The Dictyostelium Centrosome
Abstract
:1. Introduction
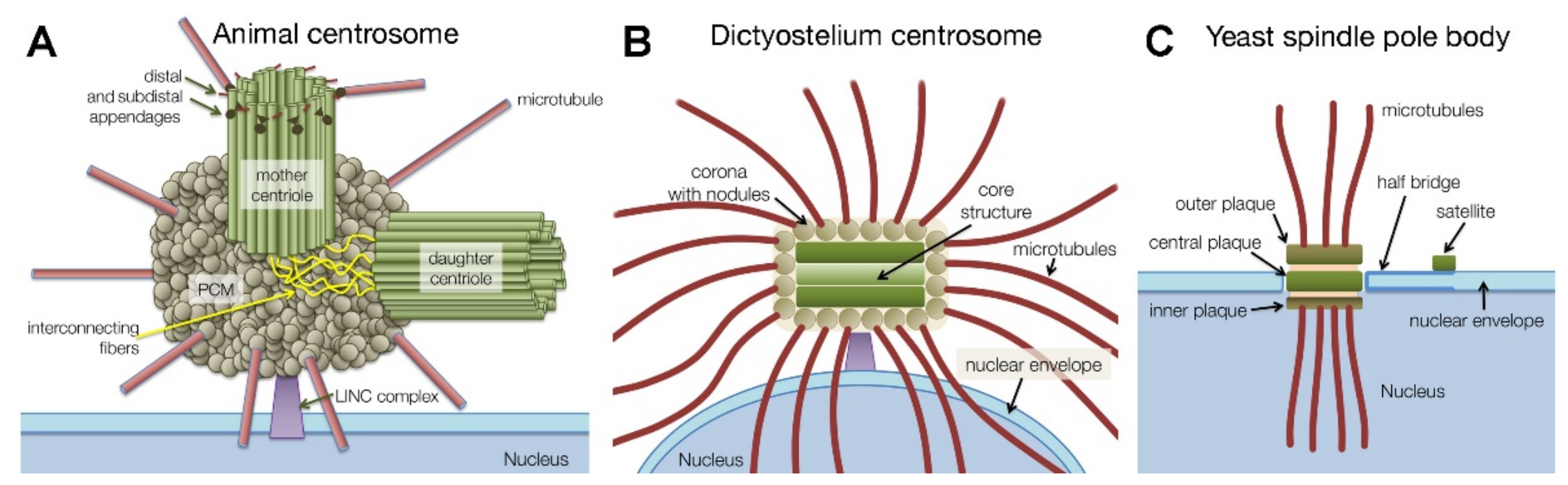
1.1. Centrosome Types and Centrosome Duplication
1.2. Centrosome Functions
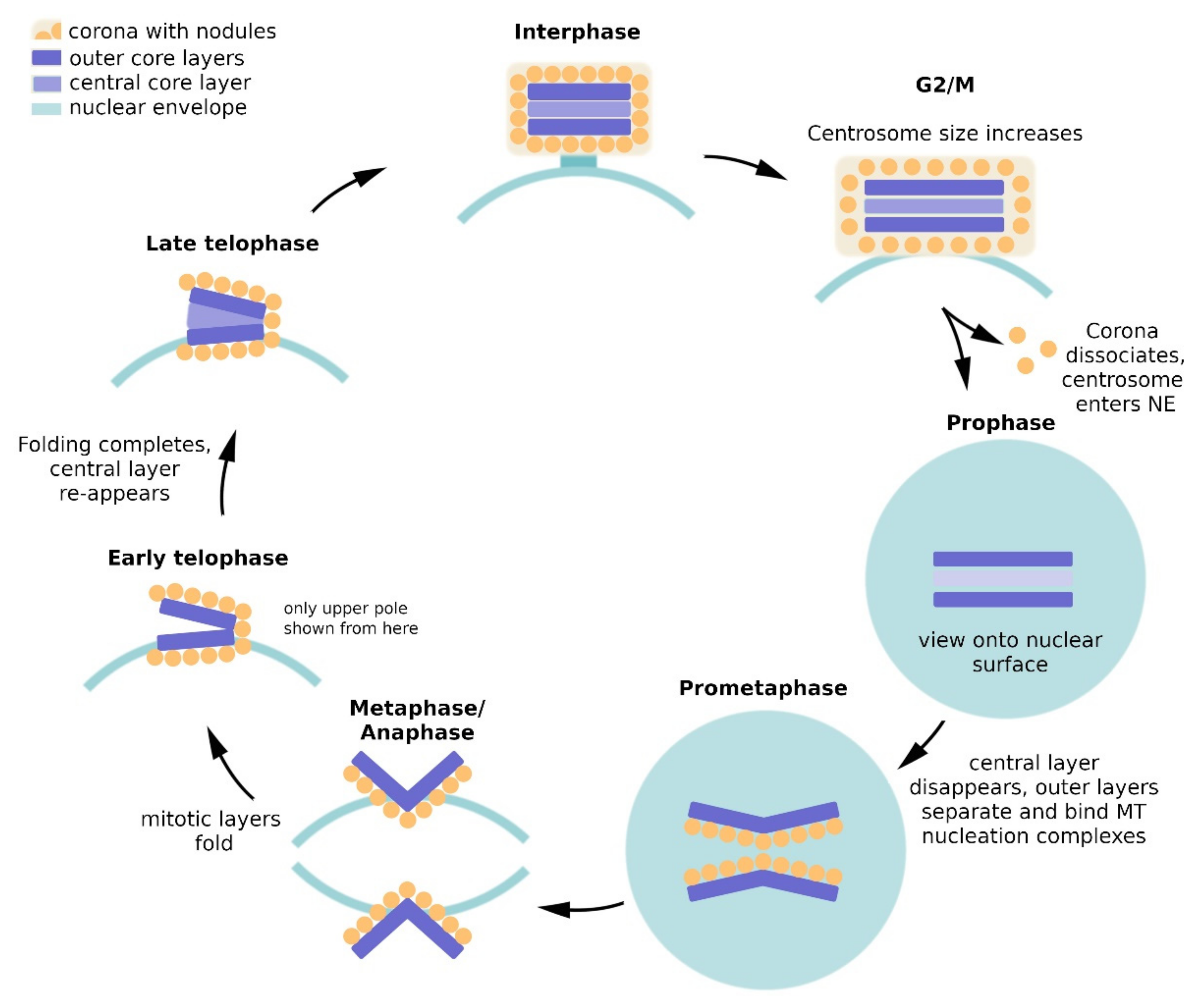
2. Dictyostelium Centrosome Composition and Topology
2.1. Composition of the Corona
2.1.1. γ-Tubulin and Its Interactors
2.1.2. Centrosomal Microtubule-Associated Proteins
2.1.3. Further Prominent Corona Components
2.2. Composition of the Layered Core
2.2.1. Outer Core Layers
2.2.2. Central Core Layer
3. Regulation of Centrosome Duplication and Mitotic Spindle Organization
3.1. Regulatory Kinases
3.2. Mitotic Spindle Organization and Spindle Elongation
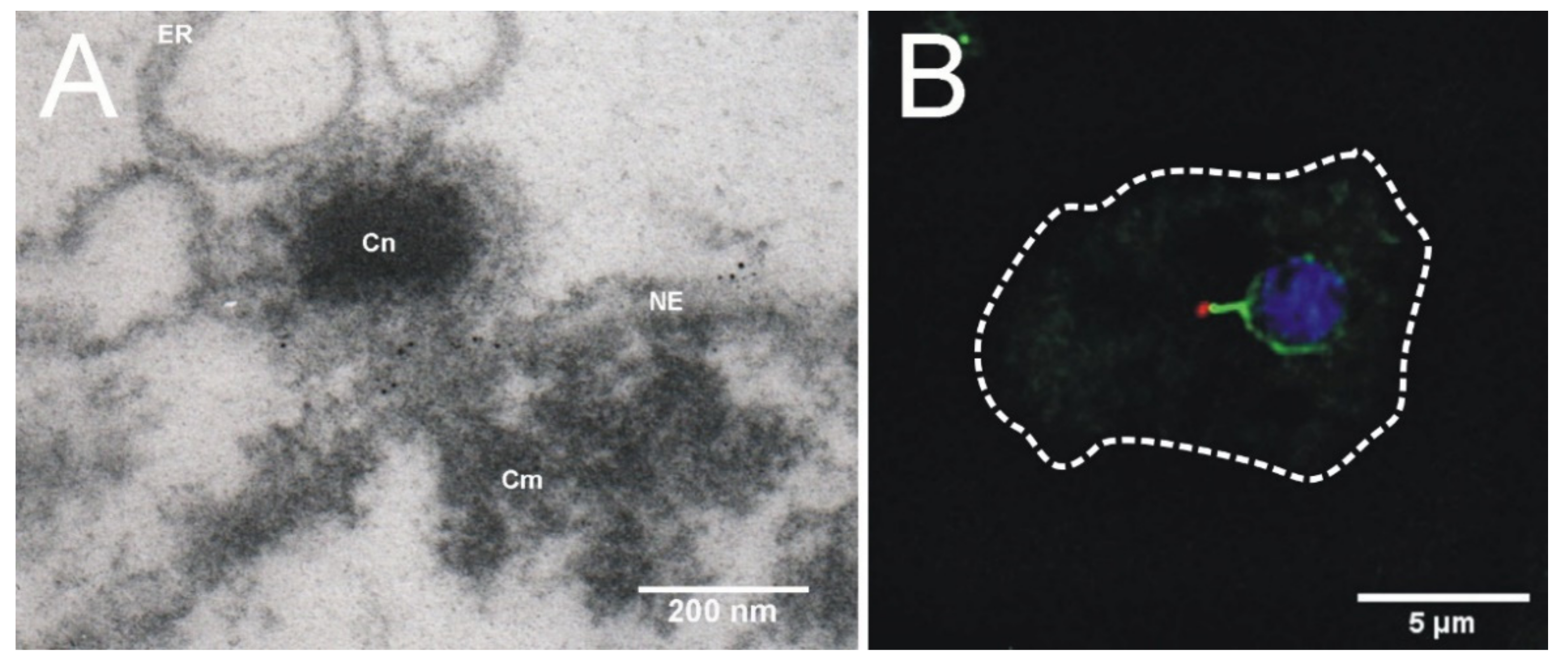
4. Centrosome-Nucleus Attachment
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flemming, W. Zellsubstanz, Kern Und Zelltheilung; Vogel: Praha, Czechia, 1882. [Google Scholar]
- Boveri, T. Ueber Den Antheil Des Spermatozoon an Der Theilung Des Eies. Sitzungsber. Ges. Morph. Physiol. München 1887, 3, 151–164. [Google Scholar]
- Van Beneden, E.; Neyt, A. Nouvelle Recherches Sur La Fécondation et La Division Mitosique Chez l’Ascaride Mégalocéphale; Wilhelm Engelmann: Lemgo, Germany, 1887; pp. 214–295. [Google Scholar]
- Gräf, R. Comparative Biology of Centrosomal Structures in Eukaryotes. Cells 2018, 7, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakley, C.E.; Oakley, B.R. Identification of Gamma-Tubulin, a New Member of the Tubulin Superfamily Encoded by MipA Gene of Aspergillus Nidulans. Nature 1989, 338, 662–664. [Google Scholar] [CrossRef]
- Andersen, J.S.; Wilkinson, C.J.; Mayor, T.; Mortensen, P.; Nigg, E.A.; Mann, M. Proteomic Characterization of the Human Centrosome by Protein Correlation Profiling. Nature 2003, 426, 570–574. [Google Scholar] [CrossRef]
- Carvalho-Santos, Z.; Azimzadeh, J.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Evolution: Tracing the Origins of Centrioles, Cilia, and Flagella. J. Cell Biol. 2011, 194, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Ito, D.; Bettencourt-Dias, M. Centrosome Remodelling in Evolution. Cells 2018, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Batsios, P.; Meyer, I. Evolution of Centrosomes and the Nuclear Lamina: Amoebozoan Assets. Eur. J. Cell Biol. 2015, 94, 249–256. [Google Scholar] [CrossRef]
- Pedersen, L.B.; Schroder, J.M.; Satir, P.; Christensen, S.T. The Ciliary Cytoskeleton. Compr. Physiol. 2012, 2, 779–803. [Google Scholar] [CrossRef]
- Liu, P.; Würtz, M.; Zupa, E.; Pfeffer, S.; Schiebel, E. Microtubule Nucleation: The Waltz between γ-Tubulin Ring Complex and Associated Proteins. Curr. Opin. Cell Biol. 2021, 68, 124–131. [Google Scholar] [CrossRef]
- Nigg, E.A.; Holland, A.J. Once and Only Once: Mechanisms of Centriole Duplication and Their Deregulation in Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Hayashi, K.; Nishida, E. Cyclin-Dependent Kinase 2 (Cdk2) Is Required for Centrosome Duplication in Mammalian Cells. Curr. Biol. 1999, 9, 429–432. [Google Scholar] [CrossRef] [Green Version]
- Gönczy, P.; Hatzopoulos, G.N. Centriole Assembly at a Glance. J. Cell. Sci. 2019, 132, jcs228833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moyer, T.C.; Holland, A.J. PLK4 Promotes Centriole Duplication by Phosphorylating STIL to Link the Procentriole Cartwheel to the Microtubule Wall. eLife 2019, 8, e46054. [Google Scholar] [CrossRef]
- Guichard, P.; Hamel, V.; Gönczy, P. The Rise of the Cartwheel: Seeding the Centriole Organelle. Bioessays 2018, 40, e1700241. [Google Scholar] [CrossRef]
- Joukov, V.; De Nicolo, A. Aurora-PLK1 Cascades as Key Signaling Modules in the Regulation of Mitosis. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Fang, G.; Zhang, D.; Yin, H.; Zheng, L.; Bi, X.; Yuan, L. Centlein Mediates an Interaction between C-Nap1 and Cep68 to Maintain Centrosome Cohesion. J. Cell Sci. 2014, 127, 1631–1639. [Google Scholar] [CrossRef] [Green Version]
- Au, F.K.C.; Hau, B.K.T.; Qi, R.Z. Nek2-Mediated GAS2L1 Phosphorylation and Centrosome-Linker Disassembly Induce Centrosome Disjunction. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayor, T.; Stierhof, Y.D.; Tanaka, K.; Fry, A.M.; Nigg, E.A. The Centrosomal Protein C-Nap1 Is Required for Cell Cycle-Regulated Centrosome Cohesion. J. Cell Biol. 2000, 151, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Bahe, S.; Stierhof, Y.D.; Wilkinson, C.J.; Leiss, F.; Nigg, E.A. Rootletin Forms Centriole-Associated Filaments and Functions in Centrosome Cohesion. J. Cell Biol. 2005, 171, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Graser, S.; Stierhof, Y.D.; Nigg, E.A. Cep68 and Cep215 (Cdk5rap2) Are Required for Centrosome Cohesion. J. Cell Sci. 2007, 120, 4321–4331. [Google Scholar] [CrossRef] [Green Version]
- He, R.; Huang, N.; Bao, Y.; Zhou, H.; Teng, J.; Chen, J. LRRC45 Is a Centrosome Linker Component Required for Centrosome Cohesion. Cell Rep. 2013, 4, 1100–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell Cycle Regulation by the NEK Family of Protein Kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef] [Green Version]
- Raff, J.W. Phase Separation and the Centrosome: A Fait Accompli? Trends Cell Biol. 2019, 29, 612–622. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Origin of the Cell Nucleus, Mitosis and Sex: Roles of Intracellular Coevolution. Biol. Direct 2010, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Kuriyama, R.; Sato, C.; Fukui, Y.; Nishibayashi, S. In Vitro Nucleation of Microtubules from Microtubule-Organizing Center Prepared from Cellular Slime Mold. Cell Motil. 1982, 2, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Roos, U.-P.; Guhl, B. Microtubules in interphase and mitosis of cellular slime molds. In Biomechanics of Active Movement and Deformation of Cells; Akkas, N., Ed.; Springer: Berlin/Heidelberg, Germany, 1990. [Google Scholar]
- Euteneuer, U.; Gräf, R.; Kube-Granderath, E.; Schliwa, M. Dictyostelium Gamma-Tubulin: Molecular Characterization and Ultrastructural Localization. J. Cell Sci. 1998, 111, 405–412. [Google Scholar] [CrossRef]
- Rüthnick, D.; Schiebel, E. Duplication and Nuclear Envelope Insertion of the Yeast Microtubule Organizing Centre, the Spindle Pole Body. Cells 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, M.; Schliwa, M.; Euteneuer, U. Unusual Centrosome Cycle in Dictyostelium: Correlation of Dynamic Behavior and Structural Changes. Mol. Biol. Cell 1999, 10, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Sazer, S.; Lynch, M.; Needleman, D. Deciphering the Evolutionary History of Open and Closed Mitosis. Curr. Biol. 2014, 24, R1099–R1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putzler, S.; Meyer, I.; Gräf, R. CP91 Is a Component of the Dictyostelium Centrosome Involved in Centrosome Biogenesis. Eur. J. Cell Biol. 2016, 95, 124–135. [Google Scholar] [CrossRef]
- Rieder, C.L.; Faruki, S.; Khodjakov, A. The Centrosome in Vertebrates: More than a Microtubule-Organizing Center. Trends Cell Biol. 2001, 11, 413–419. [Google Scholar] [CrossRef]
- Karsenti, E.; Vernos, I. The Mitotic Spindle: A Self-Made Machine. Science 2001, 294, 543–547. [Google Scholar] [CrossRef]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruss, O.J. Animal Female Meiosis: The Challenges of Eliminating Centrosomes. Cells 2018, 7, 73. [Google Scholar] [CrossRef] [Green Version]
