SUMO Interacting Motifs: Structure and Function
Abstract
:1. Introduction
2. SIM Structure and Regulation of SIM/SUMO Complex Formation
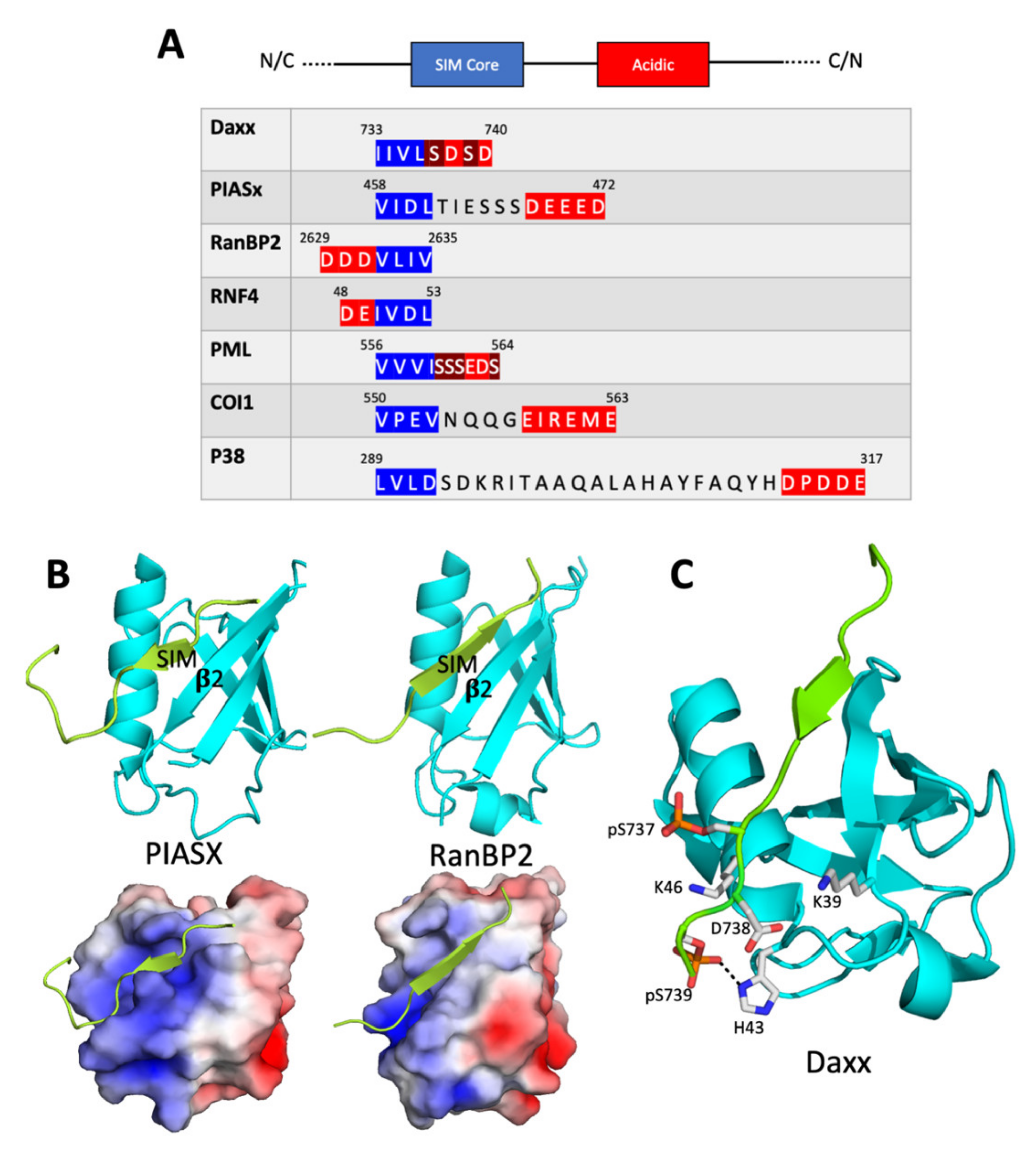
2.1. The SIM Consensus
2.2. Structure of the SUMO/SIM Complex
2.3. Regulation of SUMO/SIM Complex Formation
3. Specificity and the SUMO Interactome
4. Role for SUMO/SIM Interactions in Structure and Function of PML Bodies and Other Phase Separated Organelles
5. Role of SUMO/SIM Interactions in Histone Function
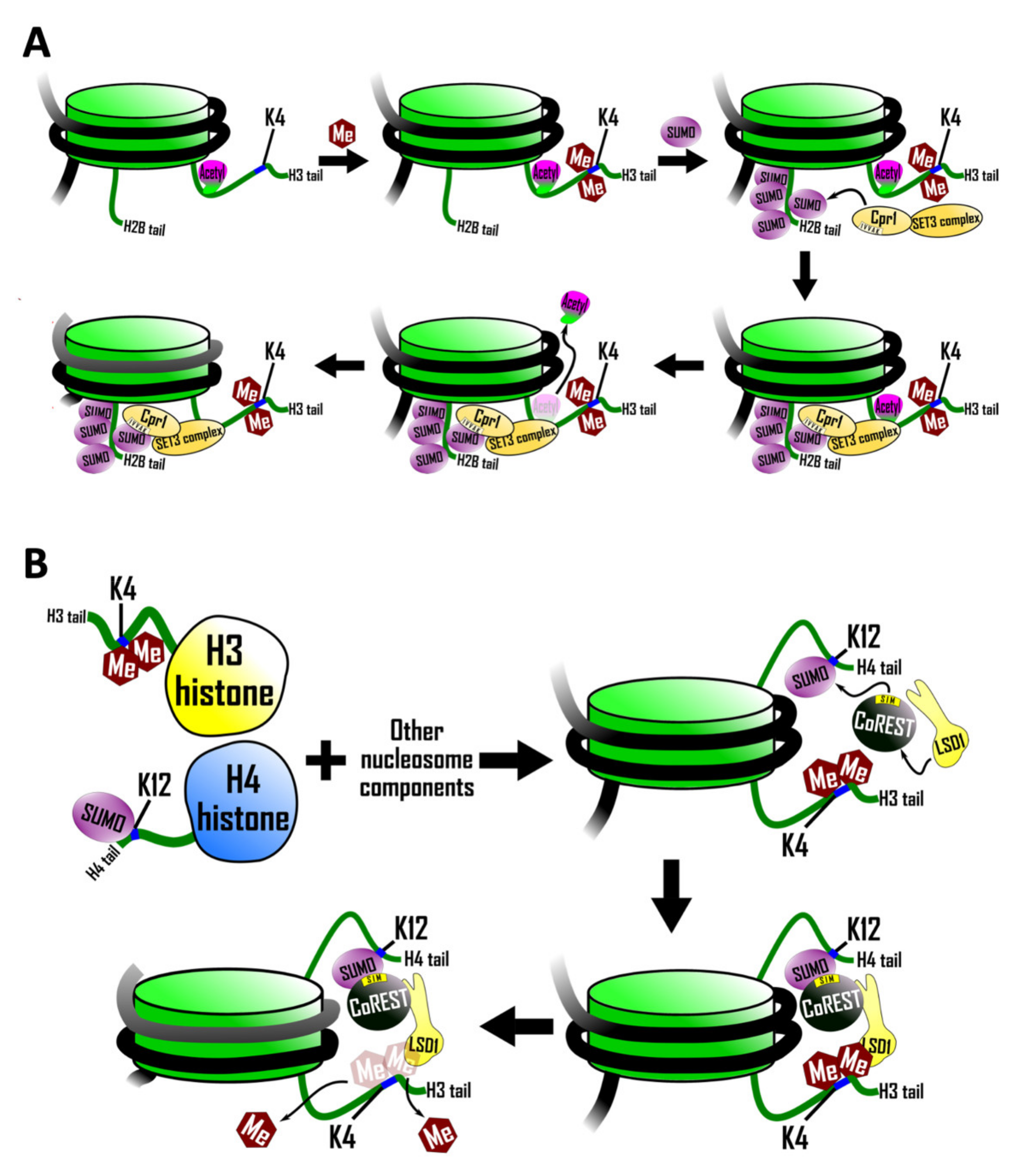
5.1. Recruitment of a SIM Containing Histone Deacetylase Complex
5.2. SIM-Mediated Crosstalk between Histone H3 and Histone H4
6. Roles for SUMO/SIM Interactions in Double Stranded DNA Break Repair and Nucleotide Excision Repair
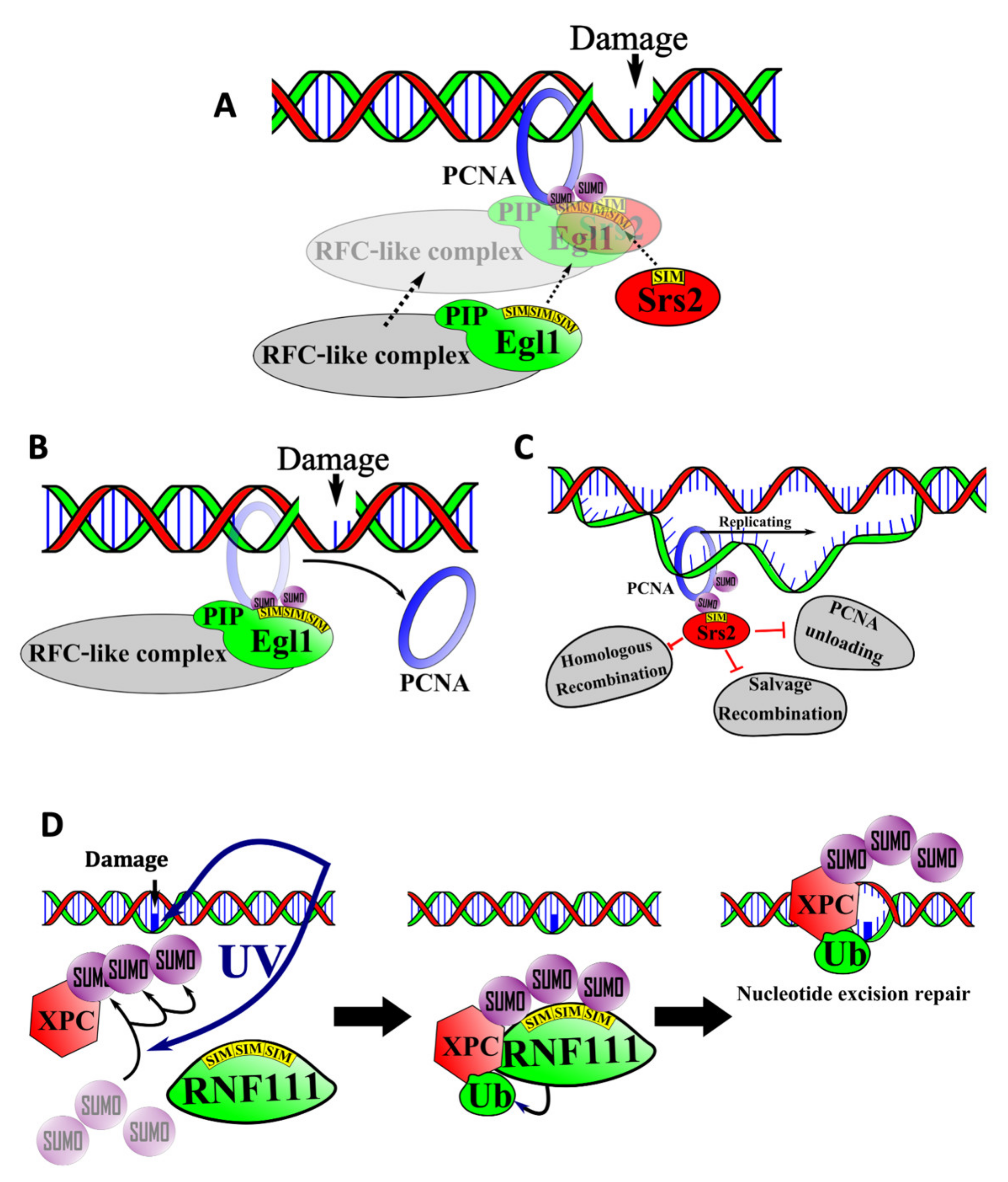
6.1. SIM-Mediated Interactions between PCNA, Elg1, and Srs2 in the Salvage Repair Pathway
6.2. SIMs in Nucleotide Excision Repair
7. SUMO and SIM in Pathogen–Host Cell Interactions
7.1. A SIM/SUMO Interaction in the Response to H. pylori Infection
7.2. SIM/SUMO Interactions in Jasmonic Acid Signaling in Plants
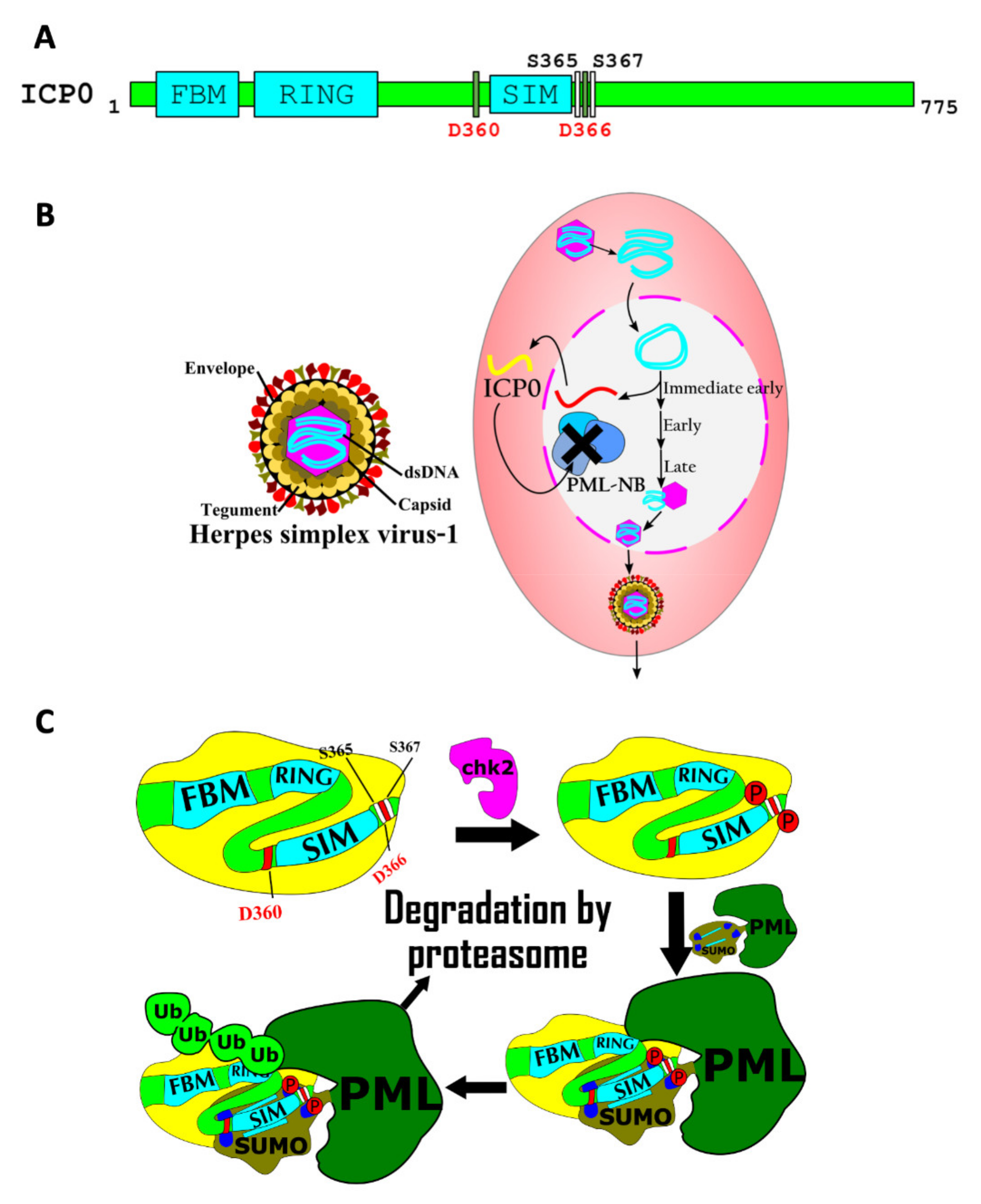
7.3. Regulation of Viral Pathogenicity by SIMs
8. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Hannoun, Z.; Greenhough, S.; Jaffray, E.; Hay, R.T.; Hay, D.C. Post-translational modification by SUMO. Toxicology 2010, 278, 288–293. [Google Scholar] [CrossRef] [Green Version]
- Yau, T.-Y.; Molina, O.; Courey, A.J. SUMOylation in development and neurodegeneration. Development 2020, 147. [Google Scholar] [CrossRef]
