SEA and GATOR 10 Years Later
Abstract
1. Introduction
2. Discovery of the SEA Complex
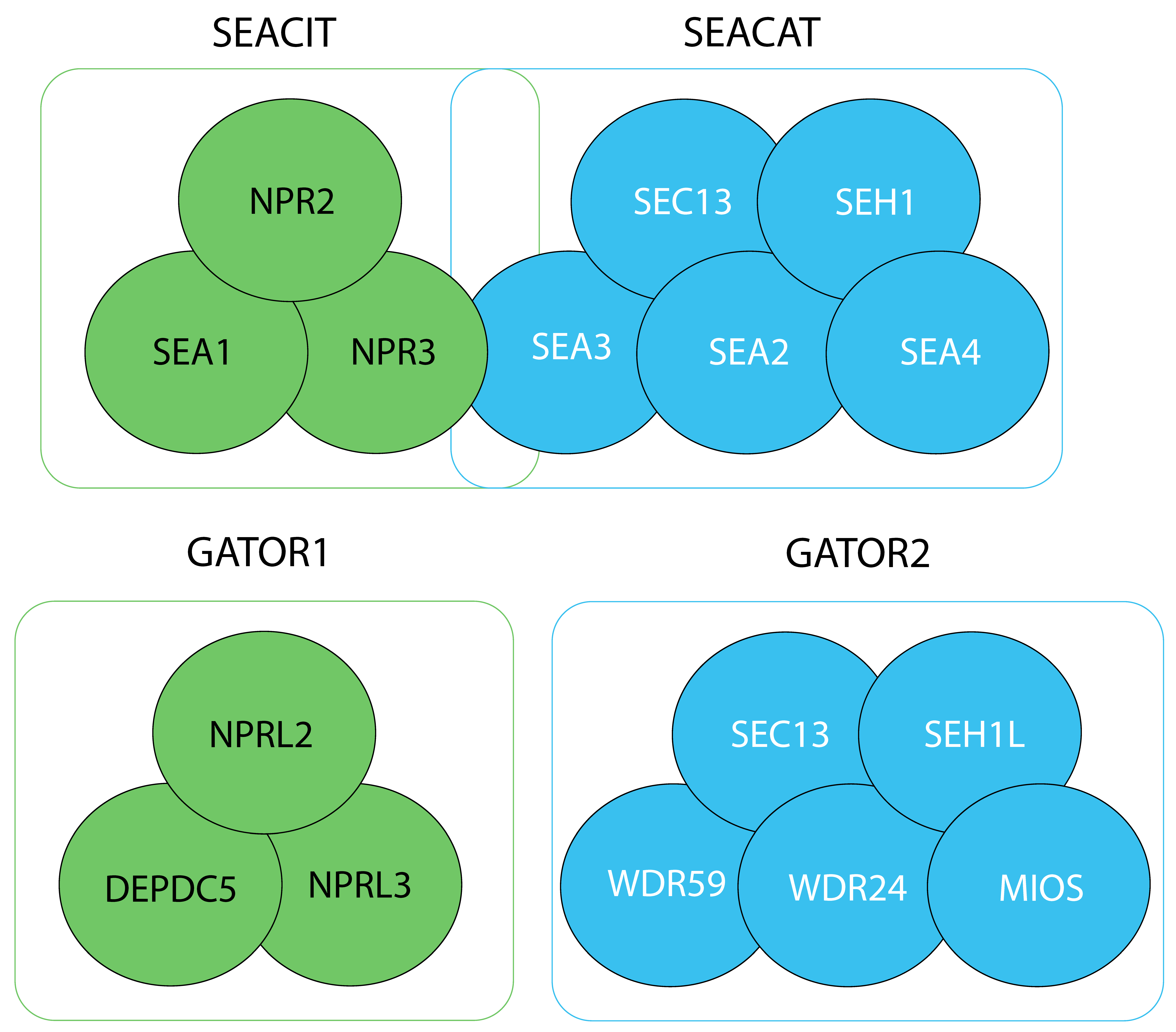
3. SEA/GATOR Nomenclature
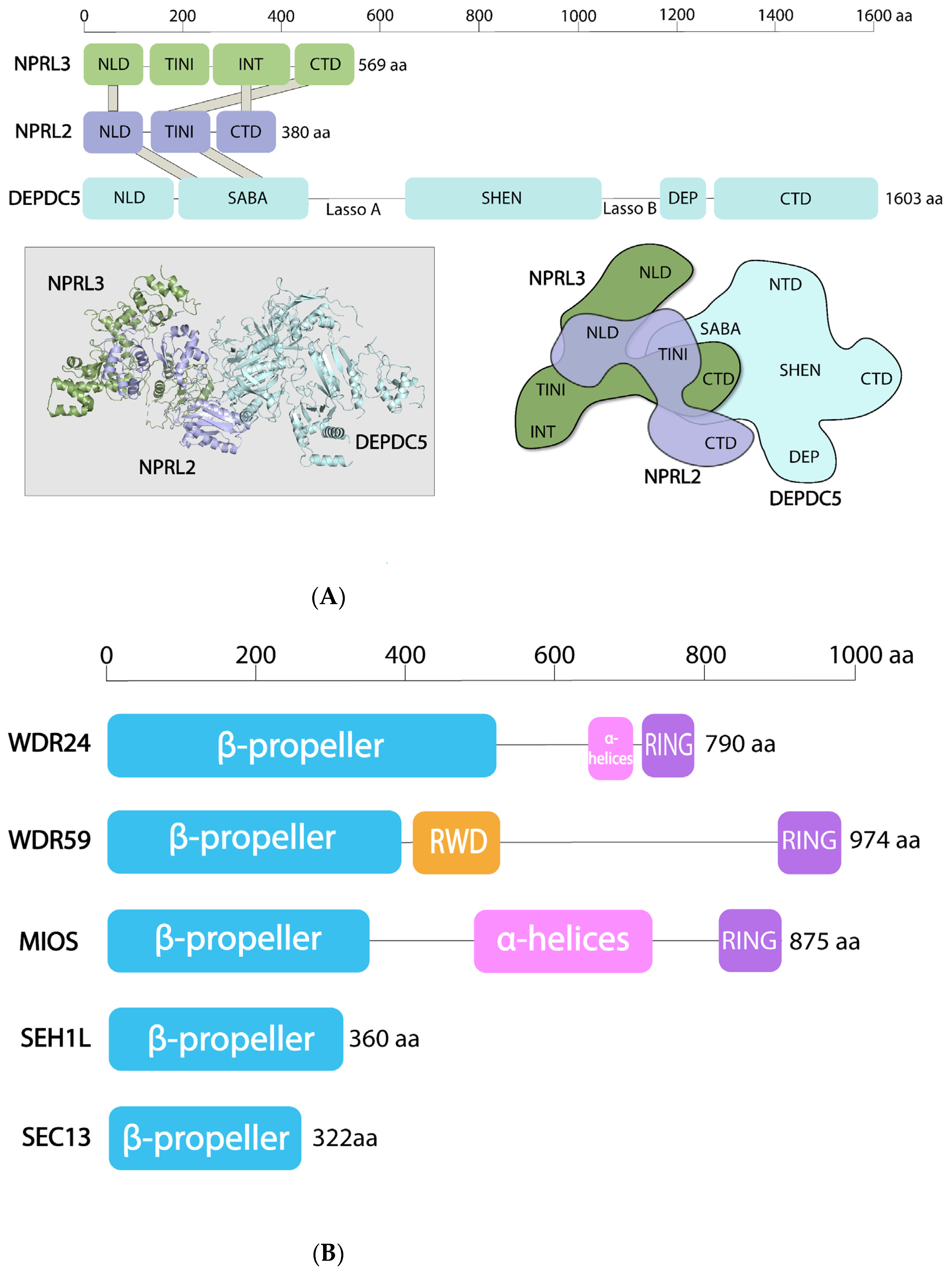
4. Structural Features of the SEA and GATOR Complexes
4.1. SEACAT/GATOR2
4.2. SEACIT/GATOR1
4.3. Posttranslational Modifications of SEA/GATOR
5. Function of the SEA and GATOR in Nutrient Sensing and Responding
5.1. Overview of Amino Acid Axis of Signaling to mTORC1
5.2. GATOR2 Interactions with Leucine Sensors SESTRINs and SAR1B and Arginine Sensor CASTOR1
5.3. GATOR1 Interaction with SAM Sensor, SAMTOR
5.4. SEACIT and Amino Acid Sensing in Yeast
5.5. SEACIT/GATOR1 as GAP for EGO/RAG
5.6. SEA/GATOR Recruitment to the Vacuolar/Lysosomal Membrane
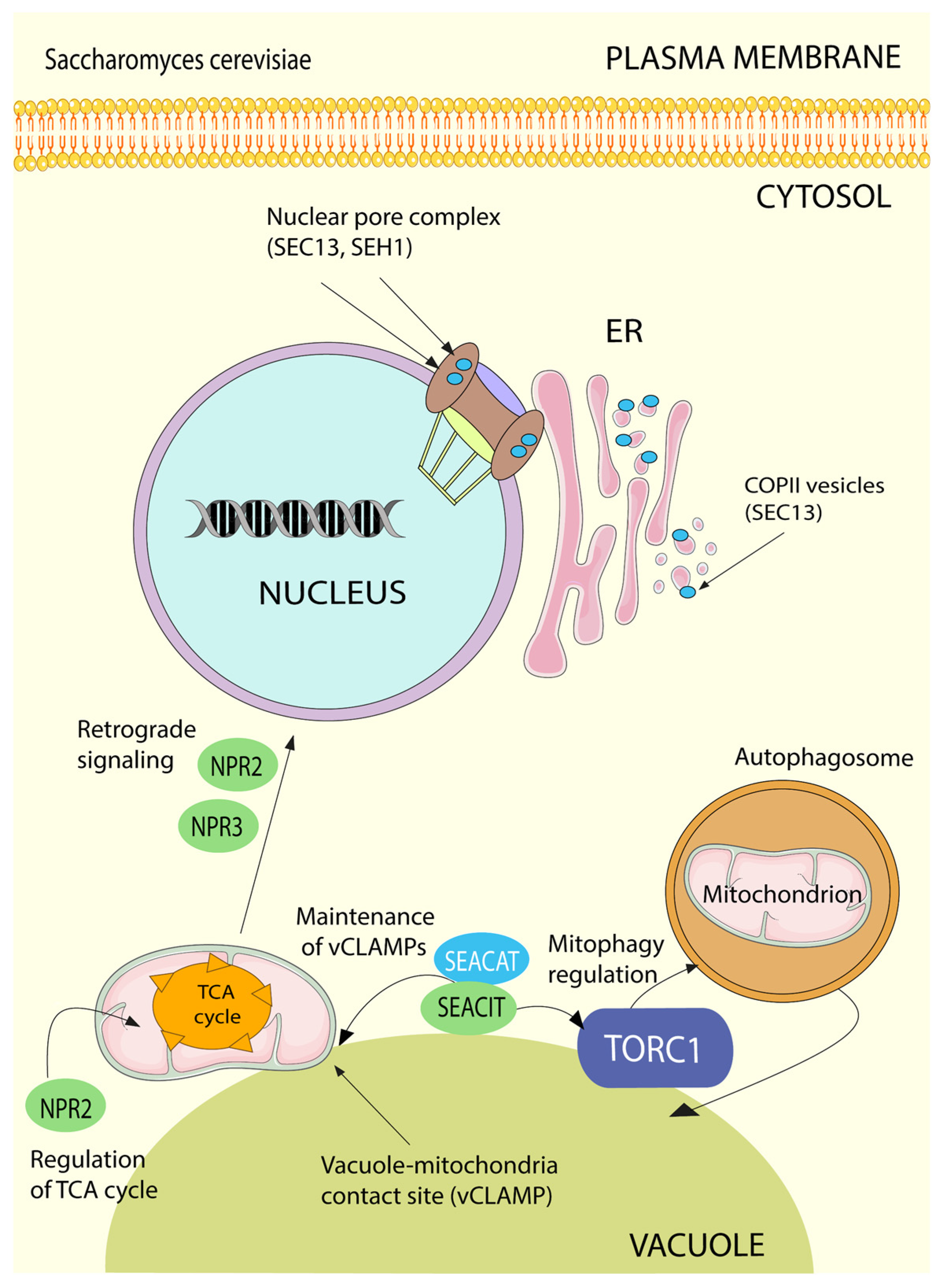
5.7. SEA/GATOR in Autophagy
6. SEA and GATOR Functions beyond Nutrient Responding
6.1. SEA/GATOR Evolution Origin
6.2. Regulation of Mitochondrial Biogenesis and Quality Control
6.3. GATOR1 and DNA Damage Response
6.4. GATOR in Cell Division and Cell Cycle Regulation
6.5. The Role of GATOR in Development
7. Deletion Phenotypes of the SEA/GATOR Components across Different Species
8. GATOR in Human Diseases
8.1. Epilepsies and Brain Malformations—DEPDC5 and Others
8.2. Cancer and Anticancer Drug Resistance—NPRL2 and Others
8.3. Cardiovascular Diseases—NPRL3
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, G.Y.; Sabatini, D.M. MTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat. Rev. Mol. Cell. Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and Metabolic Functions of MTORC1 and MTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef] [PubMed]
- Dokudovskaya, S.; Waharte, F.; Schlessinger, A.; Pieper, U.; Devos, D.P.; Cristea, I.M.; Williams, R.; Salamero, J.; Chait, B.T.; Sali, A.; et al. A Conserved Coatomer-Related Complex Containing Sec13 and Seh1 Dynamically Associates with the Vacuole in Saccharomyces Cerevisiae. Mol. Cell. Proteom. 2011, 10, M110.006478. [Google Scholar] [CrossRef]
- Dokudovskaya, S.; Rout, M.P. SEA You Later Alli-GATOR--a Dynamic Regulator of the TORC1 Stress Response Pathway. J. Cell Sci. 2015, 128, 2219–2228. [Google Scholar] [CrossRef]
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. Determining the Architectures of Macromolecular Assemblies. Nature 2007, 450, 683–694. [Google Scholar] [CrossRef]
- Dokudovskaya, S.; Rout, M.P. A Novel Coatomer-Related SEA Complex Dynamically Associates with the Vacuole in Yeast and Is Implicated in the Response to Nitrogen Starvation. Autophagy 2011, 7, 1392–1393. [Google Scholar] [CrossRef]
- Algret, R.; Dokudovskaya, S.S. The SEA Complex—the Beginning. Biopolym. Cell 2012, 28, 281–284. [Google Scholar] [CrossRef]
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. The Molecular Architecture of the Nuclear Pore Complex. Nature 2007, 450, 695–701. [Google Scholar] [CrossRef]
- Wu, X.; Tu, B.P. Selective Regulation of Autophagy by the Iml1-Npr2-Npr3 Complex in the Absence of Nitrogen Starvation. Mol. Biol. Cell 2011, 22, 4124–4133. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Davis, R.W. A Genome-Wide Screen for Regulators of TORC1 in Response to Amino Acid Starvation Reveals a Conserved Npr2/3 Complex. PLoS Genet. 2009, 5, e1000515. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, N.; Péli-Gulli, M.-P.P.; De Virgilio, C.; Peli-Gulli, M.P.; De Virgilio, C.; Péli-Gulli, M.-P.P.; De Virgilio, C. Amino Acid Deprivation Inhibits TORC1 through a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr1. Sci. Signal. 2013, 6, ra42. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A Tumor Suppressor Complex with GAP Activity for the Rag GTPases That Signal Amino Acid Sufficiency to MTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef]
- Algret, R.; Fernandez-Martinez, J.; Shi, Y.; Kim, S.J.; Pellarin, R.; Cimermancic, P.; Cochet, E.; Sali, A.; Chait, B.T.; Rout, M.P.; et al. Molecular Architecture and Function of the SEA Complex, a Modulator of the TORC1 Pathway. Mol. Cell. Proteom. 2014, 13, 2855–2870. [Google Scholar] [CrossRef]
- Panchaud, N.; Peli-Gulli, M.P.; De Virgilio, C.; Péli-Gulli, M.P.; De Virgilio, C. SEACing the GAP That NEGOCiates TORC1 Activation: Evolutionary Conservation of Rag GTPase Regulation. Cell Cycle 2013, 12, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Sabatini, D.M. The Dawn of the Age of Amino Acid Sensors for the MTORC1 Pathway. Cell Metab. 2017, 26, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Liu, Q.; Zhang, L.; Henske, E.P.; Ma, Y. TORC1 Signaling Is Governed by Two Negative Regulators in Fission Yeast. Genetics 2013, 195, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shen, H.; Sewell, A.K.; Kniazeva, M.; Han, M. A Novel Sphingolipid-TORC1 Pathway Critically Promotes Postembryonic Development in Caenorhabditis Elegans. eLife 2013, 2, e00429. [Google Scholar] [CrossRef]
- Wei, Y.; Reveal, B.; Reich, J.; Laursen, W.J.; Senger, S.; Akbar, T.; Iida-Jones, T.; Cai, W.; Jarnik, M.; Lilly, M.A. TORC1 Regulators Iml1/GATOR1 and GATOR2 Control Meiotic Entry and Oocyte Development in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E5670–E5677. [Google Scholar] [CrossRef] [PubMed]
- de Calbiac, H.; Dabacan, A.; Marsan, E.; Tostivint, H.; Devienne, G.; Ishida, S.; Leguern, E.; Baulac, S.; Muresan, R.C.; Kabashi, E.; et al. Depdc5 Knockdown Causes MTOR-Dependent Motor Hyperactivity in Zebrafish. Ann. Clin. Transl. Neurol. 2018, 5, 510–523. [Google Scholar] [CrossRef]
- Dutchak, P.A.; Laxman, S.; Estill, S.J.; Wang, C.; Wang, Y.Y.; Wang, Y.Y.; Bulut, G.B.; Gao, J.; Huang, L.J.; Tu, B.P. Regulation of Hematopoiesis and Methionine Homeostasis by MTORC1 Inhibitor NPRL2. Cell Rep. 2015, 12, 371–379. [Google Scholar] [CrossRef]
- Marsan, E.; Ishida, S.; Schramm, A.; Weckhuysen, S.; Muraca, G.; Lecas, S.; Liang, N.; Treins, C.; Pende, M.; Roussel, D.; et al. Depdc5 Knockout Rat: A Novel Model of MTORopathy. Neurobiol. Dis. 2016, 89, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Chia, K.H.; Fukuda, T.; Sofyantoro, F.; Matsuda, T.; Amai, T.; Shiozaki, K. Ragulator and GATOR1 Complexes Promote Fission Yeast Growth by Attenuating TOR Complex 1 through Rag GTPases. eLife 2017, 6, e30880. [Google Scholar] [CrossRef]
- Rousselet, G.; Simon, M.; Ripoche, P.; Buhler, J.-M.M. A Second Nitrogen Permease Regulator in Saccharomyces Cerevisiae. FEBS Lett. 1995, 359, 215–219. [Google Scholar] [CrossRef]
- Iida, T.; Lilly, M.A. Missing Oocyte Encodes a Highly Conserved Nuclear Protein Required for the Maintenance of the Meiotic Cycle and Oocyte Identity in Drosophila. Development 2004, 131, 1029–1039. [Google Scholar] [CrossRef]