- Song, J.-G.; King, M.R.; Zhang, R.; Kadzik, R.S.; Thawani, A.; Petry, S. Mechanism of How Augmin Directly Targets the γ-Tubulin Ring Complex to Microtubules. J. Cell Biol. 2018, 217, 2417–2428. [Google Scholar] [CrossRef] [Green Version]
- Green, R.A.; Paluch, E.; Oegema, K. Cytokinesis in Animal Cells. Annu. Rev. Cell Dev. Biol. 2012, 28, 29–58. [Google Scholar] [CrossRef] [Green Version]
- Fraschini, R. Cytokinesis in Eukaryotic Cells: The Furrow Complexity at a Glance. Cells 2020, 9, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromley, A.; Yeaman, C.; Rosa, J.; Redick, S.; Chen, C.T.; Mirabelle, S.; Guha, M.; Sillibourne, J.; Doxsey, S.J. Centriolin Anchoring of Exocyst and SNARE Complexes at the Midbody Is Required for Secretory-Vesicle-Mediated Abscission. Cell 2005, 123, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Bhutta, M.S.; McInerny, C.J.; Gould, G.W. ESCRT Function in Cytokinesis: Location, Dynamics and Regulation by Mitotic Kinases. Int. J. Mol. Sci. 2014, 15, 21723–21739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, E.; Sandrin, V.; Chung, H.Y.; Morham, S.G.; Gygi, S.P.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT and ALIX Proteins Interact with Proteins of the Midbody and Function in Cytokinesis. EMBO J. 2007, 26, 4215–4227. [Google Scholar] [CrossRef] [Green Version]
- Fabbro, M.; Zhou, B.B.; Takahashi, M.; Sarcevic, B.; Lal, P.; Graham, M.E.; Gabrielli, B.G.; Robinson, P.J.; Nigg, E.A.; Ono, Y.; et al. Cdk1/Erk2- and Plk1-Dependent Phosphorylation of a Centrosome Protein, Cep55, Is Required for Its Recruitment to Midbody and Cytokinesis. Dev. Cell 2005, 9, 477–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doxsey, S.; McCollum, D.; Theurkauf, W. Centrosomes in Cellular Regulation. Annu. Rev. Cell Dev. Biol. 2005, 21, 411–434. [Google Scholar] [CrossRef]
- Joukov, V.; De Nicolo, A. The Centrosome and the Primary Cilium: The Yin and Yang of a Hybrid Organelle. Cells 2019, 8, 701. [Google Scholar] [CrossRef] [Green Version]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-Resolution Microscopy Demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Arslanhan, M.D.; Gulensoy, D.; Firat-Karalar, E.N. A Proximity Mapping Journey into the Biology of the Mammalian Centrosome/Cilium Complex. Cells 2020, 9, 1390. [Google Scholar] [CrossRef]
- Galletta, B.J.; Fagerstrom, C.J.; Schoborg, T.A.; McLamarrah, T.A.; Ryniawec, J.M.; Buster, D.W.; Slep, K.C.; Rogers, G.C.; Rusan, N.M. A Centrosome Interactome Provides Insight into Organelle Assembly and Reveals a Non-Duplication Role for Plk4. Nat. Commun. 2016, 7, 12476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchins, J.R.; Toyoda, Y.; Hegemann, B.; Poser, I.; Heriche, J.K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic Analysis of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gräf, R.; Euteneuer, U.; Ueda, M.; Schliwa, M. Isolation of Nucleation-Competent Centrosomes from Dictyostelium Discoideum. Eur. J. Cell Biol. 1998, 76, 167–175. [Google Scholar] [CrossRef]
- Reinders, Y.; Schulz, I.; Gräf, R.; Sickmann, A. Identification of Novel Centrosomal Proteins in Dictyostelium Discoideum by Comparative Proteomic Approaches. J. Proteome Res. 2006, 5, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Meyer, I.; Peter, T.; Batsios, P.; Kuhnert, O.; Krüger-Genge, A.; Camurça, C.; Gräf, R. CP39, CP75 and CP91 Are Major Structural Components of the Dictyostelium Centrosome’s Core Structure. Eur. J. Cell Biol. 2017, 96, 119–130. [Google Scholar] [CrossRef]
- Pitzen, V.; Sander, S.; Baumann, O.; Gräf, R.; Meyer, I. Cep192, a Novel Missing Link between the Centrosomal Core and Corona in Dictyostelium Amoebae. Cells 2021, 10, 2384. [Google Scholar] [CrossRef]
- Gomez-Ferreria, M.A.; Rath, U.; Buster, D.W.; Chanda, S.K.; Caldwell, J.S.; Rines, D.R.; Sharp, D.J. Human Cep192 Is Required for Mitotic Centrosome and Spindle Assembly. Curr. Biol. 2007, 17, 1960–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhnert, O.; Baumann, O.; Meyer, I.; Gräf, R. CP55, a Novel Key Component of Centrosomal Organization in Dictyostelium. Cell. Mol. Life Sci. 2012, 69, 3651–3664. [Google Scholar] [CrossRef]
- Gräf, R. DdNek2, the First Non-Vertebrate Homologue of Human Nek2, Is Involved in the Formation of Microtubule-Organizing Centers. J. Cell Sci. 2002, 115, 1919–1929. [Google Scholar] [CrossRef]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A Centrosomal Function for the Human Nek2 Protein Kinase, a Member of the NIMA Family of Cell Cycle Regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, D.J.; Castilhos, B.; Immich, B.F.; Cañedo, A.D.; Henriques, J.A.P.; Lenz, G.; Saffi, J. Kin3 Protein, a NIMA-Related Kinase of Saccharomyces Cerevisiae, Is Involved in DNA Adduct Damage Response. Cell Cycle 2010, 9, 2220–2229. [Google Scholar] [CrossRef] [Green Version]
- Krien, M.J.; West, R.R.; John, U.P.; Koniaras, K.; McIntosh, J.R.; O’Connell, M.J. The Fission Yeast NIMA Kinase Fin1p Is Required for Spindle Function and Nuclear Envelope Integrity. EMBO J. 2002, 21, 1713–1722. [Google Scholar] [CrossRef] [Green Version]
- Vigneault, F.; Lachance, D.; Cloutier, M.; Pelletier, G.; Levasseur, C.; Séguin, A. Members of the Plant NIMA-Related Kinases Are Involved in Organ Development and Vascularization in Poplar, Arabidopsis and Rice. Plant J. 2007, 51, 575–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradel, L.C.; Bonhivers, M.; Landrein, N.; Robinson, D.R. NIMA-Related Kinase TbNRKC Is Involved in Basal Body Separation in Trypanosoma Brucei. J. Cell Sci. 2006, 119, 1852–1863. [Google Scholar] [CrossRef] [Green Version]
- Reininger, L.; Tewari, R.; Fennell, C.; Holland, Z.; Goldring, D.; Ranford-Cartwright, L.; Billker, O.; Doerig, C. An Essential Role for the Plasmodium Nek-2 Nima-Related Protein Kinase in the Sexual Development of Malaria Parasites. J. Biol. Chem. 2009, 284, 20858–20868. [Google Scholar] [CrossRef] [Green Version]
- Schulz, I.; Erle, A.; Gräf, R.; Krüger, A.; Lohmeier, H.; Putzler, S.; Samereier, M.; Weidenthaler, S. Identification and Cell Cycle-Dependent Localization of Nine Novel, Genuine Centrosomal Components in Dictyostelium Discoideum. Cell Motil. Cytoskeleton 2009, 66, 915–928. [Google Scholar] [CrossRef]
- Daunderer, C.; Gräf, R. Molecular Analysis of the Cytosolic Dictyostelium Gamma-Tubulin Complex. Eur. J. Cell Biol. 2002, 81, 175–184. [Google Scholar] [CrossRef]
- Knop, M.; Pereira, G.; Geissler, S.; Grein, K.; Schiebel, E. The Spindle Pole Body Component Spc97p Interacts with the Gamma-Tubulin of Saccharomyces Cerevisiae and Functions in Microtubule Organization and Spindle Pole Body Duplication. EMBO J. 1997, 16, 1550–1564. [Google Scholar] [CrossRef] [Green Version]
