SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Amino Acid Sequence Alignment and Phylogenetic Analysis
2.3. Cloning and Plasmids
2.4. Expression and Purification of Recombinant SP-1
2.5. Western Blotting and Kinetic Constant Determination
2.6. Peptide Library Preparation
2.7. Animals
2.8. Assessment of Mild Inflammation and Study Design
2.9. MPO Activity Measurement
2.10. Histological Evaluation and Scoring
2.11. Cell Cultures, SP-1 Treatment and Intestinal Permeability Assay
2.12. Cytokine Quantification
2.13. Fecal Protease Activity Measurement
2.14. Microbiota Analysis
2.15. Correlation Analyses
2.16. Statistical Analyses
3. Results
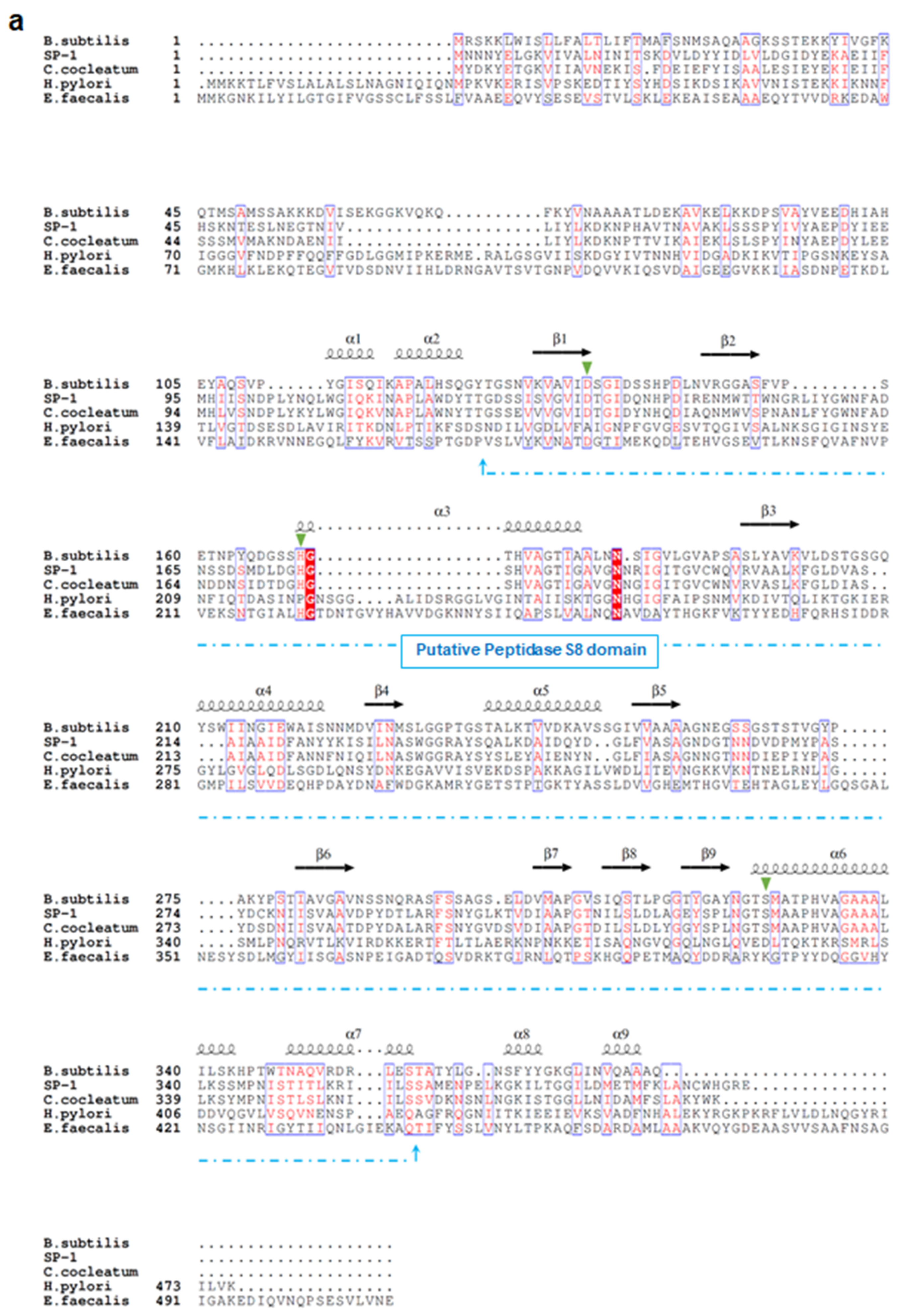
3.1. SP-1 Shares Conserved Sequence Features with Subtilisin Despite Overall Dissimilarity
3.2. Purification and Steady-State Kinetic Analysis of SP-1
3.3. Novel Substrate for SP-1
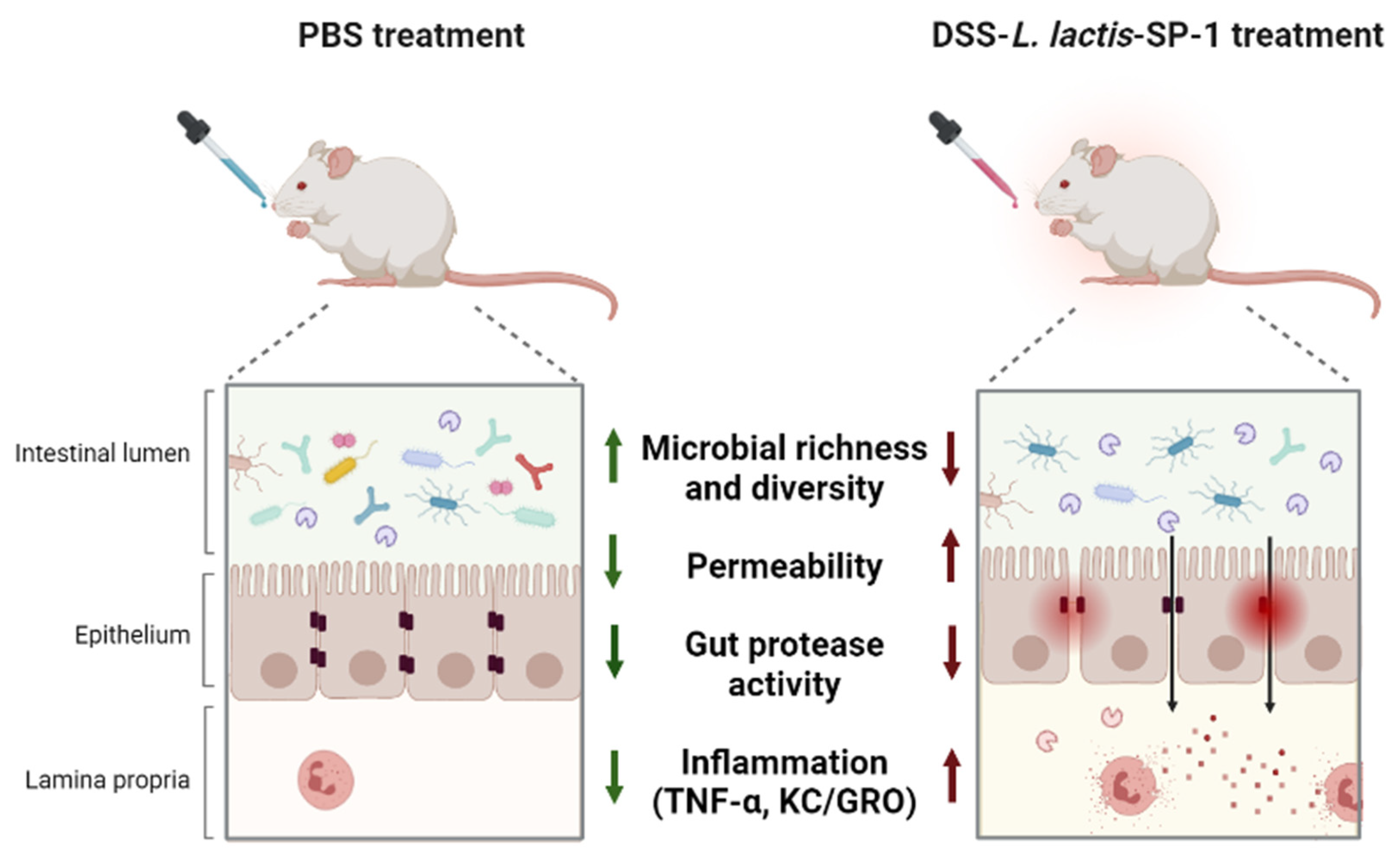
3.4. Administration of L. Lactis Secreting SP-1 Exacerbates DSS-Induced Colitis
3.5. SP-1 Increases Gut Inflammation and Fecal Protease Activity in Mice
3.6. Alteration of Gut Microbiota and Host Response
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Ananthakrishnan, A.N.; Bernstein, C.N.; Iliopoulos, D.; MacPherson, A.; Neurath, M.F.; Ali, R.A.R.; Vavricka, S.R.; Fiocchi, C. Environmental triggers in IBD: A review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 39–49. [Google Scholar] [CrossRef]
- Kaplan, G.G.; Ng, S.C. Understanding and Preventing the global increase of inflammatory bowel disease. Gastroenterology 2017, 152, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef]
- Xu, X.R.; Liu, C.Q.; Feng, B.S.; Liu, Z.J. Dysregulation of mucosal immune response in pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 3255–3264. [Google Scholar] [CrossRef]
- Solà Tapias, N.; Vergnolle, N.; Denadai-Souza, A.; Barreau, F. The interplay between genetic risk factors and proteolytic dysregulation in the pathophysiology of inflammatory bowel disease. J. Crohns. Colitis. 2020, 33, 1–33. [Google Scholar] [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; Van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.K.; Kamm, M.A.; Teo, S.M.; Inouye, M.; Wagner, J.; Kirkwood, C.D. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: A systematic review. Inflamm. Bowel. Dis. 2015, 21, 1219–1228. [Google Scholar] [PubMed] [Green Version]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A microbial signature for Crohn’s disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Okayasu, I.; Hatakeyama, S.; Yamada, M.; Ohkusa, T.; Inagaki, Y.; Nakaya, R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990, 98, 694–702. [Google Scholar] [CrossRef]
