Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes
Abstract
:1. Introduction
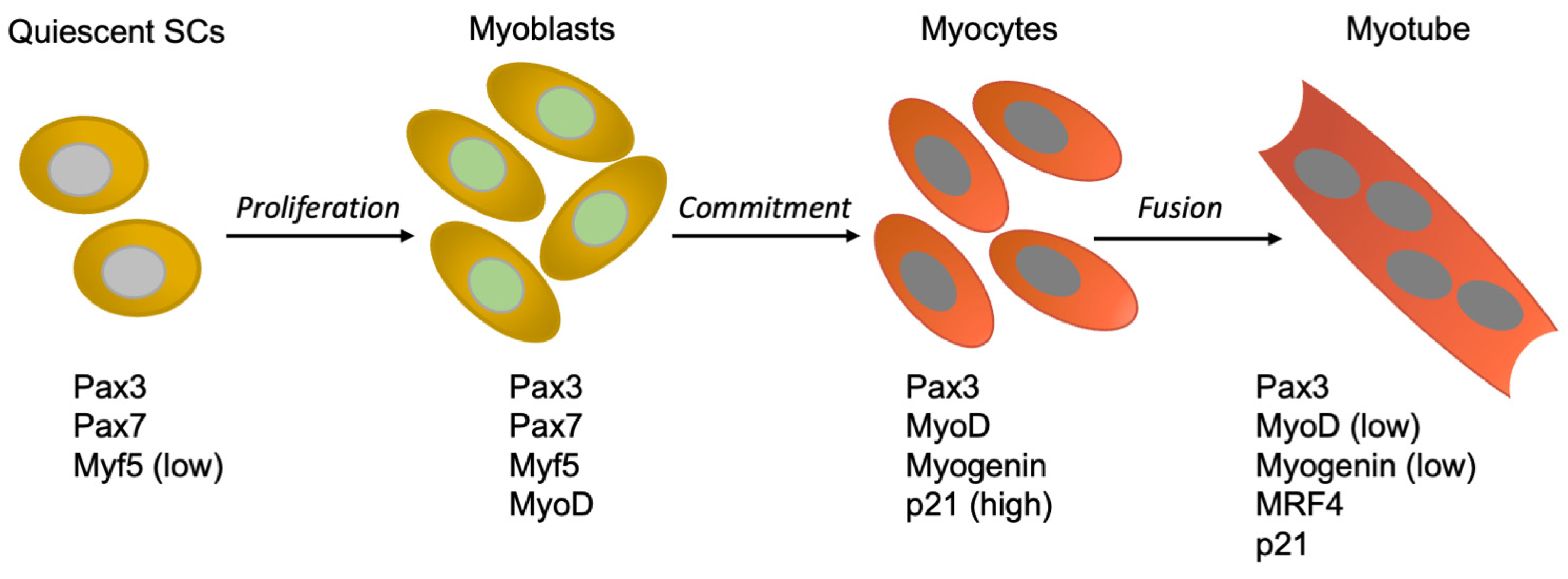
2. The Skeletal Muscle in Culture
3. The Postmitotic State in Myotubes
4. Early Attempts at Cell Cycle Reactivation
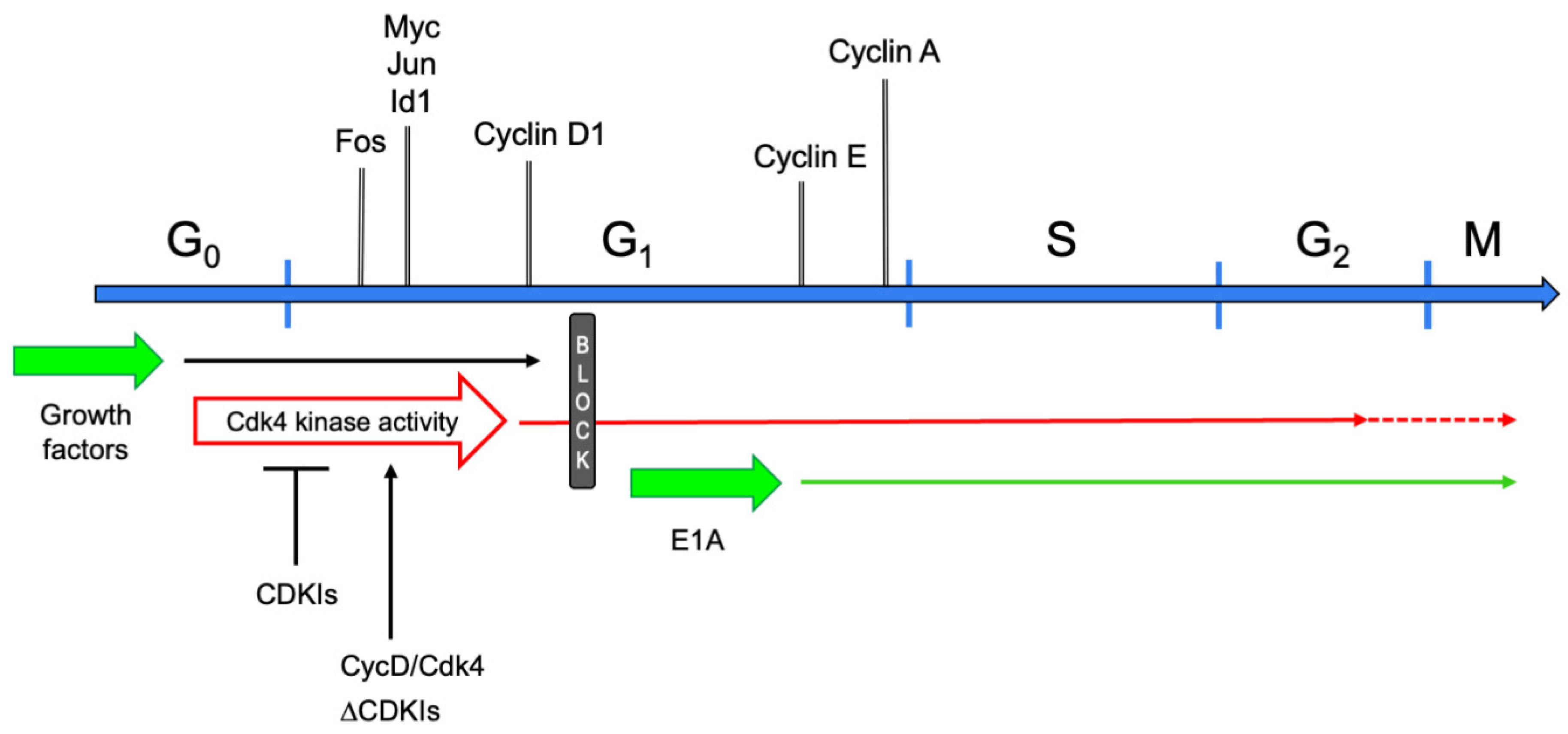
5. The Molecular Cell Cycle Era
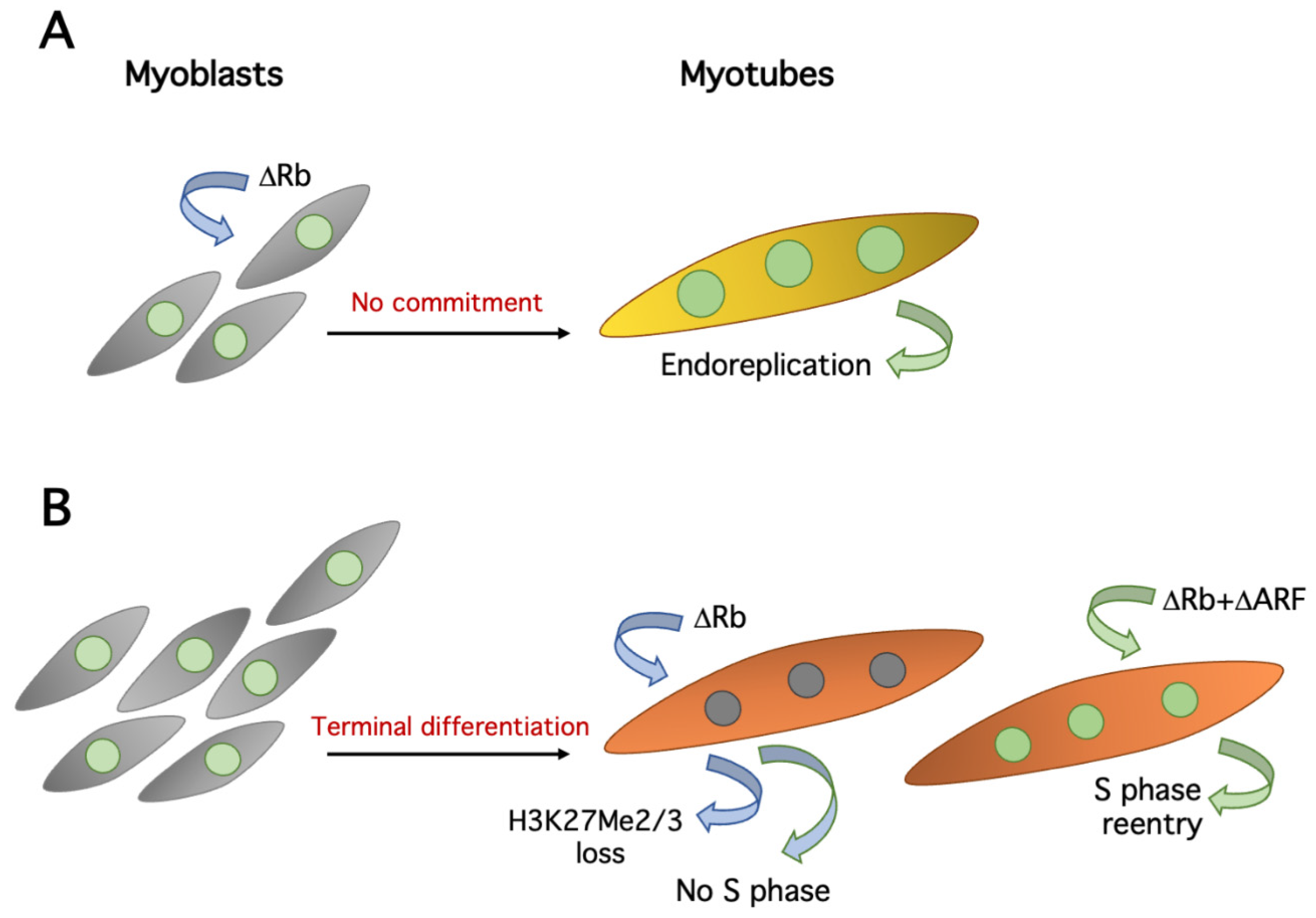
6. Maintenance of the Postmitotic State
7. Cell Cycle-Unrelated Attack Points
8. The Apoptosis Connection
9. Concluding Remarks
9.1. Lack of Molecular Understanding
9.2. Therapeutic Strategies
Author Contributions
Funding
Conflicts of Interest
References
- Johnson, J.; Mohsin, S.; Houser, S.R. Cardiomyocyte Proliferation as a Source of New Myocyte Development in the Adult Heart. Int. J. Mol. Sci. 2021, 22, 7764. [Google Scholar] [CrossRef]
- Joven, A.; Elewa, A.; Simon, A. Model systems for regeneration: Salamanders. Development 2019, 146, dev167700. [Google Scholar] [CrossRef] [Green Version]
- Asfour, H.A.; Allouh, M.Z.; Said, R.S. Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp. Biol. Med. Maywood N. J. 2018, 243, 118–128. [Google Scholar] [CrossRef]
- Rando, T.A.; Blau, H.M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994, 125, 1275–1287. [Google Scholar] [CrossRef] [Green Version]
- Musarò, A.; Carosio, S. Isolation and Culture of Satellite Cells from Mouse Skeletal Muscle. Methods Mol. Biol. 2017, 1553, 155–167. [Google Scholar] [CrossRef]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [Green Version]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef]
- Albini, S.; Puri, P.L. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: It’s time to exchange! Exp. Cell Res. 2010, 316, 3073–3080. [Google Scholar] [CrossRef] [Green Version]
- Forcales, S.V.; Albini, S.; Giordani, L.; Malecova, B.; Cignolo, L.; Chernov, A.; Coutinho, P.; Saccone, V.; Consalvi, S.; Williams, R.; et al. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2012, 31, 301–316. [Google Scholar] [CrossRef]
- Puri, P.L.; Sartorelli, V.; Yang, X.J.; Hamamori, Y.; Ogryzko, V.V.; Howard, B.H.; Kedes, L.; Wang, J.Y.; Graessmann, A.; Nakatani, Y.; et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1997, 1, 35–45. [Google Scholar] [CrossRef]
- Dilworth, F.J.; Seaver, K.J.; Fishburn, A.L.; Htet, S.L.; Tapscott, S.J. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc. Natl. Acad. Sci. USA 2004, 101, 11593–11598. [Google Scholar] [CrossRef] [Green Version]
- Baserga, R. The Biology of Cell Reproduction; Harvard University Press: Cambridge, UK; London, UK, 1985; p. 29. [Google Scholar]