- Vardy, L.; Toda, T. The Fission Yeast Gamma-Tubulin Complex Is Required in G(1) Phase and Is a Component of the Spindle Assembly Checkpoint. EMBO J. 2000, 19, 6098–6111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seltzer, V.; Janski, N.; Canaday, J.; Herzog, E.; Erhardt, M.; Evrard, J.-L.; Schmit, A.-C. Arabidopsis GCP2 and GCP3 Are Part of a Soluble Gamma-Tubulin Complex and Have Nuclear Envelope Targeting Domains. Plant J. 2007, 52, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, Z. γ-Tubulin Complex in Trypanosoma Brucei: Molecular Composition, Subunit Interdependence and Requirement for Axonemal Central Pair Protein Assembly. Mol. Microbiol. 2015, 98, 667–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maessen, S.; Wesseling, J.G.; Smits, M.A.; Konings, R.N.H.; Schoenmakers, J.G.G. The Gamma -Tubulin Gene of the Malaria Parasite Plasmodium Falciparum. Mol. Biochem. Parasitol. 1993, 60, 27–36. [Google Scholar] [CrossRef]
- Pitzen, V.; Askarzada, S.; Gräf, R.; Meyer, I. CDK5RAP2 Is an Essential Scaffolding Protein of the Corona of the Dictyostelium Centrosome. Cells 2018, 7, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, A.R.; Kilmartin, J.V.; Gergely, F. CDK5RAP2 Functions in Centrosome to Spindle Pole Attachment and DNA Damage Response. J. Cell Biol. 2010, 189, 23–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knop, M.; Schiebel, E. Receptors Determine the Cellular Localization of a Gamma-Tubulin Complex and Thereby the Site of Microtubule Formation. EMBO J. 1998, 17, 3952–3967. [Google Scholar] [CrossRef] [Green Version]
- Samejima, I.; Lourenco, P.C.; Snaith, H.A.; Sawin, K.E. Fission Yeast Mto2p Regulates Microtubule Nucleation by the Centrosomin-Related Protein Mto1p. Mol. Biol. Cell 2005, 16, 3040–3051. [Google Scholar] [CrossRef] [Green Version]
- Kuhnert, O.; Baumann, O.; Meyer, I.; Gräf, R. Functional Characterization of CP148, a Novel Key Component for Centrosome Integrity in Dictyostelium. Cell. Mol. Life Sci. 2012, 69, 1875–1888. [Google Scholar] [CrossRef]
- Doxsey, S.J.; Stein, P.; Evans, L.; Calarco, P.D.; Kirschner, M. Pericentrin, a Highly Conserved Centrosome Protein Involved in Microtubule Organization. Cell 1994, 76, 639–650. [Google Scholar] [CrossRef]
- Flory, M.R.; Morphew, M.; Joseph, J.D.; Means, A.R.; Davis, T.N. Pcp1p, an Spc110p-Related Calmodulin Target at the Centrosome of the Fission Yeast Schizosaccharomyces Pombe. Cell Growth Differ. 2002, 13, 47–58. [Google Scholar] [PubMed]
- Samereier, M.; Baumann, O.; Meyer, I.; Gräf, R. Analysis of Dictyostelium TACC Reveals Differential Interactions with CP224 and Unusual Dynamics of Dictyostelium Microtubules. Cell. Mol. Life Sci. 2011, 68, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Gergely, F.; Draviam, V.M.; Raff, J.W. The Ch-TOG/XMAP215 Protein Is Essential for Spindle Pole Organization in Human Somatic Cells. Genes Dev. 2003, 17, 336–341. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Daunderer, C.; Schliwa, M. Dictyostelium DdCP224 Is a Microtubule-Associated Protein and a Permanent Centrosomal Resident Involved in Centrosome Duplication. J. Cell Sci. 2000, 113, 1747–1758. [Google Scholar] [CrossRef]
- Cassimeris, L.; Morabito, J. TOGp, the Human Homolog of XMAP215/Dis1, Is Required for Centrosome Integrity, Spindle Pole Organization, and Bipolar Spindle Assembly. Mol. Biol. Cell 2004, 15, 1580–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.J.; Huffaker, T.C. Stu2p: A Microtubule-Binding Protein That Is an Essential Component of the Yeast Spindle Pole Body. J. Cell Biol. 1997, 139, 1271–1280. [Google Scholar] [CrossRef] [Green Version]
- Nabeshima, K.; Kurooka, H.; Takeuchi, M.; Kinoshita, K.; Nakaseko, Y.; Yanagida, M. P93dis1, Which Is Required for Sister Chromatid Separation, Is a Novel Microtubule and Spindle Pole Body-Associating Protein Phosphorylated at the Cdc2 Target Sites. Genes Dev. 1995, 9, 1572–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whittington, A.T.; Vugrek, O.; Wei, K.J.; Hasenbein, N.G.; Sugimoto, K.; Rashbrooke, M.C.; Wasteneys, G.O. MOR1 Is Essential for Organizing Cortical Microtubules in Plants. Nature 2001, 411, 610–613. [Google Scholar] [CrossRef]
- Wheeler, R.J.; Scheumann, N.; Wickstead, B.; Gull, K.; Vaughan, S. Cytokinesis in Trypanosoma Brucei Differs between Bloodstream and Tsetse Trypomastigote Forms: Implications for Microtubule-Based Morphogenesis and Mutant Analysis. Mol. Microbiol. 2013, 90, 1339–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehberg, M.; Gräf, R. Dictyostelium EB1 Is a Genuine Centrosomal Component Required for Proper Spindle Formation. Mol. Biol. Cell 2002, 13, 2301–2310. [Google Scholar] [CrossRef] [Green Version]
- Morrison, E.E.; Wardleworth, B.N.; Askham, J.M.; Markham, A.F.; Meredith, D.M. EB1, a Protein Which Interacts with the APC Tumour Suppressor, Is Associated with the Microtubule Cytoskeleton throughout the Cell Cycle. Oncogene 1998, 17, 3471–3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, K.; Richards, K.; Botstein, D. BIM1 Encodes a Microtubule-Binding Protein in Yeast. Mol. Biol. Cell 1997, 8, 2677–2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beinhauer, J.D.; Hagan, I.M.; Hegemann, J.H.; Fleig, U. Mal3, the Fission Yeast Homologue of the Human APC-Interacting Protein EB-1 Is Required for Microtubule Integrity and the Maintenance of Cell Form. J. Cell Biol. 1997, 139, 717–728. [Google Scholar] [CrossRef]
- Komaki, S.; Abe, T.; Coutuer, S.; Inzé, D.; Russinova, E.; Hashimoto, T. Nuclear-Localized Subtype of End-Binding 1 Protein Regulates Spindle Organization in Arabidopsis. J. Cell Sci. 2010, 123, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Rehberg, M.; Gräf, R. DdMoe, a Homologue of the EB1-Binding Moe1 Protein, Is a Microtubule-Associated Protein and a Permanent Centrosomal Resident in Dictyostelium. Cell Motil. Cytoskeleton 2002, 54, 185. [Google Scholar]
- Chen, C.R.; Chen, J.; Chang, E.C. A Conserved Interaction between Moe1 and Mal3 Is Important for Proper Spindle Formation in Schizosaccharomyces Pombe. Mol. Biol. Cell 2000, 11, 4067–4077. [Google Scholar] [CrossRef] [Green Version]
- Blau-Wasser, R.; Euteneuer, U.; Xiong, H.; Gassen, B.; Schleicher, M.; Noegel, A.A. CP250, a Novel Acidic Coiled-Coil Protein of the Dictyostelium Centrosome, Affects Growth, Chemotaxis, and the Nuclear Envelope. Mol. Biol. Cell 2009, 20, 4348–4361. [Google Scholar] [CrossRef] [Green Version]
- Fry, A.M.; Mayor, T.; Meraldi, P.; Stierhof, Y.D.; Tanaka, K.; Nigg, E.A. C-Nap1, a Novel Centrosomal Coiled-Coil Protein and Candidate Substrate of the Cell Cycle-Regulated Protein Kinase Nek2. J. Cell Biol. 1998, 141, 1563–1574. [Google Scholar] [CrossRef] [Green Version]
- Daunderer, C.; Schliwa, M.; Gräf, R. Dictyostelium Centrin-Related Protein (DdCrp), the Most Divergent Member of the Centrin Family, Possesses Only Two EF Hands and Dissociates from the Centrosome during Mitosis. Eur. J. Cell Biol. 2001, 80, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Middendorp, S.; Kuntziger, T.; Abraham, Y.; Holmes, S.; Bordes, N.; Paintrand, M.; Paoletti, A.; Bornens, M. A Role for Centrin 3 in Centrosome Reproduction. J. Cell Biol. 2000, 148, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Baum, P.; Furlong, C.; Byers, B. Yeast Gene Required for Spindle Pole Body Duplication: Homology of Its Product with Ca2+-Binding Proteins. Proc. Natl. Acad. Sci. USA 1986, 83, 5512–5516. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, A.; Bordes, N.; Haddad, R.; Schwartz, C.L.; Chang, F.; Bornens, M. Fission Yeast Cdc31p Is a Component of the Half-Bridge and Controls SPB Duplication. Mol. Biol. Cell 2003, 14, 2793–2808. [Google Scholar] [CrossRef]