- Bertuzzi, M.; Tang, D.; Calligaris, R.; Vlachouli, C.; Finaurini, S.; Sanges, R.; Goldwurm, S.; Catalan, M.; Antonutti, L.; Manganotti, P.; et al. A Human Minisatellite Hosts an Alternative Transcription Start Site for NPRL3 Driving Its Expression in a Repeat Number-dependent Manner. Hum. Mutat. 2020, 41, 807–824. [Google Scholar] [CrossRef]
- Zhang, J.; Shen, Y.; Yang, Z.; Yang, F.; Li, Y.; Yu, B.; Chen, W.; Gan, J. A Splicing Variation in NPRL2 Causing Familial Focal Epilepsy with Variable Foci: Additional Cases and Literature Review. J. Hum. Genet. 2021. Online ahead of print. [Google Scholar] [CrossRef]
- Lee, C.; Goldberg, J. Structure of Coatomer Cage Proteins and the Relationship among COPI, COPII, and Clathrin Vesicle Coats. Cell 2010, 142, 123–132. [Google Scholar] [CrossRef]
- Fukuda, T.; Sofyantoro, F.; Tai, Y.T.; Chia, K.H.; Matsuda, T.; Murase, T.; Morozumi, Y.; Tatebe, H.; Kanki, T.; Shiozaki, K. Tripartite Suppression of Fission Yeast TORC1 Signaling by the GATOR1-Sea3 Complex, the TSC Complex, and Gcn2 Kinase. eLife 2021, 10, e60969. [Google Scholar] [CrossRef]
- Shen, K.; Huang, R.K.; Brignole, E.J.; Condon, K.J.; Valenstein, M.L.; Chantranupong, L.; Bomaliyamu, A.; Choe, A.; Hong, C.; Yu, Z.; et al. Architecture of the Human GATOR1 and GATOR1–Rag GTPases Complexes. Nature 2018, 556, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Tafur, L.; Kefauver, J.; Loewith, R. Structural Insights into TOR Signaling. Genes 2020, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Dokudovskaya, S.; Alber, F.; Williams, R.; Chait, B.T.; Sali, A.; Rout, M.P. Components of Coated Vesicles and Nuclear Pore Complexes Share a Common Molecular Architecture. PLoS Biol. 2004, 2, e380. [Google Scholar] [CrossRef]
- Balderhaar, H.J.K.; Ungermann, C. CORVET and HOPS Tethering Complexes-Coordinators of Endosome and Lysosome Fusion. J. Cell Sci. 2013, 126, 1307–1316. [Google Scholar] [CrossRef]
- Rout, M.P.; Field, M.C. The Evolution of Organellar Coat Complexes and Organization of the Eukaryotic Cell. Annu. Rev. Biochem. 2017, 86, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Field, M.C.; Sali, A.; Rout, M.P. Evolution: On a Bender—BARs, ESCRTs, COPs, and Finally Getting Your Coat. J. Cell Biol. 2011, 193, 963–972. [Google Scholar] [CrossRef]
- Fath, S.; Mancias, J.D.; Bi, X.; Goldberg, J. Structure and Organization of Coat Proteins in the COPII Cage. Cell 2007, 129, 1325–1336. [Google Scholar] [CrossRef]
- Senger, S.; Csokmay, J.; Akbar, T.; Jones, T.I.; Sengupta, P.; Lilly, M.A.; Tanveer, A.; Jones, T.I.; Sengupta, P.; Lilly, M.A. The Nucleoporin Seh1 Forms a Complex with Mio and Serves an Essential Tissue-Specific Function in Drosophila Oogenesis. Development 2011, 138, 2133–2142. [Google Scholar] [CrossRef]
- Hesketh, G.G.; Papazotos, F.; Pawling, J.; Rajendran, D.; Knight, J.D.R.; Martinez, S.; Taipale, M.; Schramek, D.; Dennis, J.W.; Gingras, A.-C. The GATOR–Rag GTPase Pathway Inhibits MTORC1 Activation by Lysosome-Derived Amino Acids. Science 2020, 370, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic Identification of Novel Protein Domain Families Associated with Nuclear Functions. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Nameki, N.; Yoneyama, M.; Koshiba, S.; Tochio, N.; Inoue, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Harada, T.; Saito, K.; et al. Solution Structure of the RWD Domain of the Mouse GCN2 Protein. Protein Sci. 2004, 13, 2089–2100. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Willems, A.R.; Sa, D.; Kuras, L.; Thomas, D.; Craig, K.L.; Tyers, M. Cdc53 Is a Scaffold Protein for Multiple Cdc34/Skp1/F Box Protein Complexes That Regulate Cell Division and Methionine Biosynthesis in Yeast. Genes Dev. 1998, 12, 692–705. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Hughes, J.R.; Babbs, C.; Sanchez-Pulido, L.; Szumska, D.; Sharpe, J.A.; Sloane-Stanley, J.A.; Morriss-Kay, G.M.; Smoot, L.B.; Roberts, A.E.; et al. Nprl3 Is Required for Normal Development of the Cardiovascular System. Mamm. Genome 2012, 23, 404–415. [Google Scholar] [CrossRef]
- Levine, T.P.; Daniels, R.D.; Wong, L.H.; Gatta, A.T.; Gerondopoulos, A.; Barr, F.A. Discovery of New Longin and Roadblock Domains That Form Platforms for Small GTPases in Ragulator and TRAPP-II. Small GTPases 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nookala, R.K.; Langemeyer, L.; Pacitto, A.; Donaldson, J.C.; Ochoa-montan, B.; Blaszczyk, B.K.; Chirgadze, D.Y.; Barr, F.A.; Bazan, J.F.; Blundell, T.L.; et al. Crystal Structure of Folliculin Reveals a HidDENN Function in Genetically Inherited Renal Cancer. Open Biol. 2012, 2, 120071. [Google Scholar] [CrossRef]
- Consonni, S.V.; Maurice, M.M.; Bos, J.L. DEP Domains: Structurally Similar but Functionally Different. Nat. Rev. Mol. Cell Biol. 2014, 15, 357–362. [Google Scholar] [CrossRef]
- Caron, A.; Briscoe, D.M.; Richard, D.; Laplante, M. DEPTOR at the Nexus of Cancer, Metabolism, and Immunity. Physiol. Rev. 2018, 98, 1765–1803. [Google Scholar] [CrossRef]
- Albuquerque, C.P.; Smolka, M.B.; Payne, S.H.; Bafna, V.; Eng, J.; Zhou, H. A Multidimensional Chromatography Technology for In-Depth Phosphoproteome Analysis. Mol. Cell. Proteom. 2008, 7, 1389–1396. [Google Scholar] [CrossRef]
- Breitkreutz, A.; Choi, H.; Sharom, J.R.; Boucher, L.; Neduva, V.; Larsen, B.; Lin, Z.Y.; Breitkreutz, B.J.; Stark, C.; Liu, G.; et al. A Global Protein Kinase and Phosphatase Interaction Network in Yeast. Science 2010, 328, 1043–1046. [Google Scholar] [CrossRef]
- Hitchcock, A.L.; Auld, K.; Gygi, S.P.; Silver, P. A Subset of Membrane-Associated Proteins Is Ubiquitinated in Response to Mutations in the Endoplasmic Reticulum Degradation Machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 12735–12740. [Google Scholar] [CrossRef] [PubMed]
- Iesmantavicius, V.; Weinert, B.T.; Choudhary, C. Convergence of Ubiquitylation and Phosphorylation Signaling in Rapamycin-Treated Yeast Cells. Mol. Cell. Proteom. 2014, 13, 1979–1992. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Raucci, S.; Jaquenoud, M.; Hatakeyama, R.; Stumpe, M.; Rohr, R.; Reggiori, F.; De Virgilio, C.; Dengjel, J. Multilayered Control of Protein Turnover by TORC1 and Atg1. Cell Rep. 2019, 28, 3486–3496.e6. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Montañés, F.; Casanovas, A.; Sprenger, R.R.; Topolska, M.; Marshall, D.L.; Moreno-Torres, M.; Poad, B.L.J.; Blanksby, S.J.; Hermansson, M.; Jensen, O.N.; et al. Phosphoproteomic Analysis across the Yeast Life Cycle Reveals Control of Fatty Acyl Chain Length by Phosphorylation of the Fatty Acid Synthase Complex. Cell Rep. 2020, 32, 108024. [Google Scholar] [CrossRef]