- Nayak, A.; Müller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, M.S.; Dargemont, C.; Hay, R. SUMO-1 Conjugation in Vivo Requires Both a Consensus Modification Motif and Nuclear Targeting. J. Biol. Chem. 2001, 276, 12654–12659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, T.H.; Lin, H.-K.; Scaglioni, P.P.; Yung, T.M.; Pandolfi, P.P. The Mechanisms of PML-Nuclear Body Formation. Mol. Cell 2006, 24, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarangi, P.; Zhao, X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 2015, 40, 233–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhall, A.; Wei, S.; Fierz, B.; Woodcock, C.L.; Lee, T.-H.; Chatterjee, C. Sumoylated Human Histone H4 Prevents Chromatin Compaction by Inhibiting Long-range Internucleosomal Interactions. J. Biol. Chem. 2014, 289, 33827–33837. [Google Scholar] [CrossRef] [Green Version]
- Baba, D.; Maita, N.; Jee, J.-G.; Uchimura, Y.; Saitoh, H.; Sugasawa, K.; Hanaoka, F.; Tochio, H.; Hiroaki, H.; Shirakawa, M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nat. Cell Biol. 2005, 435, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Minty, A.; Dumont, X.; Kaghad, M.; Caput, D. Covalent Modification of p73α by SUMO-1. J. Biol. Chem. 2000, 275, 36316–36323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Durrin, L.K.; Wilkinson, T.A.; Krontiris, T.G.; Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 14373–14378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauclair, G.; Bridier-Nahmias, A.; Zagury, J.-F.; Saïb, A.; Zamborlini, A. JASSA: A comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 2015, 31, 3483–3491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Zhang, Z.; Hu, W.; Chen, Y. Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif. J. Biol. Chem. 2005, 280, 40122–40129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef]