- Lucke, K.; Miehlke, S.; Jacobs, E.; Schuppler, M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 2006, 55, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Sankarasubramanian, J.; Ahmad, R.; Avuthu, N.; Singh, A.B.; Guda, C. Gut Microbiota and Metabolic Specificity in Ulcerative Colitis and Crohn’s Disease. Front. Med. 2020, 7, 606298. [Google Scholar] [CrossRef]
- Leal, J.; Gregson, D.B.; Ross, T.; Church, D.L.; Laupland, K.B. Epidemiology of Clostridium species bacteremia in Calgary, Canada, 2000-2006. J. Inf. Secur. 2008, 57, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Senda, S.; Fujiyama, Y.; Ushijima, T.; Hodohara, K.; Bamba, T.; Hosoda, S.; Kobayashi, K. Clostridium ramosum, an IgA protease-producing species and its ecology in the human intestinal tract. Microbiol. Immunol. 1985, 29, 1019–1028. [Google Scholar] [CrossRef]
- Kosowska, K.; Reinholdt, J.; Rasmussen, L.K.; Sabat, A.; Potempa, J.; Kilian, M.; Poulsen, K. The Clostridium ramosum IgA proteinase represents a novel type of metalloendopeptidase. J. Biol. Chem. 2002, 277, 11987–11994. [Google Scholar] [CrossRef] [Green Version]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgington-Mitchell, L.E. Pathophysiological roles for proteases in gastrointestinal disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steck, N.; Mueller, K.; Schemann, M.; Haller, D. Bacterial proteases in IBD and IBS. Gut 2012, 61, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Carroll, I.M.; Maharshak, N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J. Gastroenterol. 2013, 19, 7531–7543. [Google Scholar] [CrossRef]
- Frees, D.; Qazi, S.N.; Hill, P.J.; Ingmer, H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 2003, 48, 1565–1578. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Y.; Shao, H.; Ma, J.; Song, X.; Zhang, M.; Chang, Z. DegP functions as a critical protease for bacterial acid resistance. FEBS J. 2018, 285, 3525–3538. [Google Scholar] [CrossRef] [Green Version]
- Carlisle, M.D.; Srikantha, R.N.; Brogden, K.A. Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J. Innate Immun. 2009, 1, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Teng, F.; Murray, B.E. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect. Immun. 2005, 73, 1606–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steck, N.; Hoffmann, M.; Sava, I.G.; Kim, S.C.; Hahne, H.; Tonkonogy, S.L.; Mair, K.; Krueger, D.; Pruteanu, M.; Shanahan, F.; et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 2011, 141, 959–971. [Google Scholar] [CrossRef] [Green Version]