- Spielewoy, N.; Guaderrama, M.; Wohlschlegel, J.A.; Ashe, M.; Yates, J.R., 3rd; Wittenberg, C.; Yates, J.R.; Wittenberg, C. Npr2, Yeast Homolog of the Human Tumor Suppressor NPRL2, Is a Target of Grr1 Required for Adaptation to Growth on Diverse Nitrogen Sources. Eukaryot. Cell 2010, 9, 592–601. [Google Scholar] [CrossRef]
- Chen, J.; Ou, Y.; Yang, Y.; Li, W.; Xu, Y.; Xie, Y.; Liu, Y. KLHL22 Activates Amino-Acid-Dependent MTORC1 Signalling to Promote Tumorigenesis and Ageing. Nature 2018, 557, 585–589. [Google Scholar] [CrossRef]
- Ma, Y.; Silveri, L.; LaCava, J.; Dokudovskaya, S. Tumor Suppressor NPRL2 Induces ROS Production and DNA Damage Response. Sci. Rep. 2017, 7, 15311. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Guo, J.; Wang, X.; Cheng, Y.; Guan, J.; Barman, P.; Sun, M.-A.; Fu, Y.; Wei, W.; Feng, C.; et al. FKBP39 Controls Nutrient Dependent Nprl3 Expression and TORC1 Activity in Drosophila. Cell Death Dis. 2021, 12, 571. [Google Scholar] [CrossRef] [PubMed]
- Orozco, J.M.; Krawczyk, P.A.; Scaria, S.M.; Cangelosi, A.L.; Chan, S.H.; Kunchok, T.; Lewis, C.A.; Sabatini, D.M. Dihydroxyacetone Phosphate Signals Glucose Availability to MTORC1. Nat. Metab. 2020, 2, 893–901. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to MTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.-L. Regulation of TORC1 by Rag GTPases in Nutrient Response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef]
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. MTORC1 Senses Lysosomal Amino Acids through an Inside-out Mechanism That Requires the Vacuolar H(+)-ATPase. Science 2011, 334, 678–683. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator Is a GEF for the Rag GTPases That Signal Amino Acid Levels to MTORC1. Cell 2012, 150, 1196–1208. [Google Scholar] [CrossRef]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag Complex Targets MTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef]
- Thomas, J.D.; Zhang, Y.J.; Wei, Y.H.; Cho, J.H.; Morris, L.E.; Wang, H.Y.; Zheng, X.F. Rab1A Is an MTORC1 Activator and a Colorectal Oncogene. Cancer Cell 2014, 26, 754–769. [Google Scholar] [CrossRef]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.L. Metabolism. Differential Regulation of MTORC1 by Leucine and Glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef]
- Ukai, H.; Araki, Y.; Kira, S.; Oikawa, Y.; May, A.I.; Noda, T. Gtr/Ego-Independent TORC1 Activation Is Achieved through a Glutamine-Sensitive Interaction with Pib2 on the Vacuolar Membrane. PLoS Genet. 2018, 14, e1007334. [Google Scholar] [CrossRef]
- Meng, D.; Yang, Q.; Wang, H.; Melick, C.H.; Navlani, R.; Frank, A.R.; Jewell, J.L. Glutamine and Asparagine Activate MTORC1 Independently of Rag GTPases. J. Biol. Chem. 2020, 295, 2890–2899. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Li, M.O. SZT2 Dictates GATOR Control of MTORC1 Signalling. Nature 2017, 543, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Wyant, G.A.; Gu, X.; Orozco, J.M.; Shen, K.; Condon, K.J.; Petri, S.; Kedir, J.; Scaria, S.M.; et al. KICSTOR Recruits GATOR1 to the Lysosome and Is Necessary for Nutrients to Regulate MTORC1. Nature 2017, 543, 438–442. [Google Scholar] [CrossRef]
- Shen, K.; Sabatini, D.M. Ragulator and SLC38A9 Activate the Rag GTPases through Noncanonical GEF Mechanisms. Proc. Natl. Acad. Sci. USA 2018, 115, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- Tsun, Z.-Y.Y.; Bar-Peled, L.; Chantranupong, L.; Zoncu, R.; Wang, T.; Kim, C.; Spooner, E.; Sabatini, D.M. The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to MTORC1. Mol. Cell 2013, 52, 495–505. [Google Scholar] [CrossRef]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-TRNA Synthetase Is an Intracellular Leucine Sensor for the MTORC1-Signaling Pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef]
- Long, X.; Ortiz-Vega, S.; Lin, Y.; Avruch, J. Rheb Binding to Mammalian Target of Rapamycin (MTOR) Is Regulated by Amino Acid Sufficiency. J. Biol. Chem. 2005, 280, 23433–23436. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT Network at the Interface of Oncogenic Signalling and Cancer Metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Demetriades, C.; Doumpas, N.; Teleman, A.A. Regulation of TORC1 in Response to Amino Acid Starvation via Lysosomal Recruitment of TSC2. Cell 2014, 156, 786–799. [Google Scholar] [CrossRef]
- Demetriades, C.; Plescher, M.; Teleman, A.A. Lysosomal Recruitment of TSC2 Is a Universal Response to Cellular Stress. Nat. Commun. 2016, 7, 10662. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, Y.; Ting, C.-Y.; Bettedi, L.; Kim, K.; Ghaniam, E.; Lilly, M.A. The Rag GTPase Regulates the Dynamic Behavior of TSC Downstream of Both Amino Acid and Growth Factor Restriction. Dev. Cell 2020, 55, 272–288.e5. [Google Scholar] [CrossRef]
- Dubouloz, F.; Deloche, O.; Wanke, V.; Cameroni, E.; De Virgilio, C. The TOR and EGO Protein Complexes Orchestrate Microautophagy in Yeast. Mol. Cell 2005, 19, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Binda, M.; Péli-Gulli, M.-P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The Vam6 GEF Controls TORC1 by Activating the EGO Complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef]
- Bonfils, G.; Jaquenoud, M.; Bontron, S.; Ostrowicz, C.; Ungermann, C.; De Virgilio, C. Leucyl-TRNA Synthetase Controls TORC1 via the EGO Complex. Mol. Cell 2012, 46, 105–110. [Google Scholar] [CrossRef]
- Péli-Gulli, M.P.; Sardu, A.; Panchaud, N.; Raucci, S.; De Virgilio, C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Rep. 2015, 13, 1–7. [Google Scholar] [CrossRef]
- Dechant, R.; Saad, S.; Ibáñez, A.J.; Peter, M. Cytosolic PH Regulates Cell Growth through Distinct GTPases, Arf1 and Gtr1, to Promote Ras/PKA and TORC1 Activity. Mol. Cell 2014, 55, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Urano, J.; Tabancay, A.P.; Yang, W.; Tamanoi, F. The Saccharomyces Cerevisiae Rheb G-Protein Is Involved in Regulating Canavanine Resistance and Arginine Uptake. J. Biol. Chem. 2000, 275, 11198–11206. [Google Scholar] [CrossRef]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 Is a Leucine Sensor for the MTORC1 Pathway. Science 2016, 351, 43–48. [Google Scholar] [CrossRef]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural Basis for Leucine Sensing by the Sestrin2-MTORC1 Pathway. Science 2016, 351, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shimkus, K.L.; Lacko, H.A.; Kutzler, L.; Jefferson, L.S.; Kimball, S.R. Evidence for a Role for Sestrin1 in Mediating Leucine-Induced Activation of MTORC1 in Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E817–E828. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ou, Y.; Luo, R.; Wang, J.; Wang, D.; Guan, J.; Li, Y.; Xia, P.; Chen, P.R.; Liu, Y. SAR1B Senses Leucine Levels to Regulate MTORC1 Signalling. Nature 2021, 596, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kim, J.H.; Yoon, I.; Lee, C.; Fallahi Sichani, M.; Kang, J.S.; Kang, J.; Guo, M.; Lee, K.Y.; Han, G.; et al. Coordination of the Leucine-Sensing Rag GTPase Cycle by Leucyl-TRNA Synthetase in the MTORC1 Signaling Pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E5279–E5288. [Google Scholar] [CrossRef]