- Chang, P.-C.; Izumiya, Y.; Wu, C.-Y.; Fitzgerald, L.D.; Campbell, M.; Ellison, T.; Lam, K.S.; Luciw, P.A.; Kung, H.-J. Kaposi’s Sarcoma-associated Herpesvirus (KSHV) Encodes a SUMO E3 ligase That Is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 2010, 285, 5266–5273. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Naik, M.; Huang, Y.-S.; Jeng, J.-C.; Liao, P.-H.; Kuo, H.-Y.; Ho, C.-C.; Hsieh, Y.-L.; Lin, C.-H.; Huang, N.-J.; et al. Structural and Functional Roles of Daxx SIM Phosphorylation in SUMO Paralog-Selective Binding and Apoptosis Modulation. Mol. Cell 2011, 42, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Mascle, X.H.; Bourdeau, V.; Tremblay-Belzile, S.; Chaker-Margot, M.; Lussier-Price, M.; Wada, J.; Sakaguchi, K.; Aubry, M.; Ferbeyre, G.; et al. Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1. Structure 2015, 23, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.; Hu, W.; Xu, L.; Wu, H.; Wu, S.; Zhang, H.; Wang, J.; Jones, G.W.; Perrett, S. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J. Biol. Chem. 2018, 293, 17663–17675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burroughs, A.M.; Balaji, S.; Iyer, L.M.; Aravind, L. Small but versatile: The extraordinary functional and structural diversity of the β-grasp fold. Biol. Direct 2007, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Reverter, D.; Lima, C.D. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nat. Cell Biol. 2005, 435, 687–692. [Google Scholar] [CrossRef]
- Kotter, A.; Mootz, H.D.; Heuer, A. Standard Binding Free Energy of a SIM–SUMO Complex. J. Chem. Theory Comput. 2019, 15, 6403–6410. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Jardin, C.; Horn, A.H.C.; Sticht, H. Binding properties of SUMO-interacting motifs (SIMs) in yeast. J. Mol. Model. 2015, 21, 50. [Google Scholar] [CrossRef]
- Chupreta, S.; Holmstrom, S.; Subramanian, L.; Iñiguez-Lluhí, J.A. A Small Conserved Surface in SUMO Is the Critical Structural Determinant of Its Transcriptional Inhibitory Properties. Mol. Cell. Biol. 2005, 25, 4272–4282. [Google Scholar] [CrossRef] [Green Version]