- Cordeiro, M.C.; Piqueras, R.; de Oliveira, D.E.; Castresana, C. Characterization of Early Induced Genes in Arabidopsis Thaliana Responding to Bacterial Inoculation: Identification of Centrin and of a Novel Protein with Two Regions Related to Kinase Domains. FEBS Lett. 1998, 434, 387–393. [Google Scholar] [CrossRef]
- He, C.Y.; Pypaert, M.; Warren, G. Golgi Duplication in Trypanosoma Brucei Requires Centrin2. Science 2005, 310, 1196–1198. [Google Scholar] [CrossRef] [PubMed]
- Roques, M.; Stanway, R.R.; Rea, E.I.; Markus, R.; Brady, D.; Holder, A.A.; Guttery, D.S.; Tewari, R. Plasmodium Centrin PbCEN-4 Localizes to the Putative MTOC and Is Dispensable for Malaria Parasite Proliferation. Biol. Open 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonce, M.P. Identification of a Microtubule-Binding Domain in a Cytoplasmic Dynein Heavy Chain. J. Biol. Chem. 1997, 272, 19714–19718. [Google Scholar] [CrossRef] [Green Version]
- Rehberg, M.; Kleylein-Sohn, J.; Faix, J.; Ho, T.H.; Schulz, I.; Gräf, R. Dictyostelium LIS1 Is a Centrosomal Protein Required for Microtubule/Cell Cortex Interactions, Nucleus/Centrosome Linkage, and Actin Dynamics. Mol. Biol. Cell 2005, 16, 2759–2771. [Google Scholar] [CrossRef] [Green Version]
- Vaisberg, E.A.; Koonce, M.P.; McIntosh, J.R. Cytoplasmic Dynein Plays a Role in Mammalian Mitotic Spindle Formation. J. Cell Biol. 1993, 123, 849–858. [Google Scholar] [CrossRef] [Green Version]
- Eshel, D.; Urrestarazu, L.A.; Vissers, S.; Jauniaux, J.C.; van Vliet-Reedijk, J.C.; Planta, R.J.; Gibbons, I.R. Cytoplasmic Dynein Is Required for Normal Nuclear Segregation in Yeast. Proc. Natl. Acad. Sci. USA 1993, 90, 11172–11176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrueta, L.; Tirnauer, J.S.; Schuyler, S.C.; Pellman, D.; Bierer, B.E. The APC-Associated Protein EB1 Associates with Components of the Dynactin Complex and Cytoplasmic Dynein Intermediate Chain. Curr. Biol. 1999, 9, 425–428. [Google Scholar] [CrossRef] [Green Version]
- Adhiambo, C.; Forney, J.D.; Asai, D.J.; LeBowitz, J.H. The Two Cytoplasmic Dynein-2 Isoforms in Leishmania Mexicana Perform Separate Functions. Mol. Biochem. Parasitol. 2005, 143, 216–225. [Google Scholar] [CrossRef]
- Daher, W.; Pierrot, C.; Kalamou, H.; Pinder, J.C.; Margos, G.; Dive, D.; Franke-Fayard, B.; Janse, C.J.; Khalife, J. Plasmodium Falciparum Dynein Light Chain 1 Interacts with Actin/Myosin during Blood Stage Development. J. Biol. Chem. 2010, 285, 20180–20191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hestermann, A.; Gräf, R. The XMAP215-Family Protein DdCP224 Is Required for Cortical Interactions of Microtubules. BMC Cell Biol. 2004, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askham, J.M.; Vaughan, K.T.; Goodson, H.V.; Morrison, E.E. Evidence That an Interaction between EB1 and P150(Glued) Is Required for the Formation and Maintenance of a Radial Microtubule Array Anchored at the Centrosome. Mol. Biol. Cell 2002, 13, 3627–3645. [Google Scholar] [CrossRef] [Green Version]
- Muhua, L.; Karpova, T.S.; Cooper, J.A. A Yeast Actin-Related Protein Homologous to That in Vertebrate Dynactin Complex Is Important for Spindle Orientation and Nuclear Migration. Cell 1994, 78, 669–679. [Google Scholar] [CrossRef]
- Gordon, J.L.; Sibley, L.D. Comparative Genome Analysis Reveals a Conserved Family of Actin-like Proteins in Apicomplexan Parasites. BMC Genom. 2005, 6, 179. [Google Scholar] [CrossRef] [Green Version]
- Reiner, O.; Carrozzo, R.; Shen, Y.; Wehnert, M.; Faustinella, F.; Dobyns, W.B.; Caskey, C.T.; Ledbetter, D.H. Isolation of a Miller-Dieker Lissencephaly Gene Containing G Protein Beta-Subunit-like Repeats. Nature 1993, 364, 717–721. [Google Scholar] [CrossRef]
- Lee, W.L.; Oberle, J.R.; Cooper, J.A. The Role of the Lissencephaly Protein Pac1 during Nuclear Migration in Budding Yeast. J. Cell Biol. 2003, 160, 355–364. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Kaller, M.; Nellen, W.; Gräf, R.; De Lozanne, A. Dictyostelium Aurora Kinase Has Properties of Both Aurora A and Aurora B Kinases. Eukaryot. Cell 2008, 7, 894–905. [Google Scholar] [CrossRef] [Green Version]
- Hochegger, H.; Hégarat, N.; Pereira-Leal, J.B. Aurora at the Pole and Equator: Overlapping Functions of Aurora Kinases in the Mitotic Spindle. Open Biol. 2013, 3, 120185. [Google Scholar] [CrossRef] [Green Version]
- García-Rodríguez, L.J.; Kasciukovic, T.; Denninger, V.; Tanaka, T.U. Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-Orientation in Budding Yeast. Curr. Biol. 2019, 29, 1536–1544.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, J.; Paris, J.; Willer, M.; Philippe, M.; Hagan, I.M. The, S. Pombe Aurora-Related Kinase Ark1 Associates with Mitotic Structures in a Stage Dependent Manner and Is Required for Chromosome Segregation. J. Cell Sci. 2001, 114, 4371–4384. [Google Scholar] [CrossRef]
- Fassolari, M.; Alonso, G.D. Aurora Kinase Protein Family in Trypanosoma Cruzi: Novel Role of an AUK-B Homologue in Kinetoplast Replication. PLoS Negl. Trop. Dis. 2019, 13, e0007256. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, T.G.; Doerig, C.; Reininger, L. Nima- and Aurora-Related Kinases of Malaria Parasites. Biochim. Biophys. Acta 2013, 1834, 1336–1345. [Google Scholar] [CrossRef]
- Golsteyn, R.M.; Schultz, S.J.; Bartek, J.; Ziemiecki, A.; Ried, T.; Nigg, E.A. Cell Cycle Analysis and Chromosomal Localization of Human Plk1, a Putative Homologue of the Mitotic Kinases Drosophila Polo and Saccharomyces Cerevisiae Cdc5. J. Cell Sci. 1994, 107, 1509–1517. [Google Scholar] [CrossRef]
- Ohkura, H.; Hagan, I.M.; Glover, D.M. The Conserved Schizosaccharomyces Pombe Kinase Plo1, Required to Form a Bipolar Spindle, the Actin Ring, and Septum, Can Drive Septum Formation in G1 and G2 Cells. Genes Dev. 1995, 9, 1059–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurasawa, Y.; An, T.; Li, Z. Polo-like Kinase in Trypanosomes: An Odd Member out of the Polo Family. Open Biol. 2020, 10, 200189. [Google Scholar] [CrossRef] [PubMed]
- Schulz, I.; Baumann, O.; Samereier, M.; Zoglmeier, C.; Gräf, R. Dictyostelium Sun1 Is a Dynamic Membrane Protein of Both Nuclear Membranes and Required for Centrosomal Association with Clustered Centromeres. Eur. J. Cell Biol. 2009, 88, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Rivero, F.; Euteneuer, U.; Mondal, S.; Mana-Capelli, S.; Larochelle, D.; Vogel, A.; Gassen, B.; Noegel, A.A. Dictyostelium Sun-1 Connects the Centrosome to Chromatin and Ensures Genome Stability. Traffic 2008, 9, 708–724. [Google Scholar] [CrossRef]
- Fridkin, A.; Mills, E.; Margalit, A.; Neufeld, E.; Lee, K.K.; Feinstein, N.; Cohen, M.; Wilson, K.L.; Gruenbaum, Y. Matefin, a Caenorhabditis Elegans Germ Line-Specific SUN-Domain Nuclear Membrane Protein, Is Essential for Early Embryonic and Germ Cell Development. Proc. Natl. Acad. Sci. USA 2004, 101, 6987–6992. [Google Scholar] [CrossRef] [Green Version]