- Tiainen, M.; Pajalunga, D.; Ferrantelli, F.; Soddu, S.; Salvatori, G.; Sacchi, A.; Crescenzi, M. Terminally differentiated skeletal myotubes are not confined in G0, but can enter G1 upon growth factor stimulation. Cell. Growth. Differ. 1996, 7, 1039–1050. [Google Scholar]
- Endo, T.; Nadal-Ginard, B. Transcriptional and posttranscriptional control of c-myc during myogenesis: Its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol. Cell. Biol. 1986, 6, 1412–1421. [Google Scholar] [CrossRef] [Green Version]
- Fogel, M.; Defendi, V. Infection of muscle cultures from various species with oncogenic DNA viruses (SV40 and polyoma). Proc. Natl. Acad. Sci. USA 1967, 58, 967–973. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, D.; Gershon, D. Multinucleated muscle fibres: Induction of DNA synthesis and mitosis by polyoma virus infection. Nature 1967, 215, 421–424. [Google Scholar] [CrossRef]
- Gruen, R.; Graessmann, M.; Graessmann, A.; Fogel, M. Infection of human cells with polyoma virus. Virology 1974, 58, 290–293. [Google Scholar] [CrossRef]
- Endo, T.; Nadal-Ginard, B. SV40 large T antigen induces reentry of terminally differentiated myotubes into the cell cycle. In Cellular and Molecular Biology of Muscle Development; Stockdale, F., Kedes, L., Eds.; Alan R. Liss, Inc.: New York, NY, USA, 1989; pp. 95–104. [Google Scholar]
- Endo, T.; Goto, S. Retinoblastoma gene product Rb accumulates during myogenic differentiation and is deinduced by the expression of SV40 large T antigen. J. Biochem. 1992, 112, 427–430. [Google Scholar] [CrossRef]
- Endo, T.; Nadal-Ginard, B. Reversal of myogenic terminal differentiation by SV40 large T antigen results in mitosis and apoptosis. J. Cell Sci. 1998, 111, 1081–1093. [Google Scholar] [CrossRef]
- Connolly, J.A.; Kiosses, B.W.; Kalnins, V.I. Centrioles are lost as embryonic myoblasts fuse into myotubes in vitro. Eur. J. Cell Biol. 1986, 39, 341–345. [Google Scholar]
- Musa, H.; Orton, C.; Morrison, E.E.; Peckham, M. Microtubule assembly in cultured myoblasts and myotubes following nocodazole induced microtubule depolymerisation. J. Muscle Res. Cell Motil. 2003, 24, 301–308. [Google Scholar] [CrossRef]
- Crescenzi, M.; Soddu, S.; Tato, F. Mitotic cycle reactivation in terminally differentiated cells by adenovirus infection. J. Cell Physiol. 1995, 162, 26–35. [Google Scholar] [CrossRef]
- Shi, Q.; King, R.W. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 2005, 437, 1038–1042. [Google Scholar] [CrossRef]