- Maharshak, N.; Huh, E.Y.; Paiboonrungruang, C.; Shanahan, M.; Thurlow, L.; Herzog, J.; Djukic, Z.; Orlando, R.; Pawlinski, R.; Ellermann, M.; et al. Enterococcus faecalis Gelatinase Mediates Intestinal Permeability via Protease-Activated Receptor 2. Infect. Immun. 2015, 83, 2762–2770. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Cummings, J.H.; Allison, C. Protein degradation by human intestinal bacteria. J. Gen. Microbiol. 1986, 132, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- Macfarlane, G.T.; Allison, C.; Gibson, S.A.; Cummings, J.H. Contribution of the microflora to proteolysis in the human large intestine. J. Appl. Bacteriol. 1988, 64, 37–46. [Google Scholar] [CrossRef]
- Harrer, A.; Boehm, M.; Backert, S.; Tegtmeyer, N. Overexpression of serine protease HtrA enhances disruption of adherens junctions, paracellular transmigration and type IV secretion of CagA by Helicobacter pylori. Gut Pathog. 2017, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Gibold, L.; Garenaux, E.; Dalmasso, G.; Gallucci, C.; Cia, D.; Mottet-Auselo, B.; Faïs, T.; Darfeuille-Michaud, A.; Nguyen, H.T.T.; Barnich, N.; et al. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell Microbiol. 2016, 18, 617–631. [Google Scholar] [CrossRef]
- Pontarollo, G.; Acquasaliente, L.; Peterle, D.; Frasson, R.; Artusi, I.; De Filippis, V. Non-canonical proteolytic activation of human prothrombin by subtilisin from Bacillus subtilis may shift the procoagulant-anticoagulant equilibrium toward thrombosis. J. Biol. Chem. 2017, 292, 15161–15179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Métoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; MetaHIT Consortium; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef] [PubMed]
- de Ruyter, P.G.; Kuipers, O.P.; de Vos, W.M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 1996, 62, 3662–3667. [Google Scholar] [CrossRef] [Green Version]
- van Asseldonk, M.; de Vos, W.M.; Simons, G. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and a heterologous alpha-amylase. Mol. Gen. Genet. 1993, 240, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, M.; Gruba, N.; Grzywa, R.; Giełdoń, A.; Bąchor, R.; Brzozowski, K.; Sieńczyk, M.; Dieter, J.; Szewczuk, Z.; Rolka, K.; et al. PEGylated substrates of NSP4 protease: A tool to study protease specificity. Sci. Rep. 2016, 6, 22856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hojo, K.; Maeda, M.; Iguchi, S.; Smith, T.; Okamoto, H.; Kawasaki, K. Amino acids and peptides. V. Facile preparation of p-nitroanilide analogs by the solid-phase method. Chem. Pharm. Bull. 2010, 48, 1740–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sole, N.A.; Barany, G. Optimization of solidphase synthesis of [Ala8]-dynorphin-A. J. Org. Chem. 1992, 57, 5399–5403. [Google Scholar] [CrossRef]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Investig. 1993, 69, 238–249. [Google Scholar]
- Krawisz, J.E.; Sharon, P.; Stenson, W.F. Quantitative assay for acute intestinal inflammation based on myeloperoxidase activity. Assessment of inflammation in rat and hamster models. Gastroenterology 1984, 87, 1344–1350. [Google Scholar] [CrossRef]
