholar] [CrossRef]
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional Culture System of Cancer Cells Combined with Biomaterials for Drug Screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef]
- Kaushik, G.; Ponnusamy, M.P.; Batra, S.K. Concise Review: Current Status of Three-Dimensional Organoids as Preclinical Models. Stem Cells 2018, 36, 1329–1340. [Google Scholar] [CrossRef]
- Hernández-Sapiéns, M.A.; Reza-Zaldívar, E.E.; Cevallos, R.R.; Márquez-Aguirre, A.L.; Gazarian, K.; Canales-Aguirre, A.A. A Three-Dimensional Alzheimer’s Disease Cell Culture Model Using iPSC-Derived Neurons Carrying A246E Mutation in PSEN1. Front. Cell. Neurosci. 2020, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Olmer, R.; Haase, A.; Merkert, S.; Cui, W.; Palecek, J.; Ran, C.; Kirschning, A.; Scheper, T.; Glage, S.; Miller, K.; et al. Long term expansion of undifferentiated human iPS and ES cells in suspension culture using a defined medium. Stem Cell Res. 2010, 5, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Ueda, Y.; Kadari, A.; Günther, K.; Ergün, S.; Heron, A.; Schnitzler, A.C.; Rook, M.; Edenhofer, F. Scalable stirred suspension culture for the generation of billions of human induced pluripotent stem cells using single-use bioreactors. J. Tissue Eng. Regen. Med. 2018, 12, e1076–e1087. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.F.; Heng, B.C.; Ge, Z.; Lu, K.; Rufaihah, A.J.; Fan, V.T.; Yeo, J.F.; Cao, T. Comparison of osteogenesis of human embryonic stem cells within 2D and 3D culture systems. Scand. J. Clin. Lab. Investig. 2008, 68, 58–67. [Google Scholar] [CrossRef]
- Fong, A.H.; Romero-López, M.; Heylman, C.M.; Keating, M.; Tran, D.; Sobrino, A.; Tran, A.Q.; Pham, H.H.; Fimbres, C.; Gershon, P.D.; et al. Three-Dimensional Adult Cardiac Extracellular Matrix Promotes Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Tissue Eng. Part A 2016, 22, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Sacchetto, C.; Vitiello, L.; de Windt, L.J.; Rampazzo, A.; Calore, M. Modeling Cardiovascular Diseases with hiPSC-Derived Cardiomyocytes in 2D and 3D Cultures. Int. J. Mol. Sci. 2020, 21, 3404. [Google Scholar] [CrossRef]
- Jeziorowska, D.; Fontaine, V.; Jouve, C.; Villard, E.; Dussaud, S.; Akbar, D.; Letang, V.; Cervello, P.; Itier, J.M.; Pruniaux, M.P.; et al. Differential Sarcomere and Electrophysiological Maturation of Human iPSC-Derived Cardiac Myocytes in Monolayer vs. Aggregation-Based Differentiation Protocols. Int. J. Mol. Sci. 2017, 18, 1173. [Google Scholar] [CrossRef]
- Haraguchi, Y.; Matsuura, K.; Shimizu, T.; Yamato, M.; Okano, T. Simple suspension culture system of human iPS cells maintaining their pluripotency for cardiac cell sheet engineering. J. Tissue Eng. Regen. Med. 2015, 9, 1363–1375. [Google Scholar] [CrossRef]
- Hamad, S.; Derichsweiler, D.; Papadopoulos, S.; Nguemo, F.; Šarić, T.; Sachinidis, A.; Brockmeier, K.; Hescheler, J.; Boukens, B.J.; Pfannkuche, K. Generation of human induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer and scalable 3D suspension bioreactor cultures with reduced batch-to-batch variations. Theranostics 2019, 9, 7222–7238. [Google Scholar] [CrossRef]
- Choi, K.-D.; Vodyanik, M.A.; Slukvin, I.I. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J. Clin. Investig. 2009, 119, 2818–2829. [Google Scholar] [CrossRef]