- Crescenzi, M.; Soddu, S.; Sacchi, A.; Tato’, F. Adenovirus infection induces reentry into the cell cycle of terminally differentiated skeletal muscle cells. Ann. N. Y. Acad. Sci. 1995, 752, 9–18. [Google Scholar] [CrossRef]
- Latella, L.; Sacchi, A.; Crescenzi, M. Long-term fate of terminally differentiated skeletal muscle cells following E1A-initiated cell cycle reactivation. Cell Death Differ. 2000, 7, 145–154. [Google Scholar] [CrossRef]
- Webster, K.A.; Muscat, G.E.; Kedes, L. Adenovirus E1A products suppress myogenic differentiation and inhibit transcription from muscle-specific promoters. Nature 1988, 332, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Sandmöller, A.; Meents, H.; Arnold, H.H. A novel E1A domain mediates skeletal-muscle-specific enhancer repression independently of pRb and p300 binding. Mol. Cell. Biol. 1996, 16, 5846–5856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiainen, M.; Spitkovsky, D.; Jansen-Dürr, P.; Sacchi, A.; Crescenzi, M. Expression of E1A in terminally differentiated muscle cells reactivates the cell cycle and suppresses tissue-specific genes by separable mechanisms. Mol. Cell. Biol. 1996, 16, 5302–5312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, T.; Bober, E.; Arnold, H.H. Inhibition of muscle differentiation by the adenovirus E1a protein: Repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev. 1992, 6, 888–902. [Google Scholar] [CrossRef] [Green Version]
- Caruso, M.; Martelli, F.; Giordano, A.; Felsani, A. Regulation of MyoD gene transcription and protein function by the transforming domains of the adenovirus E1A oncoprotein. Oncogene 1993, 8, 267–278. [Google Scholar]
- Whyte, P.; Buchkovich, K.J.; Horowitz, J.M.; Friend, S.H.; Raybuck, M.; Weinberg, R.A.; Harlow, E. Association between an oncogene and an anti-oncogene: The adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 1988, 334, 124–129. [Google Scholar] [CrossRef]
- Ludlow, J.W.; DeCaprio, J.A.; Huang, C.M.; Lee, W.H.; Paucha, E.; Livingston, D.M. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell 1989, 56, 57–65. [Google Scholar] [CrossRef]
- Dyson, N.; Howley, P.M.; Münger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef]
- Dyson, N.; Buchkovich, K.; Whyte, P.; Harlow, E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell 1989, 58, 249–255. [Google Scholar] [CrossRef]
- Liu, X.; Marmorstein, R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007, 21, 2711–2716. [Google Scholar] [CrossRef] [Green Version]