- Chantret, I.; Rodolosse, A.; Barbat, A.; Dussaulx, E.; Brot-Laroche, E.; Zweibaum, A.; Rousset, M. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: Evidence for glucose-dependent negative regulation. J. Cell Sci. 1994, 107, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Godon, J.J.; Zumstein, E.; Dabert, P.; Habouzit, F.; Moletta, R. Microbial 16S rDNA diversity in an anaerobic digester. Water Sci. Technol. 1997, 36, 49–55. [Google Scholar] [CrossRef]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2017, 34, 1287–1294. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons using rank sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Fischer, S.; Kumar, N.; Clavel, T. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017, 5, e2836. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, T.; Ng, S.C. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef]
- Loubinoux, J.; Bronowicki, J.P.; Pereira, I.A.; Mougenel, J.L.; Faou, A.E. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 2002, 40, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Golińska, E.; Tomusiak, A.; Gosiewski, T.; Więcek, G.; Machul, A.; Mikołajczyk, D.; Bulanda, M.; Heczko, P.B.; Strus, M. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J. Gastroenterol. 2013, 19, 3562–3572. [Google Scholar] [CrossRef]
- Vester-Andersen, M.K.; Mirsepasi-Lauridsen, H.C.; Prosberg, M.V.; Mortensen, C.O.; Träger, C.; Skovsen, K.; Thorkilgaard, T.; Nøjgaard, C.; Vind, I.; Krogfelt, K.A.; et al. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci. Rep. 2019, 9, 13473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banaszkiewicz, A.; Kądzielska, J.; Gawrońska, A.; Pituch, H.; Obuch-Woszczatyński, P.; Albrecht, P.; Młynarczyk, G.; Radzikowski, A. Enterotoxigenic Clostridium perfringens infection and pediatric patients with inflammatory bowel disease. J. Crohns. Colitis 2014, 8, 276–281. [Google Scholar] [CrossRef] [Green Version]
- Cottrell, G.S.; Amadesi, S.; Pikios, S.; Camerer, E.; Willardsen, J.A.; Murphy, B.R.; Caughey, G.H.; Wolters, P.J.; Coughlin, S.R.; Peterson, A.; et al. Protease-activated receptor 2, dipeptidyl peptidase I, and proteases mediate Clostridium difficile toxin A enteritis. Gastroenterology 2007, 132, 2422–2437. [Google Scholar] [CrossRef] [Green Version]
- Meldal, M.; Svendsen, I.; Breddam, K.; Auzanneau, F.I. Portionmixing peptide libraries of quenched fluorogenic substrates for complete subsite mapping of endoprotease specificity. Proc. Natl. Acad. Sci. USA 1994, 91, 3314–3318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milgotina, E.I.; Shcheglov, A.S.; Lapa, G.B.; Chestukhina, G.G.; Voyushina, T.L. Enzymatic synthesis of chromogenic substrates for Glu, Asp-specific proteinases. J. Pept. Res. 2001, 58, 12–16. [Google Scholar] [CrossRef]