- Jaspersen, S.L.; Giddings, T.H.; Winey, M. Mps3p Is a Novel Component of the Yeast Spindle Pole Body That Interacts with the Yeast Centrin Homologue Cdc31p. J. Cell Biol. 2002, 159, 945–956. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. The Product of the Spindle Formation Gene Sad1+ Associates with the Fission Yeast Spindle Pole Body and Is Essential for Viability. J. Cell Biol. 1995, 129, 1033–1047. [Google Scholar] [CrossRef] [Green Version]
- Varas, J.; Graumann, K.; Osman, K.; Pradillo, M.; Evans, D.E.; Santos, J.L.; Armstrong, S.J. Absence of SUN1 and SUN2 Proteins in Arabidopsis Thaliana Leads to a Delay in Meiotic Progression and Defects in Synapsis and Recombination. Plant J. 2015, 81, 329–346. [Google Scholar] [CrossRef] [Green Version]
- Tikhonenko, I.; Magidson, V.; Gräf, R.; Khodjakov, A.; Koonce, M.P. A Kinesin-Mediated Mechanism That Couples Centrosomes to Nuclei. Cell. Mol. Life Sci. 2013, 70, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piddini, E.; Schmid, J.A.; de Martin, R.; Dotti, C.G. The Ras-like GTPase Gem Is Involved in Cell Shape Remodelling and Interacts with the Novel Kinesin-like Protein KIF9. EMBO J. 2001, 20, 4076–4087. [Google Scholar] [CrossRef] [Green Version]
- Lakshmikanth, G.S.; Warrick, H.M.; Spudich, J.A. A Mitotic Kinesin-like Protein Required for Normal Karyokinesis, Myosin Localization to the Furrow, and Cytokinesis in Dictyostelium. Proc. Natl. Acad. Sci. USA 2004, 101, 16519–16524. [Google Scholar] [CrossRef] [Green Version]
- Mrug, M.; Li, R.; Cui, X.; Schoeb, T.R.; Churchill, G.A.; Guay-Woodford, L.M. Kinesin Family Member 12 Is a Candidate Polycystic Kidney Disease Modifier in the Cpk Mouse. J. Am. Soc. Nephrol. 2005, 16, 905–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhardt, N.; Redolfi, J.; Antonin, W. Interaction of Nup53 with Ndc1 and Nup155 Is Required for Nuclear Pore Complex Assembly. J. Cell Sci. 2014, 127, 908–921. [Google Scholar] [CrossRef] [Green Version]
- Fahrenkrog, B.; Hübner, W.; Mandinova, A.; Panté, N.; Keller, W.; Aebi, U. The Yeast Nucleoporin Nup53p Specifically Interacts with Nic96p and Is Directly Involved in Nuclear Protein Import. Mol. Biol. Cell 2000, 11, 3885–3896. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.Q.; Du, X.; Liu, J.; Balasubramanian, M.K.; Balasundaram, D. Identification of Genes Encoding Putative Nucleoporins and Transport Factors in the Fission Yeast Schizosaccharomyces Pombe: A Deletion Analysis. Yeast 2004, 21, 495–509. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Hara-Nishimura, I. The Molecular Architecture of the Plant Nuclear Pore Complex. J. Exp. Bot. 2013, 64, 823–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, R.; Stumpf, M.; Wehrstedt, R.; Sukumaran, S.K.; Karow, M.A.; Marko, M.; Noegel, A.A.; Eichinger, L. The Regulatory Subunit Phr2AB of Dictyostelium Discoideum Phosphatase PP2A Interacts with the Centrosomal Protein CEP161, a CDK5RAP2 Ortholog. Genes Cells 2018, 23, 923–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, J.-H.; McAvoy, T.; Rakhilin, S.V.; Nishi, A.; Greengard, P.; Nairn, A.C. Protein Kinase A Activates Protein Phosphatase 2A by Phosphorylation of the B56delta Subunit. Proc. Natl. Acad. Sci. USA 2007, 104, 2979–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Burke, D.J. Cdc55p, the B-Type Regulatory Subunit of Protein Phosphatase 2A, Has Multiple Functions in Mitosis and Is Required for the Kinetochore/Spindle Checkpoint in Saccharomyces Cerevisiae. Mol. Cell Biol. 1997, 17, 620–626. [Google Scholar] [CrossRef] [Green Version]
- Grallert, A.; Boke, E.; Hagting, A.; Hodgson, B.; Connolly, Y.; Griffiths, J.R.; Smith, D.L.; Pines, J.; Hagan, I.M. A PP1-PP2A Phosphatase Relay Controls Mitotic Progression. Nature 2015, 517, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corum, J.W.; Hartung, A.J.; Stamey, R.T.; Rundle, S.J. Characterization of DNA Sequences Encoding a Novel Isoform of the 55 KDa B Regulatory Subunit of the Type 2A Protein Serine/Threonine Phosphatase of Arabidopsis Thaliana. Plant Mol. Biol. 1996, 31, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Visweshwaran, S.P.; Thomason, P.A.; Guerois, R.; Vacher, S.; Denisov, E.V.; Tashireva, L.A.; Lomakina, M.E.; Lazennec-Schurdevin, C.; Lakisic, G.; Lilla, S.; et al. The Trimeric Coiled-Coil HSBP1 Protein Promotes WASH Complex Assembly at Centrosomes. EMBO J. 2018, 37, e97706. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Souza, L.; Asselbergh, B.; De Winter, V.; Goethals, S.; Timmerman, V.; Janssens, S. HSPB1 Facilitates the Formation of Non-Centrosomal Microtubules. PLoS ONE 2013, 8, e66541. [Google Scholar] [CrossRef] [Green Version]
- Hsu, S.-F.; Lai, H.-C.; Jinn, T.-L. Cytosol-Localized Heat Shock Factor-Binding Protein, AtHSBP, Functions as a Negative Regulator of Heat Shock Response by Translocation to the Nucleus and Is Required for Seed Development in Arabidopsis. Plant Physiol. 2010, 153, 773–784. [Google Scholar] [CrossRef] [Green Version]
- Sayeed, S.K.; Shah, V.; Chaubey, S.; Singh, M.; Alampalli, S.V.; Tatu, U.S. Identification of Heat Shock Factor Binding Protein in Plasmodium Falciparum. Malar. J. 2014, 13, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastner, P.M.; Schleicher, M.; Müller-Taubenberger, A. The NDR Family Kinase NdrA of Dictyostelium Localizes to the Centrosome and Is Required for Efficient Phagocytosis. Traffic 2011, 12, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Hergovich, A.; Cornils, H.; Hemmings, B.A. Mammalian NDR Protein Kinases: From Regulation to a Role in Centrosome Duplication. Biochim. Biophys. Acta 2008, 1784, 3–15. [Google Scholar] [CrossRef]
- Hergovich, A.; Stegert, M.R.; Schmitz, D.; Hemmings, B.A. NDR Kinases Regulate Essential Cell Processes from Yeast to Humans. Nat. Rev. Mol. Cell Biol. 2006, 7, 253–264. [Google Scholar] [CrossRef]
- Zhou, P.-M.; Liang, Y.; Mei, J.; Liao, H.-Z.; Wang, P.; Hu, K.; Chen, L.-Q.; Zhang, X.-Q.; Ye, D. The Arabidopsis AGC Kinases NDR2/4/5 Interact with MOB1A/1B and Play Important Roles in Pollen Development and Germination. Plant J. 2021, 105, 1035–1052. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Benz, C.; Grimaldi, R.; Stockdale, C.; Wyatt, P.; Frearson, J.; Hammarton, T.C. Nuclear DBF-2-Related Kinases Are Essential Regulators of Cytokinesis in Bloodstream Stage Trypanosoma Brucei. J. Biol. Chem. 2010, 285, 15356–15368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller-Taubenberger, A.; Kastner, P.M.; Schleicher, M.; Bolourani, P.; Weeks, G. Regulation of a LATS-Homolog by Ras GTPases Is Important for the Control of Cell Division. BMC Cell Biol. 2014, 15, 25. [Google Scholar] [CrossRef] [Green Version]
- Talevich, E.; Tobin, A.B.; Kannan, N.; Doerig, C. An Evolutionary Perspective on the Kinome of Malaria Parasites. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Müller-Taubenberger, A.; Ishikawa-Ankerhold, H.C.; Kastner, P.M.; Burghardt, E.; Gerisch, G. The STE Group Kinase SepA Controls Cleavage Furrow Formation in Dictyostelium. Cell Motil. Cytoskeleton 2009, 66, 929–939. [Google Scholar] [CrossRef]
- Arasada, R.; Pollard, T.D. Contractile Ring Stability in S. Pombe Depends on F-BAR Protein Cdc15p and Bgs1p Transport from the Golgi Complex. Cell Rep. 2014, 8, 1533–1544. [Google Scholar] [CrossRef] [Green Version]