- White, M.K.; Khalili, K. Interaction of retinoblastoma protein family members with large T-antigen of primate polyomaviruses. Oncogene 2006, 25, 5286–5293. [Google Scholar] [CrossRef] [Green Version]
- Sacco, A.; Siepi, F.; Crescenzi, M. HPV E7 expression in skeletal muscle cells distinguishes initiation of the postmitotic state from its maintenance. Oncogene 2003, 22, 4027–4034. [Google Scholar] [CrossRef] [Green Version]
- Latella, L.; Sacco, A.; Pajalunga, D.; Tiainen, M.; Macera, D.; D’Angelo, M.; Felici, A.; Sacchi, A.; Crescenzi, M. Reconstitution of cyclin D1-associated kinase activity drives terminally differentiated cells into the cell cycle. Mol. Cell Biol. 2001, 21, 5631–5643. [Google Scholar] [CrossRef] [Green Version]
- Lukas, J.; Herzinger, T.; Hansen, K.; Moroni, M.C.; Resnitzky, D.; Helin, K.; Reed, S.I.; Bartek, J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997, 11, 1479–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connell-Crowley, L.; Elledge, S.J.; Harper, J.W. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr. Biol. 1998, 8, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.M.; Wei, Q.; Zhao, X.; Paterson, B.M. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999, 18, 926–933. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.M.; Zhao, X.; Wei, Q.; Paterson, B.M. Direct inhibition of G1 cdk kinase activity by MyoD promotes myoblast cell cycle withdrawal and terminal differentiation. EMBO J. 1999, 18, 6983–6993. [Google Scholar] [CrossRef] [Green Version]
- Parker, S.B.; Eichele, G.; Zhang, P.; Rawls, A.; Sands, A.T.; Bradley, A.; Olson, E.N.; Harper, J.W.; Elledge, S.J. p53-independent expression of p21Cip1 in muscle and other terminally differentiated cells. Science 1995, 267, 1024–1027. [Google Scholar] [CrossRef]
- Missero, C.; Di Cunto, F.; Kiyokawa, H.; Koff, A.; Dotto, G.P. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996, 10, 3065–3075. [Google Scholar] [CrossRef] [Green Version]
- Durand, B.; Gao, F.-B.; Raff, M. Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. EMBO J. 1997, 16, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Phelps, D.E.; Xiong, Y. Regulation of cyclin-dependent kinase 4 during adipogenesis involves switching of cyclin D subunits and concurrent binding of p18INK4c and p27Kip1. Cell Growth Differ. 1998, 9, 595–610. [Google Scholar]