- Kim, S.; Yoon, I.; Son, J.; Park, J.; Kim, K.; Lee, J.-H.; Park, S.-Y.; Kang, B.S.; Han, J.M.; Hwang, K.Y.; et al. Leucine-Sensing Mechanism of Leucyl-TRNA Synthetase 1 for MTORC1 Activation. Cell Rep. 2021, 35, 109031. [Google Scholar] [CrossRef]
- Saxton, R.A.; Chantranupong, L.; Knockenhauer, K.E.; Schwartz, T.U.; Sabatini, D.M. Mechanism of Arginine Sensing by CASTOR1 Upstream of MTORC1. Nature 2016, 536, 229–233. [Google Scholar] [CrossRef]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the MTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef]
- Wang, S.; Tsun, Z.Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Metabolism. Lysosomal Amino Acid Transporter SLC38A9 Signals Arginine Sufficiency to MTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; De Arajo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 Is a Component of the Lysosomal Amino Acid Sensing Machinery That Controls MTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef]
- Jung, J.W.; Macalino, S.J.Y.; Cui, M.; Kim, J.E.; Kim, H.-J.; Song, D.-G.; Nam, S.H.; Kim, S.; Choi, S.; Lee, J.W. Transmembrane 4 L Six Family Member 5 Senses Arginine for MTORC1 Signaling. Cell Metab. 2019, 29, 1306–1319.e7. [Google Scholar] [CrossRef] [PubMed]
- Parmigiani, A.; Nourbakhsh, A.; Ding, B.; Wang, W.; Kim, Y.C.; Akopiants, K.; Guan, K.L.; Karin, M.; Budanov, A.V. Sestrins Inhibit MTORC1 Kinase Activation through the GATOR Complex. Cell Rep. 2014, 9, 1281–1291. [Google Scholar] [CrossRef]
- Kowalsky, A.H.; Namkoong, S.; Mettetal, E.; Park, H.-W.; Kazyken, D.; Fingar, D.C.; Lee, J.H. The GATOR2–MTORC2 Axis Mediates Sestrin2-Induced AKT Ser/Thr Kinase Activation. J. Biol. Chem. 2020, 295, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Gai, Z.; Wang, Q.; Yang, C.; Wang, L.; Deng, W.; Wu, G. Structural Mechanism for the Arginine Sensing and Regulation of CASTOR1 in the MTORC1 Signaling Pathway. Cell Discov. 2016, 2, 16051. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Ro, S.H.; Kim, M.; Park, H.W.; Semple, I.A.; Park, H.; Cho, U.S.; Wang, W.; Guan, K.L.; Karin, M.; et al. Sestrin2 Inhibits MTORC1 through Modulation of GATOR Complexes. Sci. Rep. 2015, 5, 9502. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Wolfson, R.L.; Orozco, J.M.; Saxton, R.A.; Scaria, S.M.; Bar-Peled, L.; Spooner, E.; Isasa, M.; Gygi, S.P.; Sabatini, D.M. The Sestrins Interact with GATOR2 to Negatively Regulate the Amino-Acid-Sensing Pathway Upstream of MTORC1. Cell Rep. 2014, 9, 1–8. [Google Scholar] [CrossRef]
- Xia, J.; Wang, R.; Zhang, T.; Ding, J. Structural Insight into the Arginine-Binding Specificity of CASTOR1 in Amino Acid-Dependent MTORC1 Signaling. Cell Discov. 2016, 2, 16035. [Google Scholar] [CrossRef] [PubMed]
- Suryawan, A.; Davis, T.A. Amino Acid- and Insulin-Induced Activation of MTORC1 in Neonatal Piglet Skeletal Muscle Involves Sestrin2-GATOR2, Rag A/C-MTOR, and RHEB-MTOR Complex Formation. J. Nutr. 2018, 148, 825–833. [Google Scholar] [CrossRef]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR Is an S-Adenosylmethionine Sensor for the MTORC1 Pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Rathore, R.; Caldwell, K.E.; Schutt, C.; Brashears, C.B.; Prudner, B.C.; Ehrhardt, W.R.; Leung, C.H.; Lin, H.; Daw, N.C.; Beird, H.C.; et al. Metabolic Compensation Activates Pro-Survival MTORC1 Signaling upon 3-Phosphoglycerate Dehydrogenase Inhibition in Osteosarcoma. Cell Rep. 2021, 34, 108678. [Google Scholar] [CrossRef]
- Son, S.M.; Park, S.J.; Lee, H.; Siddiqi, F.; Lee, J.E.; Menzies, F.M.; Rubinsztein, D.C. Leucine Signals to MTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metab. 2019, 29, 192–201.e7. [Google Scholar] [CrossRef]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine Inhibits Autophagy and Promotes Growth by Inducing the SAM-Responsive Methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef]
- Laxman, S.; Sutter, B.M.; Shi, L.; Tu, B.P. Npr2 Inhibits TORC1 to Prevent Inappropriate Utilization of Glutamine for Biosynthesis of Nitrogen-Containing Metabolites. Sci. Signal. 2014, 7, ra120. [Google Scholar] [CrossRef] [PubMed]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen Source Activates TOR (Target of Rapamycin) Complex 1 via Glutamine and Independently of Gtr/Rag Proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef]
- Chen, X.; Wang, G.; Zhang, Y.; Dayhoff-Brannigan, M.; Diny, N.L.; Zhao, M.; He, G.; Sing, C.N.; Metz, K.A.; Stolp, Z.D.; et al. Whi2 Is a Conserved Negative Regulator of TORC1 in Response to Low Amino Acids. PLoS Genet. 2018, 14, e1007592. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Hardwick, J.M. Whi2: A New Player in Amino Acid Sensing. Curr. Genet. 2019, 65, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Choi, J.-H.; Wang, P.; Go, C.D.; Hesketh, G.G.; Gingras, A.-C.; Jafarnejad, S.M.; Sonenberg, N. Mitochondrial Threonyl-TRNA Synthetase TARS2 Is Required for Threonine-Sensitive MTORC1 Activation. Mol. Cell 2021, 81, 398–407.e4. [Google Scholar] [CrossRef]
- Wittinghofer, A.; Vetter, I.R. Structure-Function Relationships of the G Domain, a Canonical Switch Motif. Annu. Rev. Biochem. 2011, 80, 943–971. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Valenstein, M.L.; Gu, X.; Sabatini, D.M. Arg-78 of Nprl2 Catalyzes GATOR1-Stimulated GTP Hydrolysis by the Rag GTPases. J. Biol. Chem. 2019, 294, 2970–5944. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Jiang, C.; Chen, L.; Jin, J.; Wei, J.; Zhao, L.; Chen, M.; Pan, W.; Xu, Y.; Chu, H.; et al. The Ubiquitination of RagA GTPase by RNF152 Negatively Regulates MTORC1 Activation. Mol. Cell 2015, 58, 804–818. [Google Scholar] [CrossRef]