- Yanagimachi, M.D.; Niwa, A.; Tanaka, T.; Honda-Ozaki, F.; Nishimoto, S.; Murata, Y.; Yasumi, T.; Ito, J.; Tomida, S.; Oshima, K.; et al. Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS ONE 2013, 8, e59243. [Google Scholar] [CrossRef]
- Ohta, R.; Niwa, A.; Taniguchi, Y.; Suzuki, N.M.; Toga, J.; Yagi, E.; Saiki, N.; Nishinaka-Arai, Y.; Okada, C.; Watanabe, A.; et al. Laminin-guided highly efficient endothelial commitment from human pluripotent stem cells. Sci. Rep. 2016, 6, 35680. [Google Scholar] [CrossRef]
- Cevallos, R.R.; Rodríguez-Martínez, G.; Gazarian, K. Wnt/β-Catenin/TCF Pathway Is a Phase-Dependent Promoter of Colony Formation and Mesendodermal Differentiation During Human Somatic Cell Reprogramming. Stem Cells 2018, 36, 683–695. [Google Scholar] [CrossRef]
- Ramirez-Ramirez, D.; Vadillo, E. Early Differentiation of Human CD11c(+)NK Cells with gammadelta T Cell Activation Properties Is Promoted by Dialyzable Leukocyte Extracts. J. Immunol. Res. 2016, 2016, 4097642. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Gadue, P.; Huber, T.L.; Paddison, P.J.; Keller, G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 16806–16811. [Google Scholar] [CrossRef]
- Loh, K.M.; Chen, A.; Koh, P.W.; Deng, T.Z.; Sinha, R.; Tsai, J.M.; Barkal, A.A.; Shen, K.Y.; Jain, R.; Morganti, R.M.; et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 2016, 166, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Nostro, M.C.; Cheng, X.; Keller, G.M.; Gadue, P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2008, 2, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Traver, D.; Willert, K. The role of Wnt signaling in hematopoietic stem cell development. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Sturgeon, C.M.; Ditadi, A.; Awong, G.; Kennedy, M.; Keller, G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 554–561. [Google Scholar] [CrossRef]
- Sumi, T.; Tsuneyoshi, N.; Nakatsuji, N.; Suemori, H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development 2008, 135, 2969–2979. [Google Scholar] [CrossRef]
- Clarke, R.L.; Yzaguirre, A.D.; Yashiro-Ohtani, Y.; Bondue, A.; Blanpain, C.; Pear, W.S.; Speck, N.A.; Keller, G. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol. 2013, 15, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Kyba, M. Hemogenic endothelium in a dish. Blood 2013, 121, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Nakajima-Takagi, Y.; Osawa, M.; Oshima, M.; Takagi, H.; Miyagi, S.; Endoh, M.; Endo, T.A.; Takayama, N.; Eto, K.; Toyoda, T.; et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood 2013, 121, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Nobuhisa, I.; Saito, K.; Gerel, M.; Itabashi, A.; Harada, K.; Osawa, M.; Endo, T.A.; Iwama, A.; Taga, T. Sox17-mediated expression of adherent molecules is required for the maintenance of undifferentiated hematopoietic cluster formation in midgestation mouse embryos. Differentiation 2020, 115, 53–61. [Google Scholar] [CrossRef]
- Sakurai, H.; Era, T.; Jakt, L.M.; Okada, M.; Nakai, S.; Nishikawa, S.; Nishikawa, S. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells 2006, 24, 575–586. [Google Scholar] [CrossRef]
- Wang, P.; Rodriguez, R.T.; Wang, J.; Ghodasara, A.; Kim, S.K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell 2011, 8, 335–346. [Google Scholar] [CrossRef]