- Tourigny, M.R.; Ursini-Siegel, J.; Lee, H.; Toellner, K.M.; Cunningham, A.F.; Franklin, D.S.; Ely, S.; Chen, M.; Qin, X.F.; Xiong, Y.; et al. CDK inhibitor p18(INK4c) is required for the generation of functional plasma cells. Immunity 2002, 17, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Buttitta, L.A.; Katzaroff, A.J.; Perez, C.L.; de la Cruz, A.; Edgar, B.A. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 2007, 12, 631–643. [Google Scholar] [CrossRef] [Green Version]
- Oesterle, E.C.; Chien, W.M.; Campbell, S.; Nellimarla, P.; Fero, M.L. p27 (Kip1) is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell Cycle 2011, 10, 1237–1248. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Wang, J.; Andres, V.; Smith, R.C.; Walsh, K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 1995, 15, 3823–3829. [Google Scholar] [CrossRef] [Green Version]
- Halevy, O.; Novitch, B.G.; Spicer, D.B.; Skapek, S.X.; Rhee, J.; Hannon, G.J.; Beach, D.; Lassar, A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 1995, 267, 1018–1021. [Google Scholar] [CrossRef]
- Zabludoff, S.D.; Csete, M.; Wagner, R.; Yu, X.; Wold, B.J. p27Kip1 is expressed transiently in developing myotomes and enhances myogenesis. Cell Growth Differ. 1998, 9, 1–11. [Google Scholar]
- Zhang, P.; Wong, C.; Liu, D.; Finegold, M.; Harper, J.W.; Elledge, S.J. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999, 13, 213–224. [Google Scholar] [CrossRef]
- Messina, G.; Blasi, C.; La Rocca, S.A.; Pompili, M.; Calconi, A.; Grossi, M. p27Kip1 acts downstream of N-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Mol. Biol. Cell 2005, 16, 1469–1480. [Google Scholar] [CrossRef] [Green Version]
- Pajalunga, D.; Mazzola, A.; Salzano, A.M.; Biferi, M.G.; De Luca, G.; Crescenzi, M. Critical requirement for cell cycle inhibitors in sustaining nonproliferative states. J. Cell Biol. 2007, 176, 807–818. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, D.; Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977, 270, 725–727. [Google Scholar] [CrossRef]
- Blau, H.M.; Pavlath, G.K.; Hardeman, E.C.; Chiu, C.-P.; Silberstein, L.; Webster, S.G.; Miller, S.C.; Webster, C. Plasticity of the differentiated state. Science 1985, 230, 758–766. [Google Scholar] [CrossRef] [Green Version]
- Cenciarelli, C.; De Santa, F.; Puri, P.L.; Mattei, E.; Ricci, L.; Bucci, F.; Felsani, A.; Caruso, M. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol. Cell Biol. 1999, 19, 5203–5217. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Schneider, J.W.; Condorelli, G.; Kaushal, S.; Mahdavi, V.; Nadal-Ginard, B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 1993, 72, 309–324. [Google Scholar] [CrossRef]