- Newman, H.A.; Meluh, P.B.; Lu, J.; Vidal, J.; Carson, C.; LaGesse, E.; Gray, J.J.; Boeke, J.D.; Matunis, M.J. A high throughput mutagenic analysis of yeast sumo structure and function. PLoS Genet. 2017, 13, e1006612. [Google Scholar] [CrossRef]
- Lussier-Price, M.; Mascle, X.H.; Cappadocia, L.; Kamada, R.; Sakaguchi, K.; Wahba, H.M.; Omichinski, J.G. Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins. Structure 2020, 28, 573–585.e5. [Google Scholar] [CrossRef] [PubMed]
- Mascle, X.H.; Gagnon, C.; Wahba, H.M.; Lussier-Price, M.; Cappadocia, L.; Sakaguchi, K.; Omichinski, J.G. Acetylation of SUMO1 Alters Interactions with the SIMs of PML and Daxx in a Protein-Specific Manner. Structure 2020, 28, 157–168.e5. [Google Scholar] [CrossRef]
- González-Prieto, R.; Eifler-Olivi, K.; Claessens, L.A.; Willemstein, E.; Xiao, Z.; Ormeno, C.M.T.; Ovaa, H.; Ulrich, H.D.; Vertegaal, A.C. Global non-covalent SUMO interaction networks reveal SUMO-dependent stabilization of the non-homologous end joining complex. Cell Rep. 2021, 34, 108691. [Google Scholar] [CrossRef]
- Tatham, M.; Jaffray, E.; Vaughan, O.A.; Desterro, J.; Botting, C.H.; Naismith, J.; Hay, R. Polymeric Chains of SUMO-2 and SUMO-3 Are Conjugated to Protein Substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001, 276, 35368–35374. [Google Scholar] [CrossRef] [Green Version]
- Abed, M.; Barry, K.; Kenyagin, D.; Koltun, B.; Phippen, T.M.; Delrow, J.J.; Parkhurst, S.; Orian, A. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011, 30, 1289–1301. [Google Scholar] [CrossRef] [Green Version]