- Galat, Y.; Elcheva, I.; Dambaeva, S.; Katukurundage, D.; Beaman, K.; Iannaccone, P.M.; Galat, V. Application of small molecule CHIR99021 leads to the loss of hemangioblast progenitor and increased hematopoiesis of human pluripotent stem cells. Exp. Hematol. 2018, 65, 38–48.e1. [Google Scholar] [CrossRef]
- Kang, H.; Mesquitta, W.-T.; Jung, H.S.; Moskvin, O.V.; Thomson, J.A.; Slukvin, I.I. GATA2 Is Dispensable for Specification of Hemogenic Endothelium but Promotes Endothelial-to-Hematopoietic Transition. Stem Cell Rep. 2018, 11, 197–211. [Google Scholar] [CrossRef]
- Slukvin, I.I. Deciphering the hierarchy of angiohematopoietic progenitors from human pluripotent stem cells. Cell Cycle 2013, 12, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Cordes, S.; Zou, J.; Yu, S.J.; Guitart, X.; Hong, S.G.; Dang, V.; Kang, E.; Donaires, F.S.; Hassan, S.A.; et al. GATA2 deficiency and human hematopoietic development modeled using induced pluripotent stem cells. Blood Adv. 2018, 2, 3553–3565. [Google Scholar] [CrossRef] [PubMed]
- Oszajca, K.; Szemraj, J.; Wyrzykowski, D.; Chrzanowska, B.; Salamon, A.; Przewratil, P. Single-nucleotide polymorphisms of VEGF-A and VEGFR-2 genes and risk of infantile hemangioma. Int. J. Dermatol. 2018, 57, 1201–1207. [Google Scholar] [CrossRef] [PubMed]


| Study | hPSCs | Culture System | Day(s) Hematopoietic Commitment | Hematopoietic Wave | Lineage Cells | Wnt Activation |
|---|---|---|---|---|---|---|
| Yanagimachi et al., 2013 | hESCs, hiPCSs | 2D/Monolayer | 6 | Primitive | Monocytes, Dendritic Cells | No |
| Sturgeon et al., 2014 | hESCs, HiPSCs | 3D/EB | 6 | Primitive definitive | Monocytes, Lymphoid Lineages | Yes |
| Ruiz et al., 2019 | hiPCSs | 2D/Monolayer | 5–9 | Primitive | Primitive Hematopoietic Cells | Yes |
| Niwa et al., 2011 | hiPSCs | 2D/Monolayer | 6 | Primitive | Monocytic Lineages | No |
| Kennedy et al., 2007 | hESCs | 3D/EB | 5–6 | Primitive | Early Hematopoietic Progenitors | No |
| Galat et al., 2017 | hiPSCs | 2D/Monolayer | 5 | Primitive | Monocytic lineages | Yes |
| Ditadi & Sturgeon, 2015 | hESCs | 3D/EB | 8–15 | Primitive, Definitive progenitors | Hemogenic Endothelium and early hematopoietic progenitors | Yes |
| Knorr et al., 2013 | hIPSC, hESCs | 3D/EB | 11 | Definitive | Nk, Lymhpoid | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora-Roldan, G.A.; Ramirez-Ramirez, D.; Pelayo, R.; Gazarian, K. Assessment of the Hematopoietic Differentiation Potential of Human Pluripotent Stem Cells in 2D and 3D Culture Systems. Cells 2021, 10, 2858. https://doi.org/10.3390/cells10112858
Mora-Roldan GA, Ramirez-Ramirez D, Pelayo R, Gazarian K. Assessment of the Hematopoietic Differentiation Potential of Human Pluripotent Stem Cells in 2D and 3D Culture Systems. Cells. 2021; 10(11):2858. https://doi.org/10.3390/cells10112858
Chicago/Turabian StyleMora-Roldan, German Atzin, Dalia Ramirez-Ramirez, Rosana Pelayo, and Karlen Gazarian. 2021. "Assessment of the Hematopoietic Differentiation Potential of Human Pluripotent Stem Cells in 2D and 3D Culture Systems" Cells 10, no. 11: 2858. https://doi.org/10.3390/cells10112858
APA StyleMora-Roldan, G. A., Ramirez-Ramirez, D., Pelayo, R., & Gazarian, K. (2021). Assessment of the Hematopoietic Differentiation Potential of Human Pluripotent Stem Cells in 2D and 3D Culture Systems. Cells, 10(11), 2858. https://doi.org/10.3390/cells10112858