- Kouzarides, T. Transcriptional regulation by the retinoblastoma protein. Trends Cell Biol. 1993, 3, 211–213. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Sorrentino, V.; Pepperkok, R.; Davis, R.L.; Ansorge, W.; Philipson, L. Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature 1990, 345, 813–815. [Google Scholar] [CrossRef]
- Crescenzi, M.; Fleming, T.P.; Lassar, A.B.; Weintraub, H.; Aaronson, S.A. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc. Natl. Acad. Sci. USA 1990, 87, 8442–8446. [Google Scholar] [CrossRef] [Green Version]
- Li, F.Q.; Coonrod, A.; Horwitz, M. Selection of a dominant negative retinoblastoma protein (RB) inhibiting satellite myoblast differentiation implies an indirect interaction between MyoD and RB. Mol. Cell Biol. 2000, 20, 5129–5139. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.W.; Gu, W.; Zhu, L.; Mahdavi, V.; Nadal-Ginard, B. Reversal of terminal differentiation mediated by p107 in Rb-/- muscle cells. Science 1994, 264, 1467–1471. [Google Scholar] [CrossRef]
- Novitch, B.G.; Mulligan, G.J.; Jacks, T.; Lassar, A.B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 1996, 135, 441–456. [Google Scholar] [CrossRef]
- Okazaki, K.; Holtzer, H. Myogenesis: Fusion, myosin synthesis, and the mitotic cycle. Proc. Natl. Acad. Sci. USA 1966, 56, 1484–1490. [Google Scholar] [CrossRef] [Green Version]
- Zacksenhaus, E.; Jiang, Z.; Chung, D.; Marth, J.D.; Phillips, R.A.; Gallie, B.L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996, 10, 3051–3064. [Google Scholar] [CrossRef] [Green Version]
- Sage, J.; Miller, A.L.; Perez-Mancera, P.A.; Wysocki, J.M.; Jacks, T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 2003, 424, 223–228. [Google Scholar] [CrossRef]
- Huh, M.S.; Parker, M.H.; Scime, A.; Parks, R.; Rudnicki, M.A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 2004, 166, 865–876. [Google Scholar] [CrossRef] [Green Version]
- Camarda, G.; Siepi, F.; Pajalunga, D.; Bernardini, C.; Rossi, R.; Montecucco, A.; Meccia, E.; Crescenzi, M. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J. Cell Biol. 2004, 167, 417–423. [Google Scholar] [CrossRef]
- Blais, A.; van Oevelen, C.J.; Margueron, R.; Acosta-Alvear, D.; Dynlacht, B.D. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol. 2007, 179, 1399–1412. [Google Scholar] [CrossRef] [Green Version]
- Pajcini, K.V.; Corbel, S.Y.; Sage, J.; Pomerantz, J.H.; Blau, H.M. Transient Inactivation of Rb and ARF Yields Regenerative Cells from Postmitotic Mammalian Muscle. Cell Stem Cell 2010, 7, 198–213. [Google Scholar] [CrossRef] [Green Version]
