Mapping the Proximity Interaction Network of STIM1 Reveals New Mechanisms of Cytoskeletal Regulation
Abstract
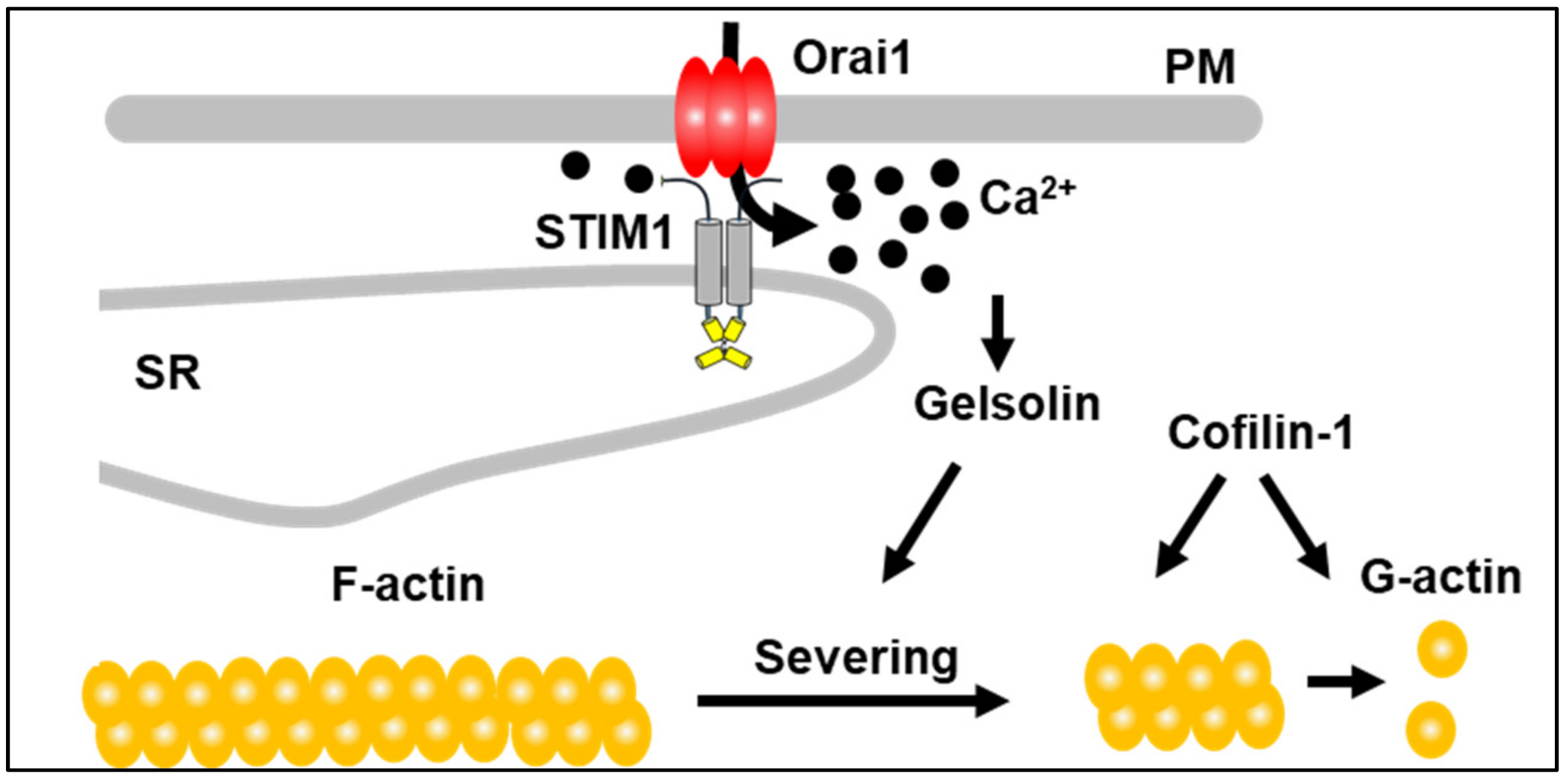
:1. Introduction
2. Materials and Methods
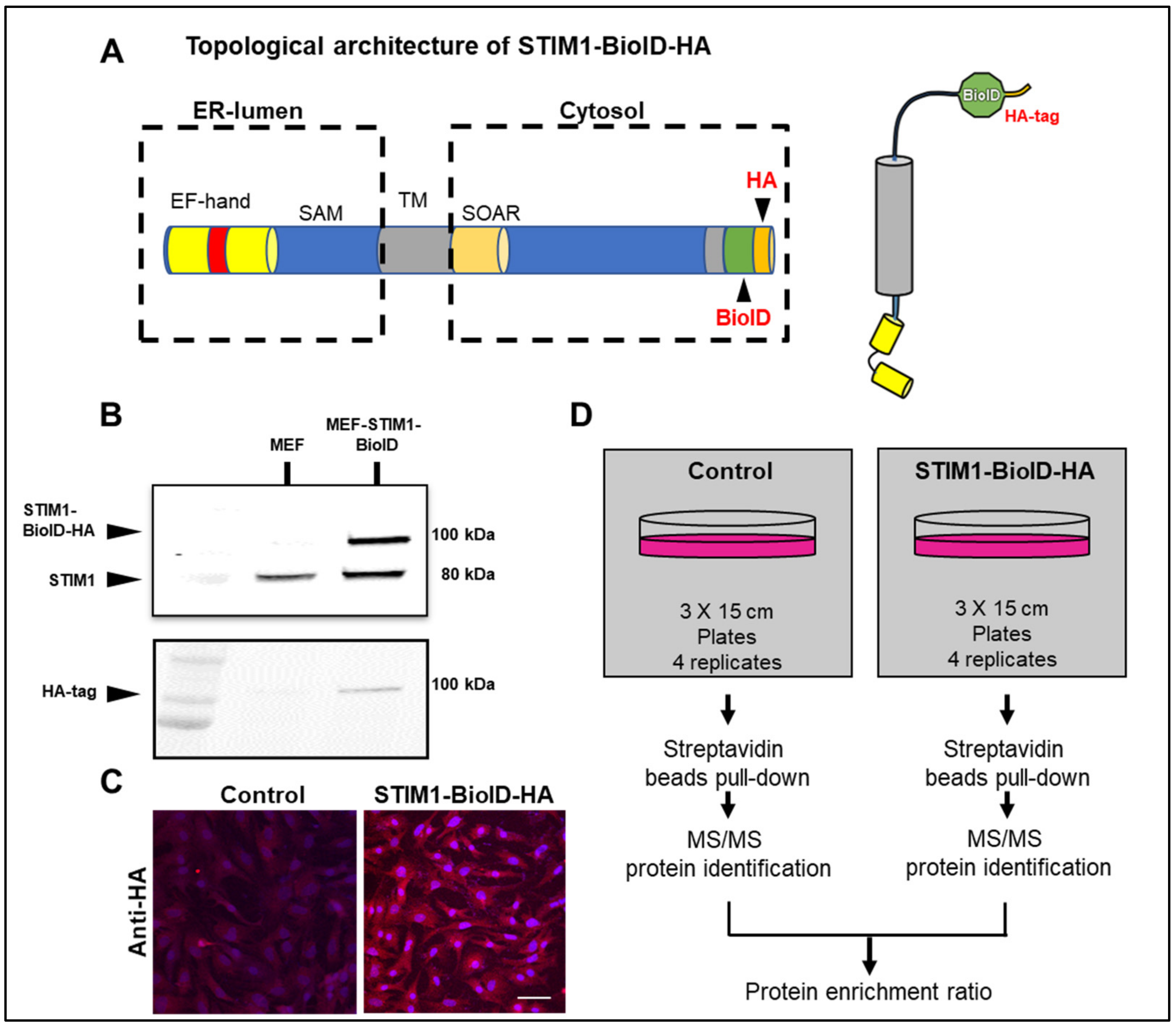
2.1. Molecular Cloning and Generation of Stable Cell Lines
2.2. Neonatal Cardiomyocytes Isolation
2.3. Mass Spectrometry
2.4. Immunoblots and Immunofluorescence
2.5. Determination of F-actin/G-actin Ratio
2.6. Data Analysis
3. Results
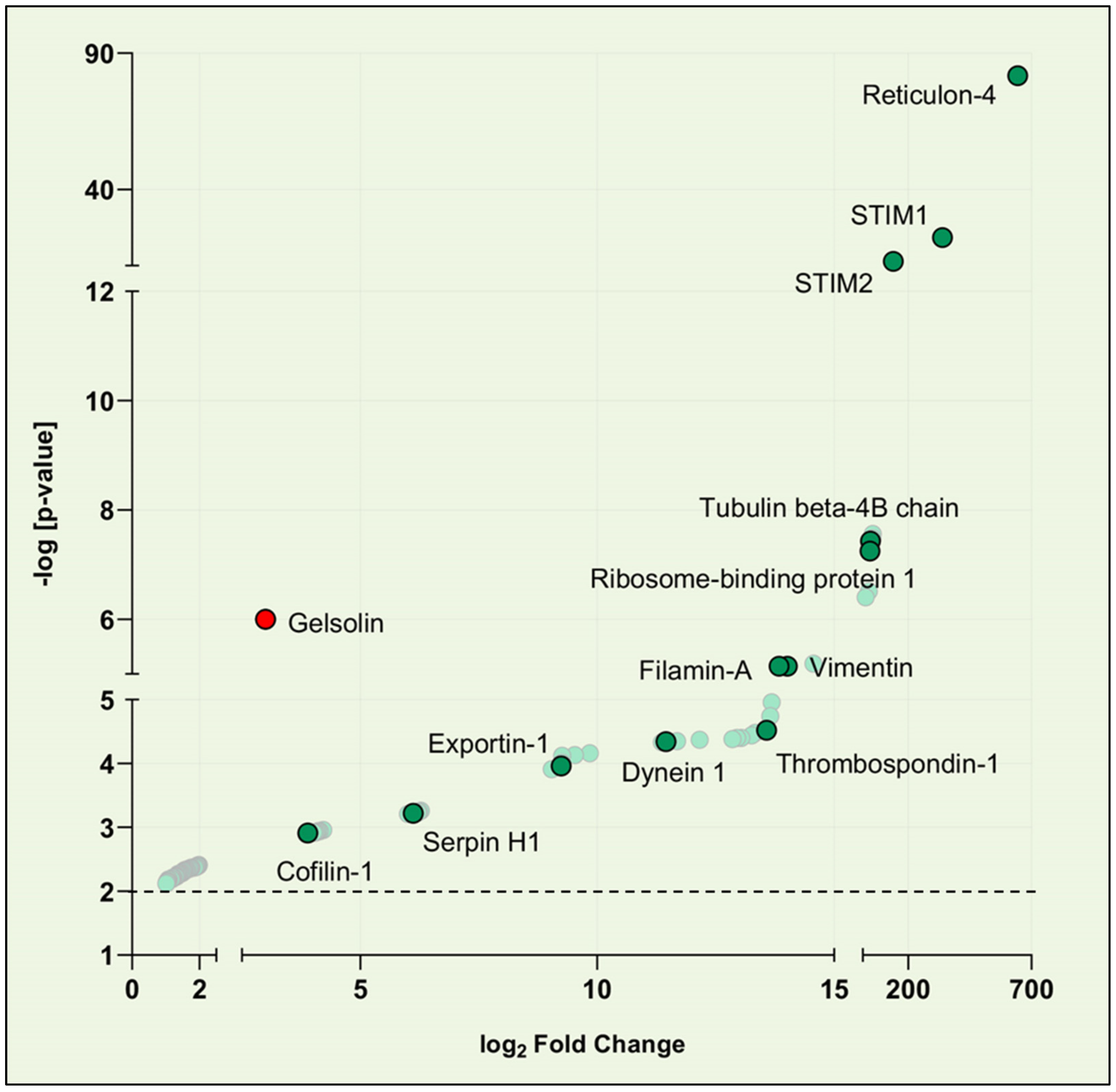
3.1. Identification of STIM1 Interactome
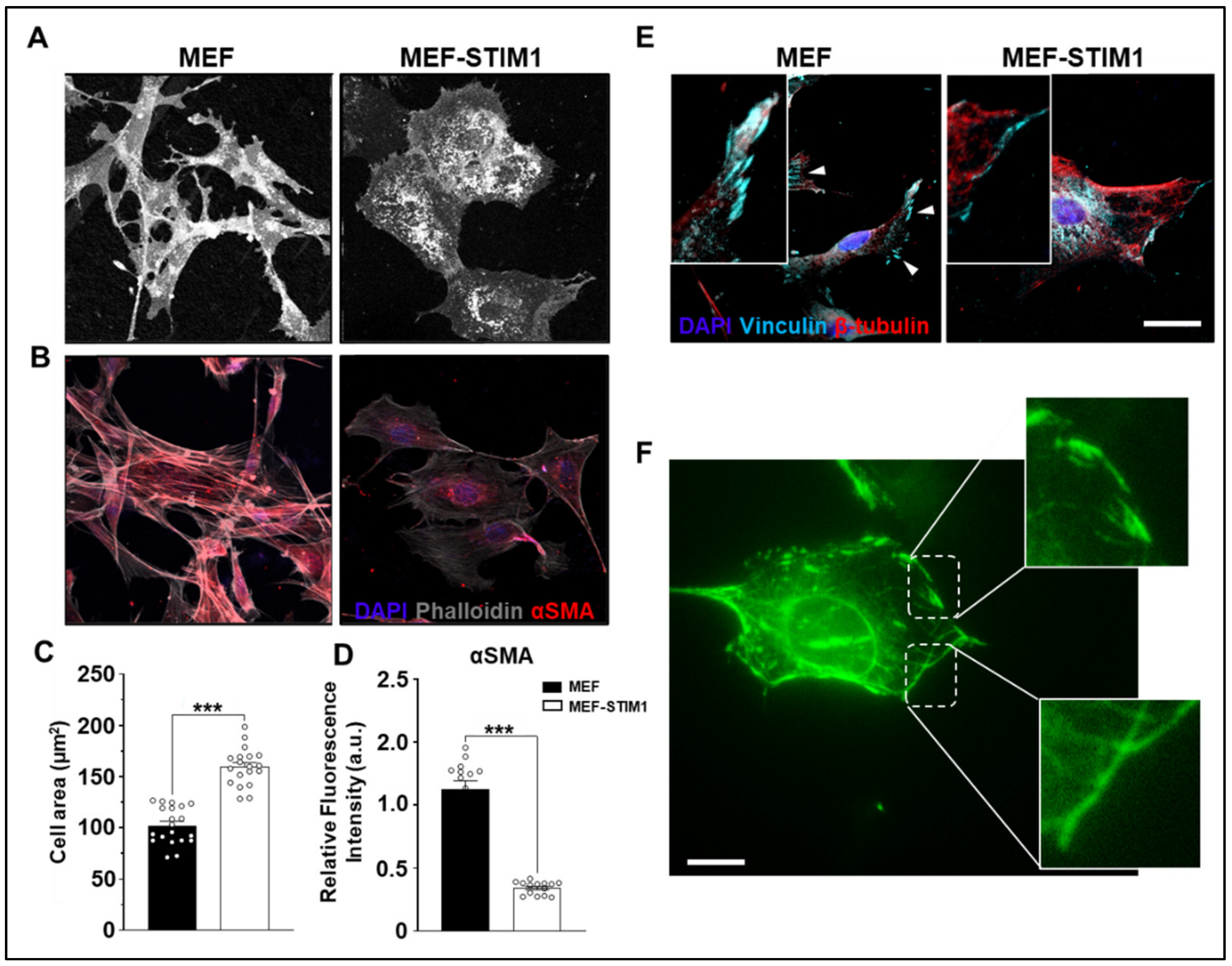
3.2. STIM1 Overexpression Alters Cell Size and Perturbs the Cytoskeleton Network
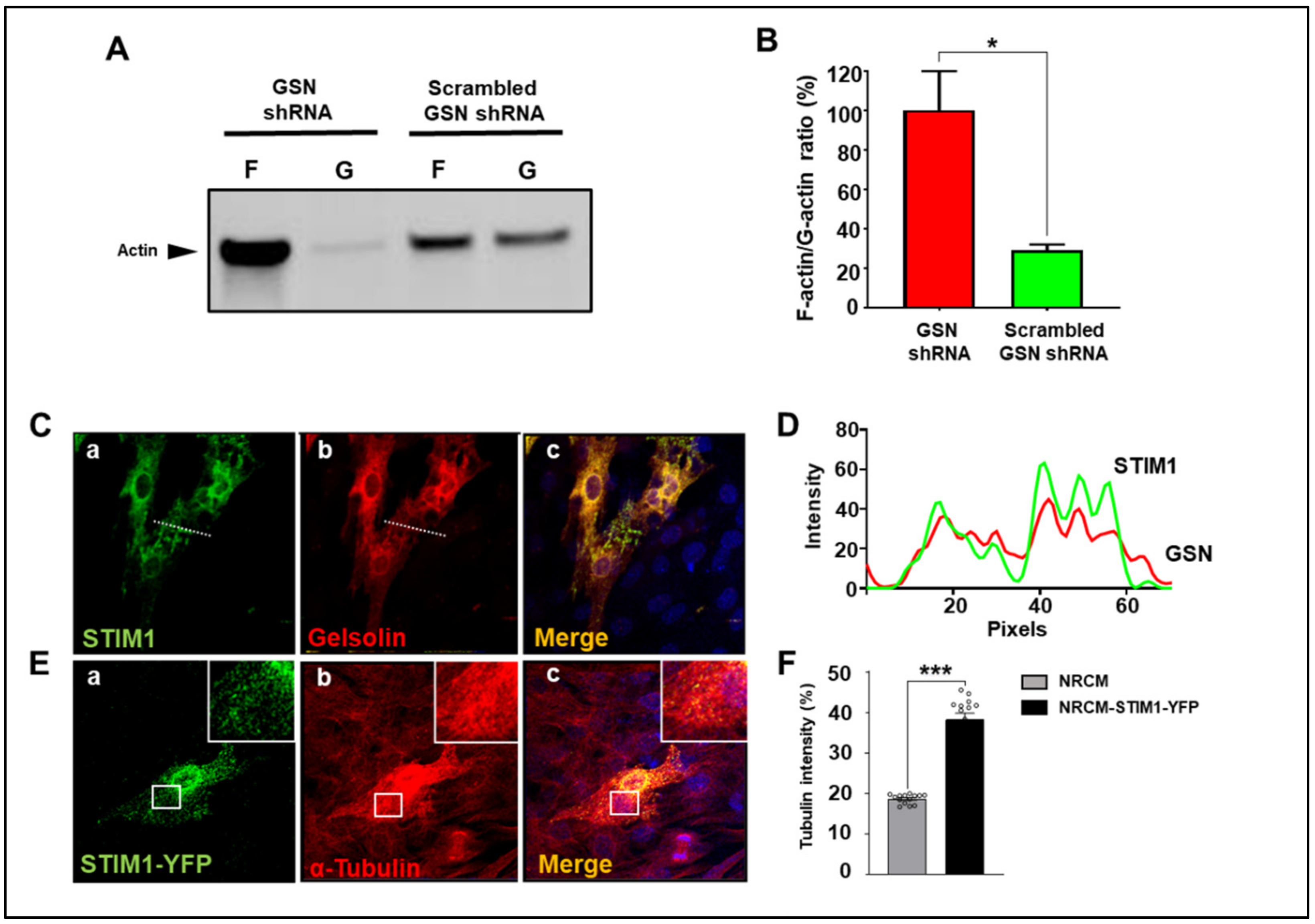
3.3. STIM1 and GSN Actively Remodel Actin in Neonatal Cardiomyocytes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E., Jr.; Meyer, T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005, 15, 1235–1241. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.T.; Manji, S.S.; Parker, N.J.; Hancock, M.S.; Van Stekelenburg, L.; Eid, J.P.; Senior, P.V.; Kazenwadel, J.S.; Shandala, T.; Saint, R.; et al. Identification and characterization of the STIM (stromal interaction molecule) gene family: Coding for a novel class of transmembrane proteins. Biochem. J. 2001, 357, 673–685. [Google Scholar] [CrossRef]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Decuypere, J.P.; Monaco, G.; Kiviluoto, S.; Oh-hora, M.; Luyten, T.; De