- Fankhauser, C.; Simanis, V. The Cdc7 Protein Kinase Is a Dosage Dependent Regulator of Septum Formation in Fission Yeast. EMBO J. 1994, 13, 3011–3019. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Shannon, K.B.; Shou, W.; Deshaies, R.J.; Li, R. The Tem1 Small GTPase Controls Actomyosin and Septin Dynamics during Cytokinesis. J. Cell Sci. 2001, 114, 1379–1386. [Google Scholar] [CrossRef]
- Balasubramanian, M.K.; McCollum, D.; Chang, L.; Wong, K.C.; Naqvi, N.I.; He, X.; Sazer, S.; Gould, K.L. Isolation and Characterization of New Fission Yeast Cytokinesis Mutants. Genetics 1998, 149, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Champion, A.; Jouannic, S.; Guillon, S.; Mockaitis, K.; Krapp, A.; Picaud, A.; Simanis, V.; Kreis, M.; Henry, Y. AtSGP1, AtSGP2 and MAP4K Alpha Are Nucleolar Plant Proteins That Can Complement Fission Yeast Mutants Lacking a Functional SIN Pathway. J. Cell Sci. 2004, 117, 4265–4275. [Google Scholar] [CrossRef] [Green Version]
- Rohlfs, M.; Arasada, R.; Batsios, P.; Janzen, J.; Schleicher, M. The Ste20-like Kinase SvkA of Dictyostelium Discoideum Is Essential for Late Stages of Cytokinesis. J. Cell Sci. 2007, 120, 4345–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creasy, C.L.; Chernoff, J. Cloning and Characterization of a Member of the MST Subfamily of Ste20-like Kinases. Gene 1995, 167, 303–306. [Google Scholar] [CrossRef]
- Sullivan, D.S.; Biggins, S.; Rose, M.D. The Yeast Centrin, Cdc31p, and the Interacting Protein Kinase, Kic1p, Are Required for Cell Integrity. J. Cell Biol. 1998, 143, 751–765. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Cui, X.; Yuan, X.; Yu, X.; Sun, J.; Gong, Q. The Hippo/STE20 Homolog SIK1 Interacts with MOB1 to Regulate Cell Proliferation and Cell Expansion in Arabidopsis. J. Exp. Bot. 2016, 67, 1461–1475. [Google Scholar] [CrossRef] [Green Version]
- Ward, P.; Equinet, L.; Packer, J.; Doerig, C. Protein Kinases of the Human Malaria Parasite Plasmodium Falciparum: The Kinome of a Divergent Eukaryote. BMC Genom. 2004, 5, 79. [Google Scholar] [CrossRef] [Green Version]
- Catalano, A.; O’Day, D.H. Nucleolar Localization and Identification of Nuclear/Nucleolar Localization Signals of the Calmodulin-Binding Protein Nucleomorphin during Growth and Mitosis in Dictyostelium. Histochem. Cell Biol. 2011, 135, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Damer, C.K.; O’Halloran, T.J. Spatially Regulated Recruitment of Clathrin to the Plasma Membrane during Capping and Cell Translocation. Mol. Biol. Cell 2000, 11, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Foraker, A.B.; Camus, S.M.; Evans, T.M.; Majeed, S.R.; Chen, C.Y.; Taner, S.B.; Correa, I.R.; Doxsey, S.J.; Brodsky, F.M. Clathrin Promotes Centrosome Integrity in Early Mitosis through Stabilization of Centrosomal Ch-TOG. J. Cell Biol. 2012, 198, 591–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.S.; Pishvaee, B.; Payne, G.S. A Modulatory Role for Clathrin Light Chain Phosphorylation in Golgi Membrane Protein Localization during Vegetative Growth and during the Mating Response of Saccharomyces Cerevisiae. Mol. Biol. Cell 1999, 10, 713–726. [Google Scholar] [CrossRef] [Green Version]
- De León, N.; Hoya, M.; Curto, M.-A.; Moro, S.; Yanguas, F.; Doncel, C.; Valdivieso, M.-H. The AP-2 Complex Is Required for Proper Temporal and Spatial Dynamics of Endocytic Patches in Fission Yeast. Mol. Microbiol. 2016, 100, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Van Damme, D.; Gadeyne, A.; Vanstraelen, M.; Inzé, D.; Van Montagu, M.C.E.; De Jaeger, G.; Russinova, E.; Geelen, D. Adaptin-like Protein TPLATE and Clathrin Recruitment during Plant Somatic Cytokinesis Occurs via Two Distinct Pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 615–620. [Google Scholar] [CrossRef] [Green Version]
- Kalb, L.C.; Frederico, Y.C.A.; Batista, C.M.; Eger, I.; Fragoso, S.P.; Soares, M.J. Clathrin Expression in Trypanosoma Cruzi. BMC Cell Biol. 2014, 15, 23. [Google Scholar] [CrossRef] [Green Version]
- Schmith, A.; Groth, M.; Ratka, J.; Gatz, S.; Spaller, T.; Siol, O.; Glöckner, G.; Winckler, T. Conserved Gene Regulatory Function of the Carboxy-Terminal Domain of Dictyostelid C-Module-Binding Factor. Eukaryotic Cell 2013, 12, 460–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubry, L.; Guetta, D.; Klein, G. The Arrestin Fold: Variations on a Theme. Curr. Genom. 2009, 10, 133–142. [Google Scholar] [CrossRef]
- Habourdin, C.; Klein, G.; Araki, T.; Williams, J.G.; Aubry, L. The Arrestin-Domain Containing Protein AdcA Is a Response Element to Stress. Cell Commun. Signal. 2013, 11, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molla-Herman, A.; Davis, K.M.; Mykytyn, K.; Benmerah, A. Monitoring β-Arrestin 2 Targeting to the Centrosome, Basal Body, and Primary Cilium by Fluorescence Microscopy. Methods Mol. Biol. 2019, 1957, 271–289. [Google Scholar] [CrossRef]
- Koonce, M.P.; Samso, M. Overexpression of Cytoplasmic Dynein’s Globular Head Causes a Collapse of the Interphase Microtubule Network in Dictyostelium. Mol. Biol. Cell 1996, 7, 935–948. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Daunderer, C.; Schulz, I. Molecular and Functional Analysis of the Dictyostelium Centrosome. Int. Rev. Cytol. 2004, 241, 155–202. [Google Scholar]
- Tovey, C.A.; Conduit, P.T. Microtubule Nucleation by γ-Tubulin Complexes and Beyond. Essays Biochem. 2018, 62, 765–780. [Google Scholar] [CrossRef]
- Lin, T.C.; Neuner, A.; Schiebel, E. Targeting of Gamma-Tubulin Complexes to Microtubule Organizing Centers: Conservation and Divergence. Trends Cell Biol. 2015, 25, 296–307. [Google Scholar] [CrossRef]
- Sukumaran, S.K.; Blau-Wasser, R.; Rohlfs, M.; Gallinger, C.; Schleicher, M.; Noegel, A.A. The Centrosomal Component CEP161 of Dictyostelium Discoideum Interacts with the Hippo Signaling Pathway. Cell Cycle 2015, 14, 1024–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuriyama, R.; Fisher, C.R. A Novel Mitosis-Specific Cep215 Domain Interacts with Cep192 and Phosphorylated Aurora A for Organization of Spindle Poles. J. Cell Sci. 2020, 133, jcs240267. [Google Scholar] [CrossRef]
- Fong, K.W.; Hau, S.Y.; Kho, Y.S.; Jia, Y.; He, L.; Qi, R.Z. Interaction of CDK5RAP2 with EB1 to Track Growing Microtubule Tips and to Regulate Microtubule Dynamics. Mol. Biol. Cell 2009, 20, 3660–3670. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Euteneuer, U.; Ho, T.H.; Rehberg, M. Regulated Expression of the Centrosomal Protein DdCP224 Affects Microtubule Dynamics and Reveals Mechanisms for the Control of Supernumerary Centrosome Number. Mol. Biol. Cell 2003, 14, 4067–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, K.V.; Reinders, Y.; Ho, T.H.; Sickmann, A.; Gräf, R. Identification and Isolation of Dictyostelium Microtubule-Associated Protein Interactors by Tandem Affinity Purification. Eur. J. Cell Biol. 2006, 85, 1079–1090. [Google Scholar] [CrossRef]