- Andres, V.; Walsh, K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 1996, 132, 657–666. [Google Scholar] [CrossRef] [Green Version]
- Pajalunga, D.; Puggioni, E.M.; Mazzola, A.; Leva, V.; Montecucco, A.; Crescenzi, M. DNA replication is intrinsically hindered in terminally differentiated myotubes. PLoS ONE 2010, 5, e11559. [Google Scholar] [CrossRef] [Green Version]
- Mastroyiannopoulos, N.P.; Nicolaou, P.; Anayasa, M.; Uney, J.B.; Phylactou, L.A. Down-regulation of myogenin can reverse terminal muscle cell differentiation. PLoS ONE 2012, 7, e29896. [Google Scholar] [CrossRef] [Green Version]
- Schwab, I.A.; Luger, O. Reinitiation of DNA synthesis in postmitotic nuclei of myotubes by virus-mediated fusion with embryonic fibroblasts. Differentiation 1980, 16, 93–99. [Google Scholar] [CrossRef]
- Clegg, C.H.; Hauschka, S.D. Heterokaryon analysis of muscle differentiation: Regulation of the postmitotic state. J. Cell Biol. 1987, 105, 937–947. [Google Scholar] [CrossRef] [Green Version]
- Ringertz, N.R.; Savage, R.E. Cell Hybrids; Academic Press: New York, NY, USA; San Francisco, CA, USA; London, UK, 1976. [Google Scholar]
- Rao, P.N.; Johnson, R.T. Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature 1970, 225, 159–164. [Google Scholar] [CrossRef]
- Rosania, G.R.; Chang, Y.T.; Perez, O.; Sutherlin, D.; Dong, H.; Lockhart, D.J.; Schultz, P.G. Myoseverin, a microtubule-binding molecule with novel cellular effects. Nat. Biotechnol. 2000, 18, 304–308. [Google Scholar] [CrossRef]
- Perez, O.D.; Chang, Y.T.; Rosania, G.; Sutherlin, D.; Schultz, P.G. Inhibition and reversal of myogenic differentiation by purine-based microtubule assembly inhibitors. Chem. Biol. 2002, 9, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Duckmanton, A.; Kumar, A.; Chang, Y.T.; Brockes, J.P. A single-cell analysis of myogenic dedifferentiation induced by small molecules. Chem. Biol. 2005, 12, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.W.; Williams, D.R. Novel chemically defined approach to produce multipotent cells from terminally differentiated tissue syncytia. ACS Chem. Biol. 2011, 6, 553–562. [Google Scholar] [CrossRef]
- Kim, W.H.; Jung, D.W.; Kim, J.; Im, S.H.; Hwang, S.Y.; Williams, D.R. Small Molecules That Recapitulate the Early Steps of Urodele Amphibian Limb Regeneration and Confer Multipotency. ACS Chem. Biol. 2012, 7, 732–743. [Google Scholar] [CrossRef]