Smedt, H.; Parys, J.B.; Missiaen, L.; Bultynck, G. STIM1, but not STIM2, is required for proper agonist-induced Ca2+ signaling. Cell Calcium 2010, 48, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Soboloff, J.; Rothberg, B.S.; Madesh, M.; Gill, D.L. STIM proteins: Dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012, 13, 549–565. [Google Scholar] [CrossRef] [Green Version]
- Krapivinsky, G.; Krapivinsky, L.; Stotz, S.C.; Manasian, Y.; Clapham, D.E. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc. Natl. Acad. Sci. USA 2011, 108, 19234–19239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palty, R.; Raveh, A.; Kaminsky, I.; Meller, R.; Reuveny, E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 2012, 149, 425–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigoriev, I.; Gouveia, S.M.; van der Vaart, B.; Demmers, J.; Smyth, J.T.; Honnappa, S.; Splinter, D.; Steinmetz, M.O.; Putney, J.W., Jr.; Hoogenraad, C.C.; et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 2008, 18, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Jing, J.; He, L.; Sun, A.; Quintana, A.; Ding, Y.; Ma, G.; Tan, P.; Liang, X.; Zheng, X.; Chen, L.; et al. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nat. Cell Biol. 2015, 17, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Jensen, S.C.; Noble, K.A.; Kc, B.; Roux, K.H.; Motamedchaboki, K.; Roux, K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27, 1188–1196. [Google Scholar] [CrossRef]