- Zheng, Y. G Protein Control of Microtubule Assembly. Annu. Rev. Cell Dev. Biol. 2004, 20, 867–894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ems-McClung, S.C.; Walczak, C.E. Aurora A Phosphorylates MCAK to Control Ran-Dependent Spindle Bipolarity. Mol. Biol. Cell 2008, 19, 2752–2765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thawani, A.; Kadzik, R.S.; Petry, S. XMAP215 Is a Microtubule Nucleation Factor That Functions Synergistically with the γ-Tubulin Ring Complex. Nat. Cell Biol. 2018, 20, 575–585. [Google Scholar] [CrossRef]
- Yan, X.; Habedanck, R.; Nigg, E.A. A Complex of Two Centrosomal Proteins, CAP350 and FOP, Cooperates with EB1 in MT Anchoring. Mol. Biol. Cell 2005, 17, 634–644. [Google Scholar] [CrossRef] [Green Version]
- Kalt, A.; Schliwa, M. A Novel Structural Component of the Dictyostelium Centrosome. J. Cell Sci. 1996, 109, 3103–3112. [Google Scholar] [CrossRef] [PubMed]
- Noegel, A.A.; Blau-Wasser, R.; Sultana, H.; Müller, R.; Israel, L.; Schleicher, M.; Patel, H.; Weijer, C.J. The Cyclase-Associated Protein CAP as Regulator of Cell Polarity and CAMP Signaling in Dictyostelium. Mol. Biol. Cell 2004, 15, 934–945. [Google Scholar] [CrossRef] [Green Version]
- Po’uha, S.T.; Kavallaris, M. Gamma-Actin Is Involved in Regulating Centrosome Function and Mitotic Progression in Cancer Cells. Cell Cycle 2015, 14, 3908–3919. [Google Scholar] [CrossRef] [Green Version]
- Farina, F.; Gaillard, J.; Guerin, C.; Coute, Y.; Sillibourne, J.; Blanchoin, L.; Thery, M. The Centrosome Is an Actin-Organizing Centre. Nat. Cell Biol. 2016, 18, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Brusis, N.; Daunderer, C.; Euteneuer, U.; Hestermann, A.; Schliwa, M.; Ueda, M. Comparative Structural, Molecular and Functional Aspects of the Dictyostelium Discoideum Centrosome. Curr. Top. Dev. Biol. 2000, 49, 161–185. [Google Scholar]
- Dantas, T.J.; Daly, O.M.; Morrison, C.G. Such Small Hands: The Roles of Centrins/Caltractins in the Centriole and in Genome Maintenance. Cell. Mol. Life Sci. 2012, 69, 2979–2997. [Google Scholar] [CrossRef]
- Li, S.; Sandercock, A.M.; Conduit, P.; Robinson, C.V.; Williams, R.L.; Kilmartin, J.V. Structural Role of Sfi1p-Centrin Filaments in Budding Yeast Spindle Pole Body Duplication. J. Cell Biol. 2006, 173, 867–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mana-Capelli, S.; Gräf, R.; Larochelle, D.A. Dictyostelium Discoideum CenB Is a Bona Fide Centrin Essential for Nuclear Architecture and Centrosome Stability. Eukaryot. Cell 2009, 8, 1106–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mana-Capelli, S.; Gräf, R.; Larochelle, D.A. Dictyostelium Centrin B Localization during Cell Cycle Progression. Commun. Integr. Biol. 2010, 3, 39–41. [Google Scholar] [CrossRef] [Green Version]
- Sanders, M.A.; Salisbury, J.L. Centrin Plays an Essential Role in Microtubule Severing during Flagellar Excision in Chlamydomonas Reinhardtii. J. Cell Biol. 1994, 124, 795–805. [Google Scholar] [CrossRef] [Green Version]
- Pulvermüller, A.; Giessl, A.; Heck, M.; Wottrich, R.; Schmitt, A.; Ernst, O.P.; Choe, H.W.; Hofmann, K.P.; Wolfrum, U. Calcium-Dependent Assembly of Centrin-G-Protein Complex in Photoreceptor Cells. Mol. Cell. Biol. 2002, 22, 2194–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, Y.; Maller, J.L. Calcium, Calmodulin, and CaMKII Requirement for Initiation of Centrosome Duplication in Xenopus Egg Extracts. Science 2002, 295, 499–502. [Google Scholar] [CrossRef]
- Fisk, H.A.; Mattison, C.P.; Winey, M. Human Mps1 Protein Kinase Is Required for Centrosome Duplication and Normal Mitotic Progression. Proc. Natl. Acad. Sci. USA 2003, 100, 14875–14880. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, K. Oncogenes and Tumour Suppressors Take on Centrosomes. Nat. Rev. Cancer 2007, 7, 911–924. [Google Scholar] [CrossRef]
- Spang, A.; Grein, K.; Schiebel, E. The Spacer Protein Spc110p Targets Calmodulin to the Central Plaque of the Yeast Spindle Pole Body. J. Cell Sci. 1996, 109, 2229–2237. [Google Scholar] [CrossRef]
- Psatha, M.; Koffer, A.; Erent, M.; Moss, S.E.; Bolsover, S. Calmodulin Spatial Dynamics in RBL-2H3 Mast Cells. Cell Calcium. 2004, 36, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Liu, T.; Clarke, M. Calmodulin and the Contractile Vacuole Complex in Mitotic Cells of Dictyostelium Discoideum. J. Cell Sci. 1993, 104, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Morio, T.; Urushihara, H.; Saito, T.; Ugawa, Y.; Mizuno, H.; Yoshida, M.; Yoshino, R.; Mitra, B.N.; Pi, M.; Sato, T.; et al. The Dictyostelium Developmental CDNA Project: Generation and Analysis of Expressed Sequence Tags from the First-Finger Stage of Development. DNA Res. 1998, 5, 335–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, A.M.; Descombes, P.; Twomey, C.; Bacchieri, R.; Nigg, E.A. The NIMA-Related Kinase X-Nek2B Is Required for Efficient Assembly of the Zygotic Centrosome in Xenopus Laevis. J. Cell Sci. 2000, 113, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Gräf, R. Maltose-Binding Protein as a Fusion Tag for the Localization and Purification of Cloned Proteins in Dictyostelium. Anal. Biochem. 2001, 289, 297–300. [Google Scholar] [CrossRef]
- Bullitt, E.; Rout, M.P.; Kilmartin, J.V.; Akey, C.W. The Yeast Spindle Pole Body Is Assembled around a Central Crystal of Spc42p. Cell 1997, 89, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Drennan, A.C.; Krishna, S.; Seeger, M.A.; Andreas, M.P.; Gardner, J.M.; Sether, E.K.R.; Jaspersen, S.L.; Rayment, I. Structure and Function of Spc42 Coiled-Coils in Yeast Centrosome Assembly and Duplication. Mol. Biol. Cell 2019, 30, 1505–1522. [Google Scholar] [CrossRef]
- Stafstrom, J.P.; Staehelin, L.A. Dynamics of the Nuclear Envelope and of Nuclear Pore Complexes during Mitosis in the Drosophila Embryo. Eur. J. Cell Biol. 1984, 34, 179–189. [Google Scholar] [PubMed]
- De Souza, C.P.; Osmani, A.H.; Hashmi, S.B.; Osmani, S.A. Partial Nuclear Pore Complex Disassembly during Closed Mitosis in Aspergillus Nidulans. Curr. Biol. 2004, 14, 1973–1984. [Google Scholar] [CrossRef] [Green Version]
- Ding, R.; West, R.R.; Morphew, M.; Oakley, B.R.; McIntosh, J.R. The Spindle Pole Body of Schizosaccharomyces Pombe Enters and Leaves the Nuclear Envelope as the Cell Cycle Proceeds. Mol. Biol. Cell 1997, 8, 1461–1479. [Google Scholar] [CrossRef]
- Tamm, T.; Grallert, A.; Grossman, E.P.S.; Alvarez-Tabares, I.; Stevens, F.E.; Hagan, I.M. Brr6 Drives the Schizosaccharomyces Pombe Spindle Pole Body Nuclear Envelope Insertion/Extrusion Cycle. J. Cell Biol. 2011, 195, 467–484. [Google Scholar] [CrossRef] [Green Version]
- Dinkel, H.; Van Roey, K.; Michael, S.; Davey, N.E.; Weatheritt, R.J.; Born, D.; Speck, T.; Kruger, D.; Grebnev, G.; Kuban, M.; et al. The Eukaryotic Linear Motif Resource ELM: 10 Years and Counting. Nucleic Acids Res. 2014, 42, D259–D266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, A.L.; Lee, S.; Park, J.S.; Han, S.; Jang, C.-Y.; Lim, J.-S.; Lee, M.S.; Yang, Y. Cancerous Inhibitor of Protein Phosphatase 2A (CIP2A) Protein Is Involved in Centrosome Separation through the Regulation of NIMA (Never in Mitosis Gene A)-Related Kinase 2 (NEK2) Protein Activity. J. Biol. Chem. 2014, 289, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Helps, N.R.; Luo, X.; Barker, H.M.; Cohen, P.T. NIMA-Related Kinase 2 (Nek2), a Cell-Cycle-Regulated Protein Kinase Localized to Centrosomes, Is Complexed to Protein Phosphatase 1. Biochem. J. 2000, 349, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Mardin, B.R.; Agircan, F.G.; Lange, C.; Schiebel, E. Plk1 Controls the Nek2A-PP1gamma Antagonism in Centrosome Disjunction. Curr. Biol. 2011, 21, 1145–1151. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Guan, K.-L. Hippo Signaling in Embryogenesis and Development. Trends Biochem. Sci. 2021, 46, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Kim, N.-G.; Gumbiner, B.M. Regulation of Hippo Pathway by Mitogenic Growth Factors via Phosphoinositide 3-Kinase and Phosphoinositide-Dependent Kinase-1. Proc. Natl. Acad. Sci. USA 2013, 110, 2569–2574. [Google Scholar] [CrossRef] [Green Version]