- Kiontke, S.; Langemeyer, L.; Kuhlee, A.; Schuback, S.; Raunser, S.; Ungermann, C.; Kümmel, D. Architecture and Mechanism of the Late Endosomal Rab7-like Ypt7 Guanine Nucleotide Exchange Factor Complex Mon1–Ccz1. Nat. Commun. 2017, 8, 14034. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.S.; Kang, K.H.; Kim, S.; Lee, S.; Lee, J.H.; Kim, J.W.; Byun, B.; Meadows, G.G.; Joe, C.O. Amino Acid-Dependent NPRL2 Interaction with Raptor Determines MTOR Complex 1 Activation. Cell Signal. 2016, 28, 32–41. [Google Scholar] [CrossRef]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 Is a Major Target of TORC1 in Saccharomyces Cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Sturgill, T.W.; Cohen, A.; Diefenbacher, M.; Trautwein, M.; Martin, D.E.; Hall, M.N. TOR1 and TOR2 Have Distinct Locations in Live Cells. Eukaryot. Cell 2008, 7, 1819–1830. [Google Scholar] [CrossRef]
- Betz, C.; Hall, M.N. Where Is MTOR and What Is It Doing There? J. Cell Biol. 2013, 203, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Kondo, K.; Itoh, T.; Ikari, S.; Nada, S.; Okada, M.; Noda, T. Rheb Localized on the Golgi Membrane Activates Lysosome-Localized MTORC1 at the Golgi-Lysosome Contact Site. J. Cell Sci. 2017, 131, jcs.208017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kim, J.; Alexander, A.; Cai, S.; Tripathi, D.N.; Dere, R.; Tee, A.R.; Tait-Mulder, J.; Di Nardo, A.; Han, J.M.; et al. A Tuberous Sclerosis Complex Signalling Node at the Peroxisome Regulates MTORC1 and Autophagy in Response to ROS. Nat. Cell Biol. 2013, 15, 1186–1196. [Google Scholar] [CrossRef]
- Zhang, J.; Andersen, J.; Sun, H.; Liu, X.; Sonenberg, N.; Nie, J.; Shi, Y. Aster-C Coordinates with COP I Vesicles to Regulate Lysosomal Trafficking and Activation of MTORC1. EMBO Rep. 2020, 21, e49898. [Google Scholar] [CrossRef]
- Meng, J.; Ferguson, S.M. GATOR1-Dependent Recruitment of FLCN–FNIP to Lysosomes Coordinates Rag GTPase Heterodimer Nucleotide Status in Response to Amino Acids. J. Cell Biol. 2018, 217, 2765–2776. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.S.; Roczniak-Ferguson, A.; Ferguson, S.M. Recruitment of Folliculin to Lysosomes Supports the Amino Acid-Dependent Activation of Rag GTPases. J. Cell Biol. 2013, 202, 1107–1122. [Google Scholar] [CrossRef]
- Shen, K.; Rogala, K.B.; Chou, H.-T.; Huang, R.K.; Yu, Z.; Sabatini, D.M. Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex. Cell 2019, 179, 1319–1329.e8. [Google Scholar] [CrossRef]
- Lawrence, R.E.; Fromm, S.A.; Fu, Y.; Yokom, A.L.; Kim, D.J.; Thelen, A.M.; Young, L.N.; Lim, C.-Y.; Samelson, A.J.; Hurley, J.H.; et al. Structural Mechanism of a Rag GTPase Activation Checkpoint by the Lysosomal Folliculin Complex. Science 2019, 366, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Palmieri, M.; Chaudhury, A.; Klisch, T.J.; di Ronza, A.; Neilson, J.R.; Rodney, G.G.; Sardiello, M. Src Regulates Amino Acid-Mediated MTORC1 Activation by Disrupting GATOR1-Rag GTPase Interaction. Nat. Commun. 2018, 9, 4351. [Google Scholar] [CrossRef]
- Padi, S.K.R.; Singh, N.; Bearss, J.J.; Olive, V.; Song, J.H.; Cardó-Vila, M.; Kraft, A.S.; Okumura, K. Phosphorylation of DEPDC5, a Component of the GATOR1 Complex, Releases Inhibition of MTORC1 and Promotes Tumor Growth. Proc. Natl. Acad. Sci. USA 2019, 116, 20505–20510. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Lee, S.W.; Zhang, X.; Cai, Z.; Gao, Y.; Chou, P.C.; Rezaeian, A.H.; Han, F.; Wang, C.Y.; Yao, J.C.; et al. Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent Amino-Acid-Dependent MTORC1 Hyperactivation by Recruiting GATOR1. Mol. Cell 2015, 58, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Chen, L.; Zhao, L.; Xu, Y.; Peng, X.; Wang, X.; Ding, L.; Jin, J.; Teng, H.; Wang, Y.; et al. Ubiquitination of Rheb Governs Growth Factor-Induced MTORC1 Activation. Cell Res. 2019, 29, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Graef, M.; Nunnari, J. Mitochondria Regulate Autophagy by Conserved Signalling Pathways. EMBO J. 2011, 30, 2101–2114. [Google Scholar] [CrossRef] [PubMed]
- Kira, S.; Tabata, K.; Shirahama-Noda, K.; Nozoe, A.; Yoshimori, T.; Noda, T. Reciprocal Conversion of Gtr1 and Gtr2 Nucleotide-Binding States by Npr2-Npr3 Inactivates TORC1 and Induces Autophagy. Autophagy 2014, 10, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Reveal, B.; Cai, W.; Lilly, M.A. The GATOR1 Complex Regulates Metabolic Homeostasis and the Response to Nutrient Stress in Drosophila Melanogaster. G3 2016, 6, 3859–3867. [Google Scholar] [CrossRef]
- Qi, B.; Kniazeva, M.; Han, M. A Vitamin-B2-Sensing Mechanism That Regulates Gut Protease Activity to Impact Animal’s Food Behavior and Growth. eLife 2017, 6, e26243. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Shao, L.; Chen, Z.; Hu, D.; Jiang, L.; Tang, W. NPRL2 Promotes Docetaxel Chemoresistance in Castration Resistant Prostate Cancer Cells by Regulating Autophagy through the MTOR Pathway. Exp. Cell Res. 2020, 390, 111981. [Google Scholar] [CrossRef]
- Cai, W.; Wei, Y.; Jarnik, M.; Reich, J.; Lilly, M.A. The GATOR2 Component Wdr24 Regulates TORC1 Activity and Lysosome Function. PLoS Genet. 2016, 12, e1006036. [Google Scholar] [CrossRef] [PubMed]
- Michaillat, L.; Baars, T.L.; Mayer, A. Cell-Free Reconstitution of Vacuole Membrane Fragmentation Reveals Regulation of Vacuole Size and Number by TORC1. Mol. Biol. Cell 2012, 23, 881–895. [Google Scholar] [CrossRef]
- Ma, Y.; Moors, A.; Camougrand, N.; Dokudovskaya, S. The SEACIT Complex Is Involved in the Maintenance of Vacuole–Mitochondria Contact Sites and Controls Mitophagy. Cell. Mol. Life Sci. 2019, 76, 1623–1640. [Google Scholar] [CrossRef]
- Liu, Y.; Okamoto, K. The TORC1 Signaling Pathway Regulates Respiration-Induced Mitophagy in Yeast. Biochem. Biophys. Res. Commun. 2018, 502, 76–83. [Google Scholar] [CrossRef]
- Elbaz-Alon, Y.; Rosenfeld-Gur, E.; Shinder, V.; Futerman, A.H.; Geiger, T.; Schuldiner, M. A Dynamic Interface between Vacuoles and Mitochondria in Yeast. Dev. Cell 2014, 30, 95–102. [Google Scholar] [CrossRef]
- Chong, Y.T.; Koh, J.L.Y.; Friesen, H.; Kaluarachchi Duffy, S.; Cox, M.J.; Moses, A.; Moffat, J.; Boone, C.; Andrews, B.J. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell 2015, 161, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Weill, U.; Yofe, I.; Sass, E.; Stynen, B.; Davidi, D.; Natarajan, J.; Ben-Menachem, R.; Avihou, Z.; Goldman, O.; Harpaz, N.; et al. Genome-Wide SWAp-Tag Yeast Libraries for Proteome Exploration. Nat. Methods 2018, 15, 617–622. [Google Scholar] [CrossRef]
- Orre, L.M.; Vesterlund, M.; Pan, Y.; Arslan, T.; Zhu, Y.; Fernandez Woodbridge, A.; Frings, O.; Fredlund, E.; Lehtiö, J. SubCellBarCode: Proteome-Wide Mapping of Protein Localization and Relocalization. Mol. Cell 2019, 73, 166–182.e7. [Google Scholar] [CrossRef] [PubMed]