- Abed, M.; Bitman-Lotan, E.; Orian, A. The Biology of SUMO-Targeted Ubiquitin Ligases in Drosophila Development, Immunity, and Cancer. J. Dev. Biol. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Poulsen, S.L.; Hansen, R.K.; Wagner, S.A.; Van Cuijk, L.; van Belle, G.J.C.; Streicher, W.; Wikström, M.; Choudhary, C.; Houtsmuller, A.B.; Marteijn, J.; et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 2013, 201, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Tatham, M.H.; Geoffroy, M.-C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Plechanovová, A.; Simpson, P.; Marchant, J.; Leidecker, O.; Kraatz, S.; Hay, R.T.; Matthews, S.J. Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4. Nat. Commun. 2014, 5, 4217. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Hunter, T. Poly-Small Ubiquitin-like Modifier (PolySUMO)-binding Proteins Identified through a String Search. J. Biol. Chem. 2012, 287, 42071–42083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, D.J.; Tiede, C.; Penswick, N.; Tang, A.A.-S.; Trinh, C.H.; Mandal, U.; Zajac, K.Z.; Gaule, T.; Howell, G.; Edwards, T.A.; et al. Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology. Sci. Signal. 2017, 10, 1–33. [Google Scholar] [CrossRef] [Green Version]
- Banani, S.F.; Rice, A.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.; Matunis, M.; Dejean, A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998, 17, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; Müller, S.; Ronchetti, S.; Freemont, P.; Dejean, A.; Pandolfi, P.P. Role of SUMO-1–modified PML in nuclear body formation. Blood 2000, 95, 2748–2752. [Google Scholar] [CrossRef]
- Okamoto, K.; Seimiya, H. Revisiting Telomere Shortening in Cancer. Cells 2019, 8, 107. [Google Scholar] [CrossRef] [Green Version]