- Gosalia, D.N.; Salisbury, C.M.; Ellman, J.A.; Diamond, S.L. High throughput substrate specificity profiling of serine and cysteine proteases using solution-phase fluorogenic peptide microarrays. Mol. Cell Proteomics 2005, 4, 626–636. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, S.; Popp, V.; Kindermann, M.; Gerlach, K.; Weigmann, B.; Fichtner-Feigl, S.; Neurath, M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017, 12, 1295–1309. [Google Scholar] [CrossRef]
- Martini, E.; Krug, S.M.; Siegmund, B.; Neurath, M.F.; Becker, C. Mend Your Fences: The epithelial barrier and its relationship with mucosal immunity in Inflammatory Bowel Disease. Cell Mol. Gastroenterol. Hepatol. 2017, 4, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Fukui, H. Increased intestinal permeability and decreased barrier function: Does it really influence the risk of inflammation? Inflamm. Intest. Dis. 2016, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Van Spaendonk, H.; Ceuleers, H.; Witters, L.; Patteet, E.; Joossens, J.; Augustyns, K.; Lambeir, A.-M.; De Meester, I.; De Man, J.G.; De Winter, B.Y. Regulation of intestinal permeability: The role of proteases. World J. Gastroenterol. 2017, 23, 2106–2123. [Google Scholar] [CrossRef]
- Jablaoui, A.; Kriaa, A.; Mkaouar, H.; Akermi, N.; Soussou, S.; Wysocka, M.; Wołoszyn, D.; Amouri, A.; Gargouri, A.; Maguin, E.; et al. Fecal serine protease profiling in Inflammatory Bowel Diseases. Front. Cell Infect. Microbiol. 2020, 10, 21. [Google Scholar] [CrossRef]
- Gecse, K.; Róka, R.; Ferrier, L.; Leveque, M.; Eutamene, H.; Cartier, C.; Ait-Belgnaoui, A.; Rosztoczy, A.; Izbeki, F.; Fioramonti, J.; et al. Increased faecal serine protease activity in diarrhoeic IBS patients: A colonic lumenal factor impairing colonic permeability and sensitivity. Gut 2008, 57, 591–599. [Google Scholar] [CrossRef]
- Woodmansey, E.J.; McMurdo, M.E.; Macfarlane, G.T.; Macfarlane, S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl. Environ. Microbiol. 2004, 70, 6113–6122. [Google Scholar] [CrossRef] [Green Version]
- Carroll, I.M.; Ringel-Kulka, T.; Ferrier, L.; Wu, M.C.; Siddle, J.P.; Bueno, L.; Ringel, Y. Fecal protease activity is associated with compositional alterations in the intestinal microbiota. PLoS ONE 2013, 8, e78017. [Google Scholar] [CrossRef]
- Palmela, C.; Chevarin, C.; Xu, Z.; Torres, J.; Sevrin, G.; Hirten, R.; Barnich, N.; Ng, S.C.; Colombel, J.-F. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 2018, 67, 574–587. [Google Scholar] [CrossRef]