- Karnabi, E.; Qu, Y.; Mancarella, S.; Yue, Y.; Wadgaonkar, R.; Boutjdir, M. Silencing of Cav1.2 gene in neonatal cardiomyocytes by lentiviral delivered shRNA. Biochem. Biophys. Res. Commun. 2009, 384, 409–414. [Google Scholar] [CrossRef] [Green Version]
- Parks, C.; Alam, M.A.; Sullivan, R.; Mancarella, S. STIM1-dependent Ca(2+) microdomains are required for myofilament remodeling and signaling in the heart. Sci. Rep. 2016, 6, 25372. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Birendra, K.C.; Zhu, W.; Motamedchaboki, K.; Doye, V.; Roux, K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. USA 2014, 111, E2453–E2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oertle, T.; van der Haar, M.E.; Bandtlow, C.E.; Robeva, A.; Burfeind, P.; Buss, A.; Huber, A.B.; Simonen, M.; Schnell, L.; Brosamle, C.; et al. Nogo-A inhibits neurite outgrowth and cell spreading with three discrete regions. J. Neurosci. 2003, 23, 5393–5406. [Google Scholar] [CrossRef] [PubMed]
- Voeltz, G.K.; Prinz, W.A.; Shibata, Y.; Rist, J.M.; Rapoport, T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 2006, 124, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Craene, J.O.; Coleman, J.; Estrada de Martin, P.; Pypaert, M.; Anderson, S.; Yates, J.R., 3rd; Ferro-Novick, S.; Novick, P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol. Biol. Cell 2006, 17, 3009–3020. [Google Scholar] [CrossRef] [Green Version]