- De Franceschi, N.; Wild, K.; Schlacht, A.; Dacks, J.B.; Sinning, I.; Filippini, F. Longin and GAF Domains: Structural Evolution and Adaptation to the Subcellular Trafficking Machinery: Structure and Evolution of Longin Domains. Traffic 2014, 15, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The Genetic Landscape of a Cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A Global Genetic Interaction Network Maps a Wiring Diagram of Cellular Function. Science 2016, 353, aaf1420. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, I.E.; Tokarchuk, A.V.; Zheltukhin, A.O.; Dalina, A.A.; Safronov, G.G.; Evstafieva, A.G.; Lyamzaev, K.G.; Chumakov, P.M.; Budanov, A.V. Mitochondrial Localization of SESN2. PLoS ONE 2020, 15, e0226862. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, D.; Xie, T.; Xu, L.-G. Sec13 Is a Positive Regulator of VISA-Mediated Antiviral Signaling. Virus Genes 2018, 54, 514–526. [Google Scholar] [CrossRef]
- De Falco, F.; Cutarelli, A.; Gentile, I.; Cerino, P.; Uleri, V.; Catoi, A.F.; Roperto, S. Bovine Delta Papillomavirus E5 Oncoprotein Interacts With TRIM25 and Hampers Antiviral Innate Immune Response Mediated by RIG-I-Like Receptors. Front. Immunol. 2021, 12, 658762. [Google Scholar] [CrossRef]
- Perrone, G.G.; Grant, C.M.; Dawes, I.W. Genetic and Environmental Factors Influencing Glutathione Homeostasis in Saccharomyces Cerevisiae. Mol. Biol. Cell 2005, 16, 218–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, J.; Sutter, B.M.; Shi, L.; Tu, B.P. GATOR1 Regulates Nitrogenic Cataplerotic Reactions of the Mitochondrial TCA Cycle. Nat. Chem. Biol. 2017, 13, 1179–1186. [Google Scholar] [CrossRef]
- Dutchak, P.A.; Estill-Terpack, S.J.; Plec, A.A.; Zhao, X.; Yang, C.; Chen, J.; Ko, B.; Deberardinis, R.J.; Yu, Y.; Tu, B.P. Loss of a Negative Regulator of MTORC1 Induces Aerobic Glycolysis and Altered Fiber Composition in Skeletal Muscle. Cell Rep. 2018, 23, 1907–1914. [Google Scholar] [CrossRef]
- Burger, B.J.; Rose, S.; Bennuri, S.C.; Gill, P.S.; Tippett, M.L.; Delhey, L.; Melnyk, S.; Frye, R.E. Autistic Siblings with Novel Mutations in Two Different Genes: Insight for Genetic Workups of Autistic Siblings and Connection to Mitochondrial Dysfunction. Front. Pediatr. 2017, 5, 219. [Google Scholar] [CrossRef]
- Graber, T.G.; Fry, C.S.; Brightwell, C.R.; Moro, T.; Maroto, R.; Bhattarai, N.; Porter, C.; Wakamiya, M.; Rasmussen, B.B. Skeletal Muscle–Specific Knockout of DEP Domain Containing 5 Protein Increases MTORC1 Signaling, Muscle Cell Hypertrophy, and Mitochondrial Respiration. J. Biol. Chem. 2019, 294, 4091–4102. [Google Scholar] [CrossRef]
- Guaragnella, N.; Coyne, L.P.; Chen, X.J.; Giannattasio, S. Mitochondria–Cytosol–Nucleus Crosstalk: Learning from Saccharomyces Cerevisiae. FEMS Yeast Res. 2018, 18, foy088. [Google Scholar] [CrossRef]
- Quirós, P.M.; Mottis, A.; Auwerx, J. Mitonuclear Communication in Homeostasis and Stress. Nat. Rev. Mol. Cell Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef]
- Zung, N.; Schuldiner, M. New Horizons in Mitochondrial Contact Site Research. Biol. Chem. 2020, 401, 793–809. [Google Scholar] [CrossRef] [PubMed]
- Hönscher, C.; Mari, M.; Auffarth, K.; Bohnert, M.; Griffith, J.; Geerts, W.; van der Laan, M.; Cabrera, M.; Reggiori, F.; Ungermann, C.; et al. Cellular Metabolism Regulates Contact Sites between Vacuoles and Mitochondria. Dev. Cell 2014, 30, 86–94. [Google Scholar] [CrossRef]
- Schenk, P.W.; Brok, E.; Boersma, A.W.M.; Brandsma, J.A.; Den Dulk, H.; Burger, H.; Stoter, G.; Brouwer, J.; Nooter, K. Anticancer Drug Resistance Induced by Disruption of the Saccharomyces Cerevisiae NPR2 Gene: A Novel Component Involved in Cisplatin- and Doxorubicin-Provoked Cell Kill. Mol. Pharmacol. 2003, 64, 259–268. [Google Scholar] [CrossRef]
- Ueda, K.; Kawashima, H.; Ohtani, S.; Deng, W.-G.G.; Ravoori, M.; Bankson, J.; Gao, B.; Girard, L.; Minna, J.D.; Roth, J.A.; et al. The 3p21.3 Tumor Suppressor NPRL2 Plays an Important Role in Cisplatin-Induced Resistance in Human Non-Small-Cell Lung Cancer Cells. Cancer Res. 2006, 66, 9682–9690. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Chang, J.-Y. New Insights into Mechanisms of Cisplatin Resistance: From Tumor Cell to Microenvironment. Int. J. Mol. Sci. 2019, 20, 4136. [Google Scholar] [CrossRef]
- Sritharan, S.; Sivalingam, N. A Comprehensive Review on Time-Tested Anticancer Drug Doxorubicin. Life Sci. 2021, 278, 119527. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, G.; Ueda, K.; Wang, B.; Roth, J.A.; Ji, L. NPRL2 Sensitizes Human Non-Small Cell Lung Cancer (NSCLC) Cells to Cisplatin Treatment by Regulating Key Components in the DNA Repair Pathway. PLoS ONE 2010, 5, e11994. [Google Scholar] [CrossRef]
- Ma, Y.; Vassetzky, Y.; Dokudovskaya, S. MTORC1 Pathway in DNA Damage Response. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2018, 1865, 1293–1311. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Bettedi, L.; Ting, C.-Y.; Kim, K.; Zhang, Y.; Cai, J.; Lilly, M.A. The GATOR Complex Regulates an Essential Response to Meiotic Double-Stranded Breaks in Drosophila. eLife 2019, 8, e42149. [Google Scholar] [CrossRef] [PubMed]
- Platani, M.; Trinkle-Mulcahy, L.; Porter, M.; Arockia Jeyaprakash, A.; Earnshaw, W.C. Mio Depletion Links MTOR Regulation to Aurora A and Plk1 Activation at Mitotic Centrosomes. J. Cell Biol. 2015, 210, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Platani, M.; Samejima, I.; Samejima, K.; Kanemaki, M.T.; Earnshaw, W.C. Seh1 Targets GATOR2 and Nup153 to Mitotic Chromosomes. J. Cell Sci. 2018, 131, jcs213140. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Cai, J.; Cheng, Y.; Fu, Y.; Wei, W.; Zhang, Z.; Zhuang, Z.; Hao, Y.; Lilly, M.A.; Wei, Y. The TORC1 Inhibitor Nprl2 Protects Age-Related Digestive Function in Drosophila. Aging 2019, 11, 9811–9828. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sewell, A.K.; Han, M. Intestinal Apical Polarity Mediates Regulation of TORC1 by Glucosylceramide in C. Elegans. Genes Dev. 2015, 29, 1218–1223. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, A.; Hassan-Abdi, R.; Renault, S.; Siekierska, A.; Riché, R.; Liao, M.; de Witte, P.A.M.; Yanicostas, C.; Soussi-Yanicostas, N.; Drapeau, P.; et al. Non-Canonical MTOR-Independent Role of DEPDC5 in Regulating GABAergic Network Development. Curr. Biol. 2018, 28, 1924–1937.e5. [Google Scholar] [CrossRef]
- Liu, Z.; Yan, M.; Liang, Y.; Liu, M.; Zhang, K.; Shao, D.; Jiang, R.; Li, L.; Wang, C.; Nussenzveig, D.R.; et al. Nucleoporin Seh1 Interacts with Olig2/Brd7 to Promote Oligodendrocyte Differentiation and Myelination. Neuron 2019, 102, 587–601.e7. [Google Scholar] [CrossRef]
- International Mouse Phenotyping Consortium. Available online: https://www.mousephenotype.org (accessed on 31 August 2021).