- Heaphy, C.M.; Subhawong, A.P.; Hong, S.-M.; Goggins, M.G.; Montgomery, E.A.; Gabrielson, E.; Netto, G.J.; Epstein, J.I.; Lotan, T.; Westra, W.H.; et al. Prevalence of the Alternative Lengthening of Telomeres Telomere Maintenance Mechanism in Human Cancer Subtypes. Am. J. Pathol. 2011, 179, 1608–1615. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019, 33, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Hembram, D.S.S.; Negi, H.; Biswas, P.; Tripathi, V.; Bhushan, L.; Shet, D.; Kumar, V.; Das, R. The Viral SUMO–Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2. J. Mol. Biol. 2020, 432, 1952–1977. [Google Scholar] [CrossRef] [PubMed]
- Keiten-Schmitz, J.; Röder, L.; Hornstein, E.; Müller-McNicoll, M.; Müller, S. SUMO: Glue or Solvent for Phase-Separated Ribonucleoprotein Complexes and Molecular Condensates? Front. Mol. Biosci. 2021, 8, 673038. [Google Scholar] [CrossRef]
- Pirrotta, V.; Li, H. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012, 22, 101–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jentsch, S.; Psakhye, I. Control of Nuclear Activities by Substrate-Selective and Protein-Group SUMOylation. Annu. Rev. Genet. 2013, 47, 167–186. [Google Scholar] [CrossRef]
- Psakhye, I.; Jentsch, S. Protein Group Modification and Synergy in the SUMO Pathway as Exemplified in DNA Repair. Cell 2012, 151, 807–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriks, I.A.; Lyon, D.; Su, D.; Skotte, N.H.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.-Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-Y.; Zhao, D.; Li, J.; Su, D.; Hochstrasser, M. Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 2020, 48, 12151–12168. [Google Scholar] [CrossRef]
- Dhall, A.; Weller, C.E.; Chu, A.; Shelton, P.M.M.; Chatterjee, C. Chemically Sumoylated Histone H4 Stimulates Intranucleosomal Demethylation by the LSD1–CoREST Complex. ACS Chem. Biol. 2017, 12, 2275–2280. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.; Shi, Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warbrick, E. The puzzle of PCNA’s many partners. Bioessays 2000, 22, 997–1006. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.-L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nat. Cell Biol. 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Park, S.H. Eukaryotic clamp loaders and unloaders in the maintenance of genome stability. Exp. Mol. Med. 2020, 52, 1948–1958. [Google Scholar] [CrossRef] [PubMed]
- Ben Aroya, S.; Kupiec, M. The Elg1 replication factor C-like complex: A novel guardian of genome stability. DNA Repair 2005, 4, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Parnas, O.; Zipin-Roitman, A.; Pfander, B.; Liefshitz, B.; Mazor, Y.; Ben-Aroya, S.; Jentsch, S.; Kupiec, M. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 2010, 29, 2611–2622. [Google Scholar] [CrossRef] [Green Version]
- Arbel, M.; Bronstein, A.; Sau, S.; Liefshitz, B.; Kupiec, M. Access to PCNA by Srs2 and Elg1 Controls the Choice between Alternative Repair Pathways in Saccharomyces cerevisiae. mBio 2020, 11, 11. [Google Scholar] [CrossRef]
- De Laat, W.; Jaspers, N.G.; Hoeijmakers, J.H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999, 13, 768–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.-E. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005, 33, 4023–4034. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.Y.; Hsu, P.I.; Wu, D.C.; Chen, T.C.; Jarman, A.P.; Powell, L.M.; Chen, A. SUMOs Mediate the Nuclear Transfer of p38 and p-p38 during Helicobacter Pylori Infection. Int. J. Mol. Sci. 2018, 19, 2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parikh, N.S.; Ahlawat, R. Helicobacter pylori. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Blaser, M.J.; Atherton, J.C. Helicobacter pylori persistence: Biology and disease. J. Clin. Investig. 2004, 113, 321–333. [Google Scholar] [CrossRef] [Green Version]