- Simon, H.G.; Nelson, C.; Goff, D.; Laufer, E.; Morgan, B.A.; Tabin, C. Differential expression of myogenic regulatory genes and Msx-1 during dedifferentiation and redifferentiation of regenerating amphibian limbs. Dev. Dyn. 1995, 202, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Engeler, R.; Constantinescu, S.; Kokkaliaris, K.D.; Dimitrakopoulos, C.; Schroeder, T.; Beerenwinkel, N.; Paro, R. Ectopic expression of Msx2 in mammalian myotubes recapitulates aspects of amphibian muscle dedifferentiation. Stem Cell Res. 2015, 15, 542–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odelberg, S.J.; Kollhoff, A.; Keating, M.T. Dedifferentiation of Mammalian Myotubes Induced by msx1. Cell 2000, 103, 1099–1109. [Google Scholar] [CrossRef] [Green Version]
- McGann, C.J.; Odelberg, S.J.; Keating, M.T. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc. Natl. Acad. Sci. USA 2001, 98, 13699–13704. [Google Scholar] [CrossRef] [Green Version]
- Meech, R.; Gomez, M.; Woolley, C.; Barro, M.; Hulin, J.A.; Walcott, E.C.; Delgado, J.; Makarenkova, H.P. The homeobox transcription factor Barx2 regulates plasticity of young primary myofibers. PLoS ONE 2010, 5, e11612. [Google Scholar] [CrossRef] [Green Version]
- Paliwal, P.; Conboy, I.M. Inhibitors of tyrosine phosphatases and apoptosis reprogram lineage-marked differentiated muscle to myogenic progenitor cells. Chem. Biol. 2011, 18, 1153–1166. [Google Scholar] [CrossRef] [Green Version]
- Hjiantoniou, E.; Anayasa, M.; Nicolaou, P.; Bantounas, I.; Saito, M.; Iseki, S.; Uney, J.B.; Phylactou, L.A. Twist induces reversal of myotube formation. Differentiation 2008, 76, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Loof, S.; Borg, P.; Nader, G.A.; Blau, H.M.; Simon, A. Turning terminally differentiated skeletal muscle cells into regenerative progenitors. Nat. Commun. 2015, 6, 7916. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pajalunga, D.; Crescenzi, M. Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes. Cells 2021, 10, 2753. https://doi.org/10.3390/cells10102753
Pajalunga D, Crescenzi M. Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes. Cells. 2021; 10(10):2753. https://doi.org/10.3390/cells10102753
Chicago/Turabian StylePajalunga, Deborah, and Marco Crescenzi. 2021. "Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes" Cells 10, no. 10: 2753. https://doi.org/10.3390/cells10102753
APA StylePajalunga, D., & Crescenzi, M. (2021). Restoring the Cell Cycle and Proliferation Competence in Terminally Differentiated Skeletal Muscle Myotubes. Cells, 10(10), 2753. https://doi.org/10.3390/cells10102753