- Hughes, J.; Dawson, R.; Tea, M.; McAninch, D.; Piltz, S.; Jackson, D.; Stewart, L.; Ricos, M.G.; Dibbens, L.M.; Harvey, N.L.; et al. Knockout of the Epilepsy Gene Depdc5 in Mice Causes Severe Embryonic Dysmorphology with Hyperactivity of MTORC1 Signalling. Sci. Rep. 2017, 7, 12618. [Google Scholar] [CrossRef]
- Yuskaitis, C.J.; Jones, B.M.; Wolfson, R.L.; Super, C.E.; Dhamne, S.C.; Rotenberg, A.; Sabatini, D.M.; Sahin, M.; Poduri, A. A Mouse Model of DEPDC5-Related Epilepsy: Neuronal Loss of Depdc5 Causes Dysplastic and Ectopic Neurons, Increased MTOR Signaling, and Seizure Susceptibility. Neurobiol. Dis. 2018, 111, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.-S.; Kowalsky, A.H.; Namkoong, S.; Park, S.-R.; Wu, S.; Kim, B.; James, A.; Gu, B.; Semple, I.A.; Tohamy, M.A.; et al. Concurrent Activation of Growth Factor and Nutrient Arms of MTORC1 Induces Oxidative Liver Injury. Cell Discov. 2019, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Grabiner, B.C.; Nardi, V.; Birsoy, K.K.; Possemato, R.; Shen, K.; Sinha, S.; Jordan, A.; Beck, A.H.; Sabatini, D.M. A Diverse Array of Cancer-Associated MTOR Mutations Are Hyperactivating and Can Predict Rapamycin Sensitivity. Cancer Discov. 2014, 4, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Weckhuysen, S.; Marsan, E.; Lambrecq, V.; Marchal, C.; Morin-Brureau, M.; An-Gourfinkel, I.; Baulac, M.; Fohlen, M.; Kallay Zetchi, C.; Seeck, M.; et al. Involvement of GATOR Complex Genes in Familial Focal Epilepsies and Focal Cortical Dysplasia. Epilepsia 2016, 57, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Picard, F.; Rudolf, G.; Noé, E.; Achaz, G.; Thomas, P.; Genton, P.; Mundwiller, E.; Wolff, M.; Marescaux, C.; et al. Mutations of DEPDC5 Cause Autosomal Dominant Focal Epilepsies. Nat. Genet. 2013, 45, 552–555. [Google Scholar] [CrossRef]
- Dibbens, L.M.; de Vries, B.; Donatello, S.; Heron, S.E.; Hodgson, B.L.; Chintawar, S.; Crompton, D.E.; Hughes, J.N.; Bellows, S.T.; Klein, K.M.; et al. Mutations in DEPDC5 Cause Familial Focal Epilepsy with Variable Foci. Nat. Genet. 2013, 45, 546–551. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Heron, S.E.; Regan, B.M.; Mandelstam, S.; Crompton, D.E.; Hodgson, B.L.; Licchetta, L.; Provini, F.; Bisulli, F.; Vadlamudi, L.; et al. Mutations in Mammalian Target of Rapamycin Regulator DEPDC5 Cause Focal Epilepsy with Brain Malformations. Ann. Neurol. 2014, 75, 782–787. [Google Scholar] [CrossRef]
- Nascimento, F.A.; Borlot, F.; Cossette, P.; Minassian, B.A.; Andrade, D.M. Two Definite Cases of Sudden Unexpected Death in Epilepsy in a Family with a DEPDC5 Mutation. Neurol. Genet. 2015, 1, e28. [Google Scholar] [CrossRef]
- Sim, J.C.; Scerri, T.; Fanjul-Fernández, M.; Riseley, J.R.; Gillies, G.; Pope, K.; Van Roozendaal, H.; Heng, J.I.; Mandelstam, S.A.; McGillivray, G.; et al. Familial Cortical Dysplasia Caused by Mutation in the Mammalian Target of Rapamycin Regulator NPRL3. Ann. Neurol. 2016, 79, 132–137. [Google Scholar] [CrossRef]
- Ricos, M.G.; Hodgson, B.L.; Pippucci, T.; Saidin, A.; Ong, Y.S.; Heron, S.E.; Licchetta, L.; Bisulli, F.; Bayly, M.A.; Hughes, J.; et al. Mutations in the Mammalian Target of Rapamycin Pathway Regulators NPRL2 and NPRL3 Cause Focal Epilepsy. Ann. Neurol. 2016, 79, 120–131. [Google Scholar] [CrossRef]
- Baldassari, S.; Picard, F.; Verbeek, N.E.; van Kempen, M.; Brilstra, E.H.; Lesca, G.; Conti, V.; Guerrini, R.; Bisulli, F.; Licchetta, L.; et al. The Landscape of Epilepsy-Related GATOR1 Variants. Genet. Med. 2019, 21, 398–408. [Google Scholar] [CrossRef]
- Ribierre, T.; Deleuze, C.; Bacq, A.; Baldassari, S.; Marsan, E.; Chipaux, M.; Muraca, G.; Roussel, D.; Navarro, V.; Leguern, E.; et al. Second-Hit Mosaic Mutation in MTORC1 Repressor DEPDC5 Causes Focal Cortical Dysplasia–Associated Epilepsy. J. Clin. Investig. 2018, 128, 2452–2458. [Google Scholar] [CrossRef]
- Lee, W.S.; Stephenson, S.E.M.; Howell, K.B.; Pope, K.; Gillies, G.; Wray, A.; Maixner, W.; Mandelstam, S.A.; Berkovic, S.F.; Scheffer, I.E.; et al. Second-hit DEPDC5 Mutation Is Limited to Dysmorphic Neurons in Cortical Dysplasia Type IIA. Ann. Clin. Transl. Neurol. 2019, 6, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- D’Gama, A.M.; Woodworth, M.B.; Hossain, A.A.; Bizzotto, S.; Hatem, N.E.; LaCoursiere, C.M.; Najm, I.; Ying, Z.; Yang, E.; Barkovich, A.J.; et al. Somatic Mutations Activating the MTOR Pathway in Dorsal Telencephalic Progenitors Cause a Continuum of Cortical Dysplasias. Cell Rep. 2017, 21, 3754–3766. [Google Scholar] [CrossRef]
- Chandrasekar, I.; Tourney, A.; Loo, K.; Carmichael, J.; James, K.; Ellsworth, K.A.; Dimmock, D.; Joseph, M. Hemimegalencephaly and Intractable Seizures Associated with the NPRL3 Gene Variant in a Newborn: A Case Report. Am. J. Med. Genet. 2021, 185, 2126–2130. [Google Scholar] [CrossRef]
- Ryu, C.S.; Bae, J.; Kim, I.J.; Kim, J.; Oh, S.H.; Kim, O.J.; Kim, N.K. MPG and NPRL3 Polymorphisms Are Associated with Ischemic Stroke Susceptibility and Post-Stroke Mortality. Diagnostics 2020, 10, 947. [Google Scholar] [CrossRef]
- Cocito, L.; Loeb, C. Focal Epilepsy as a Possible Sign of Transient Subclinical Ischemia. Eur. Neurol. 1989, 29, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Basel-Vanagaite, L.; Hershkovitz, T.; Heyman, E.; Raspall-Chaure, M.; Kakar, N.; Smirin-Yosef, P.; Vila-Pueyo, M.; Kornreich, L.; Thiele, H.; Bode, H.; et al. Biallelic SZT2 Mutations Cause Infantile Encephalopathy with Epilepsy and Dysmorphic Corpus Callosum. Am. J. Hum. Genet. 2013, 93, 524–529. [Google Scholar] [CrossRef]
- Baple, E.L.; Maroofian, R.; Chioza, B.A.; Izadi, M.; Cross, H.E.; Al-Turki, S.; Barwick, K.; Skrzypiec, A.; Pawlak, R.; Wagner, K.; et al. Mutations in KPTN Cause Macrocephaly, Neurodevelopmental Delay, and Seizures. Am. J. Hum. Genet. 2014, 94, 87–94. [Google Scholar] [CrossRef]
- Trivisano, M.; Rivera, M.; Terracciano, A.; Ciolfi, A.; Napolitano, A.; Pepi, C.; Calabrese, C.; Digilio, M.C.; Tartaglia, M.; Curatolo, P.; et al. Developmental and Epileptic Encephalopathy Due to SZT2 Genomic Variants: Emerging Features of a Syndromic Condition. Epilepsy Behav. 2020, 108, 107097. [Google Scholar] [CrossRef] [PubMed]
- Crino, P.B. MTOR: A Pathogenic Signaling Pathway in Developmental Brain Malformations. Trends Mol. Med. 2011, 17, 734–742. [Google Scholar] [CrossRef]
- Lim, K.C.; Crino, P.B. Focal Malformations of Cortical Development: New Vistas for Molecular Pathogenesis. Neuroscience 2013, 252, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Jiang, Q.; Li, S.; Zhu, H.; Xu, R.; Song, N.; Ding, X.; Liu, J.; Chen, M.; Song, M.; et al. Opposing Functions of β-Arrestin 1 and 2 in Parkinson’s Disease via Microglia Inflammation and Nprl3. Cell Death. Differ. 2021, 28, 1822–1836. [Google Scholar] [CrossRef]