- Jozsef, L.; Tashiro, K.; Kuo, A.; Park, E.J.; Skoura, A.; Albinsson, S.; Rivera-Molina, F.; Harrison, K.D.; Iwakiri, Y.; Toomre, D.; et al. Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry. J. Biol. Chem. 2014, 289, 9380–9395. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, N.; Oritani, K.; Saito, K.; Yokota, T.; Ichii, M.; Sudo, T.; Fujita, N.; Nakajima, K.; Okada, M.; Kanakura, Y. Identification of functional domains and novel binding partners of STIM proteins. J. Cell. Biochem. 2011, 112, 147–156. [Google Scholar] [CrossRef]
- Duquette, M.; Nadler, M.; Okuhara, D.; Thompson, J.; Shuttleworth, T.; Lawler, J. Members of the thrombospondin gene family bind stromal interaction molecule 1 and regulate calcium channel activity. Matrix Biol 2014, 37, 15–24. [Google Scholar] [CrossRef]
- Ayscough, K.R. In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 1998, 10, 102–111. [Google Scholar] [CrossRef]
- Gremm, D.; Wegner, A. Gelsolin as a calcium-regulated actin filament-capping protein. Eur. J. Biochem. 2000, 267, 4339–4345. [Google Scholar] [CrossRef]
- Bryan, J.; Coluccio, L.M. Kinetic analysis of F-actin depolymerization in the presence of platelet gelsolin and gelsolin-actin complexes. J. Cell Biol. 1985, 101, 1236–1244. [Google Scholar] [CrossRef] [Green Version]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.B.; Zhabyeyev, P.; Chen, X.; Wang, F.; Paul, M.; Fan, D.; McLean, B.A.; Basu, R.; Zhang, P.; Shah, S.; et al. PI3Kalpha-regulated gelsolin activity is a critical determinant of cardiac cytoskeletal remodeling and heart disease. Nat. Commun. 2018, 9, 5390. [Google Scholar] [CrossRef] [PubMed]