- Kotlowski, R.; Bernstein, C.N.; Sepehri, S.; Krause, D.O. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007, 56, 669–675. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chen, H.; He, H.; Du, Y.; Hu, J.; Li, Y.; Li, Y.; Zhou, Y.; Wang, H.; Chen, Y.; et al. Increased Enterococcus faecalis infection is associated with clinically active Crohn disease. Medicine 2016, 95, e5019. [Google Scholar] [CrossRef]
- Rowan, F.; Docherty, N.G.; Murphy, M.; Murphy, B.; Calvin Coffey, J.; O’Connell, P.R. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon. Rectum 2010, 53, 1530–1536. [Google Scholar] [CrossRef]
- Singh, S.; Bhatia, R.; Khare, P.; Sharma, S.; Rajarammohan, S.; Bishnoi, M.; Bhadada, S.K.; Sharma, S.S.; Kaur, J.; Kondepudi, K.K. Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci. Rep. 2020, 10, 18597. [Google Scholar] [CrossRef]







| Score | Weight Loss (%) | Stool Consistency | Visible Blood in Stool |
|---|---|---|---|
| 0 | None | Normal | No bleeding |
| 1 | 1–5 | Loose stools | Slight bleeding |
| 2 | 5–10 | ||
| 3 | 10–15 | Diarrhea | Gross bleeding |
| 4 | >15 |
| Substrate | Sequence |
|---|---|
| SP-1 | ABZ-Ileu-Met-Ala-Arg-ANB-NH2 |
| Trypsin-like | AB2-Val-Val-Ser-Lys-ANB-NH2 |
| Elastase-like | AB2-Met-Pro-Val-Ala-Trp-Glu-Tyr-(3-NO2)-NH2 |
| PR3-like | AB2-Tyr-Tyr-ABU-Asn-Glu-Pro-Tyr-(3-NO2)-NH2 |
| Substrate | KM × 10−4 (M) | kcat (s−1) | kcat/KM × 104 (M−1s−1) |
|---|---|---|---|
| ABZ-Ileu-Met-Ala-Arg-ANB-NH2 | 1.91 ± 0.3 | 2.83 ± 0.5 | 1.48 |
| AB2-Val-Val-Ser-Lys-ANB-NH2 | 6.12 ± 0.5 | 1.79 ± 0.2 | 0.29 |
| AB2-Met-Pro-Val-Ala-Trp-Glu-Tyr-(3-NO2)-NH2 | 17.7 ± 0.1 | 0.46 ± 0.08 | 0.025 |
| AB2-Tyr-Tyr-ABU-Asn-Glu-Pro-Tyr-(3-NO2)-NH2 | 14.1 ± 0.7 | 0.40 ± 0.06 | 0.027 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kriaa, A.; Jablaoui, A.; Rhimi, S.; Soussou, S.; Mkaouar, H.; Mariaule, V.; Gruba, N.; Gargouri, A.; Maguin, E.; Lesner, A.; et al. SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice. Cells 2021, 10, 2658. https://doi.org/10.3390/cells10102658
Kriaa A, Jablaoui A, Rhimi S, Soussou S, Mkaouar H, Mariaule V, Gruba N, Gargouri A, Maguin E, Lesner A, et al. SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice. Cells. 2021; 10(10):2658. https://doi.org/10.3390/cells10102658
Chicago/Turabian StyleKriaa, Aicha, Amin Jablaoui, Soufien Rhimi, Souha Soussou, Héla Mkaouar, Vincent Mariaule, Natalia Gruba, Ali Gargouri, Emmanuelle Maguin, Adam Lesner, and et al. 2021. "SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice" Cells 10, no. 10: 2658. https://doi.org/10.3390/cells10102658
APA StyleKriaa, A., Jablaoui, A., Rhimi, S., Soussou, S., Mkaouar, H., Mariaule, V., Gruba, N., Gargouri, A., Maguin, E., Lesner, A., & Rhimi, M. (2021). SP-1, a Serine Protease from the Gut Microbiota, Influences Colitis and Drives Intestinal Dysbiosis in Mice. Cells, 10(10), 2658. https://doi.org/10.3390/cells10102658