- Iffland, P.H.; Carson, V.; Bordey, A.; Crino, P.B. GATOR Opathies: The Role of Amino Acid Regulatory Gene Mutations in Epilepsy and Cortical Malformations. Epilepsia 2019, 60, 2163–2173. [Google Scholar] [CrossRef] [PubMed]
- Lerman, M.I.; Minna, J.D. The 630-Kb Lung Cancer Homozygous Deletion Region on Human Chromosome 3p21.3: Identification and Evaluation of the Resident Candidate Tumor Suppressor Genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res. 2000, 60, 6116–6133. [Google Scholar] [PubMed]
- Bertucci, F.; Ng, C.K.Y.; Patsouris, A.; Droin, N.; Piscuoglio, S.; Carbuccia, N.; Soria, J.C.; Dien, A.T.; Adnani, Y.; Kamal, M.; et al. Genomic Characterization of Metastatic Breast Cancers. Nature 2019, 569, 560–564. [Google Scholar] [CrossRef]
- Otani, S.; Takeda, S.; Yamada, S.; Sakakima, Y.; Sugimoto, H.; Nomoto, S.; Kasuya, H.; Kanazumi, N.; Nagasaka, T.; Nakao, A. The Tumor Suppressor NPRL2 in Hepatocellular Carcinoma Plays an Important Role in Progression and Can Be Served as an Independent Prognostic Factor. J. Surg. Oncol. 2009, 100, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Y.; Jiang, L.; Tang, W. Decreased Expression of NPRL2 in Renal Cancer Cells Is Associated with Unfavourable Pathological, Proliferation and Apoptotic Features. Pathol. Oncol. Res. POR 2014, 20, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, F.; Haraldson, K.; Protopopov, A.; Duh, F.M.; Geil, L.; Kuzmin, I.; Minna, J.D.; Stanbridge, E.; Braga, E.; et al. Functional Characterization of the Candidate Tumor Suppressor Gene NPRL2/G21 Located in 3p21.3C. Cancer Res. 2004, 64, 6438–6443. [Google Scholar] [CrossRef][Green Version]
- Yogurtcu, B.; Hatemi, I.; Aydin, I.; Buyru, N. NPRL2 Gene Expression in the Progression of Colon Tumors. Genet. Mol. Res. 2012, 11, 4810–4816. [Google Scholar] [CrossRef]
- Liu, M.N.; Liu, A.Y.; Pei, F.H.; Ma, X.; Fan, Y.J.; Du, Y.J.; Liu, B.R. Functional Mechanism of the Enhancement of 5-Fluorouracil Sensitivity by TUSC4 in Colon Cancer Cells. Oncol. Lett. 2015, 10, 3682–3688. [Google Scholar] [CrossRef]
- Liu, A.; Qiao, J.; He, L.; Liu, Z.; Chen, J.; Pei, F.; Du, Y. Nitrogen Permease Regulator-Like-2 Exhibited Anti-Tumor Effects and Enhanced the Sensitivity of Colorectal Cancer Cells to Oxaliplatin and 5-Fluorouracil. OTT 2019, 12, 8637–8644. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Dai, H.; Wang, E.; Lin, C.C.J.; Mo, W.; Peng, G.; Lin, S.Y. TUSC4 Functions as a Tumor Suppressor by Regulating BRCA1 Stability. Cancer Res. 2015, 75, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Nishizaki, M.; Gao, B.N.; Burbee, D.; Kondo, M.; Kamibayashi, C.; Xu, K.; Yen, N.; Atkinson, E.N.; Fang, B.L.; et al. Expression of Several Genes in the Human Chromosome 3p21.3 Homozygous Deletion Region by an Adenovirus Vector Results in Tumor Suppressor Activities in Vitro and in Vivo. Cancer Res. 2002, 62, 2715–2720. [Google Scholar] [PubMed]
- Anedchenko, E.A.; Dmitriev, A.A.; Krasnov, G.S.; Kondrat’eva, O.O.; Kopantsev, E.P.; Vinogradova, T.V.; Zinov’eva, M.V.; Zborovskaya, I.B.; Polotsky, B.E.; Sacharova, O.V.; et al. Downregulation of RBSP3/CTDSPL, NPRL2/G21, RASSF1A, ITGA9, HYAL1, and HYAL2 in Non-Small Cell Lung Cancer. Mol. Biol. 2008, 42, 859–869. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, S.; Chen, Y.; Xie, X.; Du, Z.; Jiang, L. High Expression of NPRL2 Is Linked to Poor Prognosis in Patients with Prostate Cancer. Hum. Pathol. 2018, 76, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, B. Overexpression of Nitrogen Permease Regulator Like-2 (NPRL2) Enhances Sensitivity to Irinotecan (CPT-11) in Colon Cancer Cells by Activating the DNA Damage Checkpoint Pathway. Med. Sci. Monit. 2018, 24, 1424–1433. [Google Scholar] [CrossRef]
- Cai, Y.; Xu, G.; Wu, F.; Michelini, F.; Chan, C.; Qu, X.; Selenica, P.; Ladewig, E.; Castel, P.; Cheng, Y.; et al. Genomic Alterations in PIK3CA-Mutated Breast Cancer Result in MTORC1 Activation and Limit Sensitivity to PI3Kα Inhibitors. Cancer Res. 2021, 81, 2470–2480. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Jiang, Q.; Zhu, P.; Chen, Y.; Xie, X.; Du, Z.; Jiang, L.; Tang, W. NPRL2 Enhances Autophagy and the Resistance to Everolimus in Castration-Resistant Prostate Cancer. Prostate 2019, 79, 44–53. [Google Scholar] [CrossRef]
- Pang, Y.; Xie, F.; Cao, H.; Wang, C.; Zhu, M.; Liu, X.; Lu, X.; Huang, T.; Shen, Y.; Li, K.; et al. Mutational Inactivation of MTORC1 Repressor Gene DEPDC5 in Human Gastrointestinal Stromal Tumors. Proc. Natl. Acad. Sci. USA 2019, 116, 22746–22753. [Google Scholar] [CrossRef]
- Milton, J.N.; Rooks, H.; Drasar, E.; McCabe, E.L.; Baldwin, C.T.; Melista, E.; Gordeuk, V.R.; Nouraie, M.; Kato, G.J.R.; Minniti, C.; et al. Genetic Determinants of Haemolysis in Sickle Cell Anaemia. Br. J. Haematol. 2013, 161, 270–278. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Hughes, J.R.; Garrick, D.; Lynch, M.D.; Sharpe, J.A.; Sloane-Stanley, J.A.; McGowan, S.J.; De Gobbi, M.; Hosseini, M.; Vernimmen, D.; et al. Intragenic Enhancers Act as Alternative Promoters. Mol. Cell 2012, 45, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.; Hughes, J.R.; Babbs, C.; Davies, J.O.J.; Graham, B.J.; Hanssen, L.L.P.; Kassouf, M.T.; Oudelaar, A.M.; Sharpe, J.A.; Suciu, M.C.; et al. Genetic Dissection of the α-Globin Super-Enhancer in Vivo. Nat. Genet. 2016, 48, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Gillemans, N.; Hockman, D.; Demmers, J.A.A.; Cheng, J.-F.; Hou, J.; Salminen, M.; Fisher, C.A.; Taylor, S.; Gibbons, R.J.; et al. An Evolutionarily Ancient Mechanism for Regulation of Hemoglobin Expression in Vertebrate Red Cells. Blood 2020, 136, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Everything Reptiles. Available online: https://www.everythingreptiles.com/Alligator-vs-Crocodile (accessed on 31 August 2021).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loissell-Baltazar, Y.A.; Dokudovskaya, S. SEA and GATOR 10 Years Later. Cells 2021, 10, 2689. https://doi.org/10.3390/cells10102689
Loissell-Baltazar YA, Dokudovskaya S. SEA and GATOR 10 Years Later. Cells. 2021; 10(10):2689. https://doi.org/10.3390/cells10102689
Chicago/Turabian StyleLoissell-Baltazar, Yahir A., and Svetlana Dokudovskaya. 2021. "SEA and GATOR 10 Years Later" Cells 10, no. 10: 2689. https://doi.org/10.3390/cells10102689
APA StyleLoissell-Baltazar, Y. A., & Dokudovskaya, S. (2021). SEA and GATOR 10 Years Later. Cells, 10(10), 2689. https://doi.org/10.3390/cells10102689