- Conte, E.; Pannunzio, A.; Imbrici, P.; Camerino, G.M.; Maggi, L.; Mora, M.; Gibertini, S.; Cappellari, O.; De Luca, A.; Coluccia, M.; et al. Gain-of-Function STIM1 L96V Mutation Causes Myogenesis Alteration in Muscle Cells From a Patient Affected by Tubular Aggregate Myopathy. Front. Cell Dev. Biol 2021, 9, 635063. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Shi, Y.; Chen, Y.; Sun, M.; Sader, S.; Maekawa, Y.; Arab, S.; Dawood, F.; Chen, M.; De Couto, G.; et al. Gelsolin regulates cardiac remodeling after myocardial infarction through DNase I-mediated apoptosis. Circ. Res. 2009, 104, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hojayev, B.; Jiang, N.; Wang, Z.V.; Tandan, S.; Rakalin, A.; Rothermel, B.A.; Gillette, T.G.; Hill, J.A. STIM1-dependent store-operated Ca(2)(+) entry is required for pathological cardiac hypertrophy. J. Mol. Cell Cardiol. 2012, 52, 136–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichavaram, P.; Yin, W.; Evanson, K.W.; Jaggar, J.H.; Mancarella, S. Elevated plasma catecholamines functionally compensate for the reduced myogenic tone in smooth muscle STIM1 knockout mice but with deleterious cardiac effects. Cardiovasc. Res. 2018, 114, 668–678. [Google Scholar] [CrossRef]