- Garza-Gonzalez, E.; Perez-Perez, G.I.; Maldonado-Garza, H.J.; Bosques-Padilla, F.J. A review of Helicobacter pylori diagnosis, treatment, and methods to detect eradication. World J. Gastroenterol. 2014, 20, 1438–1449. [Google Scholar] [CrossRef]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Orosa, B.; Singh, P.; Cummins, I.; Walsh, C.; Zhang, C.; Grant, M.; Roberts, M.R.; Anand, G.S.; Fitches, E.; et al. SUMO Suppresses the Activity of the Jasmonic Acid Receptor CORONATINE INSENSITIVE1. Plant Cell 2018, 30, 2099–2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.-S.; Campbell, M.; Chang, P.-C. SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLOS Pathog. 2017, 13, e1006216. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Kobayashi, K.; Kim, K.Y.; Pochampalli, M.; Izumiya, C.; Shevchenko, B.; Wang, D.-H.; Huerta, S.B.; Martinez, A.; Campbell, M.; et al. Kaposi’s Sarcoma-Associated Herpesvirus K-Rta Exhibits SUMO-Targeting Ubiquitin Ligase (STUbL) Like Activity and Is Essential for Viral Reactivation. PLOS Pathog. 2013, 9, e1003506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yau, T.-Y.; Sander, W.; Eidson, C.; Courey, A.J. SUMO Interacting Motifs: Structure and Function. Cells 2021, 10, 2825. https://doi.org/10.3390/cells10112825
Yau T-Y, Sander W, Eidson C, Courey AJ. SUMO Interacting Motifs: Structure and Function. Cells. 2021; 10(11):2825. https://doi.org/10.3390/cells10112825
Chicago/Turabian StyleYau, Tak-Yu, William Sander, Christian Eidson, and Albert J. Courey. 2021. "SUMO Interacting Motifs: Structure and Function" Cells 10, no. 11: 2825. https://doi.org/10.3390/cells10112825
APA StyleYau, T.-Y., Sander, W., Eidson, C., & Courey, A. J. (2021). SUMO Interacting Motifs: Structure and Function. Cells, 10(11), 2825. https://doi.org/10.3390/cells10112825