- Hartzell, C.A.; Jankowska, K.I.; Burkhardt, J.K.; Lewis, R.S. Calcium influx through CRAC channels controls actin organization and dynamics at the immune synapse. Elife 2016, 5. [Google Scholar] [CrossRef]
- Cooper, J.A.; Bryan, J.; Schwab, B., 3rd; Frieden, C.; Loftus, D.J.; Elson, E.L. Microinjection of Gelsolin into living cells. J. Cell Biol. 1987, 104, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Szatmari, D.; Xue, B.; Kannan, B.; Burtnick, L.D.; Bugyi, B.; Nyitrai, M.; Robinson, R.C. ATP competes with PIP2 for binding to Gelsolin. PLoS ONE 2018, 13, e0201826. [Google Scholar] [CrossRef]
- Schober, R.; Waldherr, L.; Schmidt, T.; Graziani, A.; Stilianu, C.; Legat, L.; Groschner, K.; Schindl, R. STIM1 and Orai1 regulate Ca(2+) microdomains for activation of transcription. Biochim Biophys Acta Mol. Cell Res. 2019, 1866, 1079–1091. [Google Scholar] [CrossRef]
- Southwick, F.S. Gelsolin and ADF/cofilin enhance the actin dynamics of motile cells. Proc. Natl. Acad. Sci. USA 2000, 97, 6936–6938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redondo, P.C.; Harper, M.T.; Rosado, J.A.; Sage, S.O. A role for cofilin in the activation of store-operated calcium entry by de novo conformational coupling in human platelets. Blood 2006, 107, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Schafer, C.; Rymarczyk, G.; Ding, L.; Kirber, M.T.; Bolotina, V.M. Role of molecular determinants of store-operated Ca(2+) entry (Orai1, phospholipase A2 group 6, and STIM1) in focal adhesion formation and cell migration. J. Biol. Chem. 2012, 287, 40745–40757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litwin, M.; Nowak, D.; Mazur, A.J.; Baczynska, D.; Mannherz, H.G.; Malicka-Blaszkiewicz, M. Gelsolin affects the migratory ability of human colon adenocarcinoma and melanoma cells. Life Sci. 2012, 90, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Shynlova, O.; Chow, M.; Lye, S.J. Expression and organization of basement membranes and focal adhesion proteins in pregnant myometrium is regulated by uterine stretch. Reprod. Sci. 2009, 16, 960–969. [Google Scholar] [CrossRef]
- Czarnowski, A.; Papp, S.; Szaraz, P.; Opas, M. Calreticulin affects cell adhesiveness through differential phosphorylation of insulin receptor substrate-1. Cell Mol. Biol. Lett. 2014, 19, 77–97. [Google Scholar] [CrossRef]
- Kostin, S.; Scholz, D.; Shimada, T.; Maeno, Y.; Mollnau, H.; Hein, S.; Schaper, J. The internal and external protein scaffold of the T-tubular system in cardiomyocytes. Cell Tissue Res. 1998, 294, 449–460. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, J.J.; Huang, X.Y. Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, H.; Pan, T.; Li, L.; Li, J.; Yang, H. STIM1 silencing inhibits the migration and invasion of A549 cells. Mol. Med. Rep. 2017, 16, 3283–3289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, F.C.; Seki, A.; Yang, H.W.; Hayer, A.; Carrasco, S.; Malmersjo, S.; Meyer, T. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 2014, 16, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azuma, T.; Witke, W.; Stossel, T.P.; Hartwig, J.H.; Kwiatkowski, D.J. Gelsolin is a downstream effector of rac for fibroblast motility. EMBO J. 1998, 17, 1362–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, S.; Kostin, S.; Heling, A.; Maeno, Y.; Schaper, J. The role of the cytoskeleton in heart failure. Cardiovasc. Res. 2000, 45, 273–278. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gammons, J.; Halpage, J.; Mancarella, S. Mapping the Proximity Interaction Network of STIM1 Reveals New Mechanisms of Cytoskeletal Regulation. Cells 2021, 10, 2701. https://doi.org/10.3390/cells10102701
Gammons J, Halpage J, Mancarella S. Mapping the Proximity Interaction Network of STIM1 Reveals New Mechanisms of Cytoskeletal Regulation. Cells. 2021; 10(10):2701. https://doi.org/10.3390/cells10102701
Chicago/Turabian StyleGammons, Jesse, Janith Halpage, and Salvatore Mancarella. 2021. "Mapping the Proximity Interaction Network of STIM1 Reveals New Mechanisms of Cytoskeletal Regulation" Cells 10, no. 10: 2701. https://doi.org/10.3390/cells10102701
APA StyleGammons, J., Halpage, J., & Mancarella, S. (2021). Mapping the Proximity Interaction Network of STIM1 Reveals New Mechanisms of Cytoskeletal Regulation. Cells, 10(10), 2701. https://doi.org/10.3390/cells10102701