- Mardin, B.R.; Lange, C.; Baxter, J.E.; Hardy, T.; Scholz, S.R.; Fry, A.M.; Schiebel, E. Components of the Hippo Pathway Cooperate with Nek2 Kinase to Regulate Centrosome Disjunction. Nat. Cell Biol. 2010, 12, 1166–1176. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.K.; Stumpf, M.; Salamon, S.; Ahmad, I.; Bhattacharya, K.; Fischer, S.; Muller, R.; Altmuller, J.; Budde, B.; Thiele, H.; et al. CDK5RAP2 Interaction with Components of the Hippo Signaling Pathway May Play a Role in Primary Microcephaly. Mol. Genet Genom. 2017, 292, 365–383. [Google Scholar] [CrossRef] [Green Version]
- Mukai, S.; Yabuta, N.; Yoshida, K.; Okamoto, A.; Miura, D.; Furuta, Y.; Abe, T.; Nojima, H. Lats1 Suppresses Centrosome Overduplication by Modulating the Stability of Cdc25B. Sci. Rep. 2015, 5, 16173. [Google Scholar] [CrossRef] [Green Version]
- Simanis, V. Pombe’s Thirteen—Control of Fission Yeast Cell Division by the Septation Initiation Network. J. Cell Sci. 2015, 128, 1465–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, R.J. The Cyclin-Dependent Kinase Family in the Social Amoebozoan Dictyostelium Discoideum. Cell. Mol. Life Sci. 2014, 71, 629–639. [Google Scholar] [CrossRef]
- Heald, R.; Khodjakov, A. Thirty Years of Search and Capture: The Complex Simplicity of Mitotic Spindle Assembly. J. Cell Biol. 2015, 211, 1103–1111. [Google Scholar] [CrossRef]
- Kufer, T.A.; Sillje, H.H.; Korner, R.; Gruss, O.J.; Meraldi, P.; Nigg, E.A. Human TPX2 Is Required for Targeting Aurora-A Kinase to the Spindle. J. Cell Biol. 2002, 158, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Gruss, O.J.; Wittmann, M.; Yokoyama, H.; Pepperkok, R.; Kufer, T.; Sillje, H.; Karsenti, E.; Mattaj, I.W.; Vernos, I. Chromosome-Induced Microtubule Assembly Mediated by TPX2 Is Required for Spindle Formation in HeLa Cells. Nat. Cell Biol. 2002, 4, 871–879. [Google Scholar] [CrossRef]
- Garrido, G.; Vernos, I. Non-Centrosomal TPX2-Dependent Regulation of the Aurora A Kinase: Functional Implications for Healthy and Pathological Cell Division. Front. Oncol. 2016, 6, 88. [Google Scholar] [CrossRef] [Green Version]
- Thawani, A.; Stone, H.A.; Shaevitz, J.W.; Petry, S. Spatiotemporal Organization of Branched Microtubule Networks. eLife 2019, 8, e43890. [Google Scholar] [CrossRef] [PubMed]
- Alfaro-Aco, R.; Thawani, A.; Petry, S. Biochemical Reconstitution of Branching Microtubule Nucleation. eLife 2020, 9, e49797. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, C.B.; Khodjakov, A.L. Cooperative Mechanisms of Mitotic Spindle Formation. J. Cell Sci. 2007, 120, 1717–1722. [Google Scholar] [CrossRef] [Green Version]
- Kaller, M.; Euteneuer, U.; Nellen, W. Differential Effects of Heterochromatin Protein 1 Isoforms on Mitotic Chromosome Distribution and Growth in Dictyostelium Discoideum. Eukaryot. Cell 2006, 5, 530–543. [Google Scholar] [CrossRef] [Green Version]
- King, M.C.; Drivas, T.G.; Blobel, G. A Network of Nuclear Envelope Membrane Proteins Linking Centromeres to Microtubules. Cell 2008, 134, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Samereier, M. Functional Analyses of Microtubule and Centrosome-Associated Proteins in Dictyostelium Discoideum. Ph.D. Thesis, University of Potsdam, Potsdam, Germany, 2011. Available online: https://publishup.uni-potsdam.de/frontdoor/index/index/docId/5092 (accessed on 4 October 2021).
- Rieder, C.L.; Maiato, H. Stuck in Division or Passing through: What Happens When Cells Cannot Satisfy the Spindle Assembly Checkpoint. Dev. Cell 2004, 7, 637–651. [Google Scholar] [CrossRef] [Green Version]
- Tikhonenko, I.; Irizarry, K.; Khodjakov, A.; Koonce, M.P. Organization of Microtubule Assemblies in Dictyostelium Syncytia Depends on the Microtubule Crosslinker, Ase1. Cell. Mol. Life Sci. 2016, 73, 859–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- She, Z.-Y.; Wei, Y.-L.; Lin, Y.; Li, Y.-L.; Lu, M.-H. Mechanisms of the Ase1/PRC1/MAP65 Family in Central Spindle Assembly. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2033–2048. [Google Scholar] [CrossRef] [PubMed]
- Tapley, E.C.; Starr, D.A. Connecting the Nucleus to the Cytoskeleton by SUN-KASH Bridges across the Nuclear Envelope. Curr. Opin. Cell Biol. 2013, 25, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batsios, P.; Meyer, I.; Gräf, R. Proximity-Dependent Biotin Identification (BioID) in Dictyostelium Amoebae. Methods Enzymol. 2016, 569, 23–42. [Google Scholar] [CrossRef]
- Rivero, F.; Kuspa, A.; Brokamp, R.; Matzner, M.; Noegel, A.A. Interaptin, an Actin-Binding Protein of the Alpha-Actinin Superfamily in Dictyostelium Discoideum, Is Developmentally and CAMP-Regulated and Associates with Intracellular Membrane Compartments. J. Cell Biol. 1998, 142, 735–750. [Google Scholar] [CrossRef] [Green Version]
- Batsios, P.; Gräf, R.; Koonce, M.P.; Larochelle, D.A.; Meyer, I. Nuclear Envelope Organization in Dictyostelium Discoideum. Int. J. Dev. Biol. 2019, 63, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luxton, G.W.G.; Starr, D.A. KASHing up with the Nucleus: Novel Functional Roles of KASH Proteins at the Cytoplasmic Surface of the Nucleus. Curr. Opin Cell Biol. 2014, 28, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Amoebozoa Dictyostelium | Opisthokonta Metazoa Homo sapiens | Opisthokonta Fungi S. cerevisiae | Opisthokonta Fungi S. pombe | Archaeplastida Arabidopsis thaliana | Excavata Trypanosoma spec. | SAR Plasmodium falciparum, Albugo spec. |
|---|---|---|---|---|---|---|
| Central layer(s) | ||||||
| CP91 [33] | - | - | - | - | - | - |
| CP75 [53] | - | - | - | - | - | - |
| CP39 [53] | - | - | - | - | - | - |
| Outer core layer | ||||||
| Cep192 [54] | Cep192/SPD2 [55] | - | - | - | - | - |
| CP55 [56] | - | - | - | - | - | - |
| Nek2 [57] | Nek2 [58] | Kin3p [59] | Fin1 [60] | AtNek2 [61] | TbNRKC [62] | Pfnek-2 [63] |
| CP44 [64] | - | - | - | - | - | - |
| Corona | ||||||
| γ-tubulin [65] | γ-tubulin [5] | Tub4 [66] | gtb1 [67] | γ-tubulin [68] | γ-tubulin [69] | γ-tubulin [70] |
| Spc97 [65] | GCP2 | Spc97 [66] | Alp4 [67] | GCP2 [68] | GCP2 [69] | GI: 389585322 |
| Spc98 [65] | GCP3 | Spc98 [66] | Alp6 [67] | GCP3 [68] | GCP3 [69] | GI: 389585419 |
| CDK5RAP2/Cep161 [71] | CDK5RAP2/Cep215/Cnn [72] | Spc72p [73] | Mto1/Mbo1/Mod20 [74] | - | GI: 407424972 | GI: 23479271 |
| CP148 [75] | Pericentrin/PCNT/PLP [76] | Spc110p [73] | Pcp1 [77] | - | - | GI: 325186828 |
| TACC [78] | TACC/Maskin [79] | - | Alp7/Mia1 | GI: 297312240 | - * | GI: 325183149 |
| CP224 [80] | chTOG/XMAP215 [81] | Stu2p [82] | Dis1/Alp14 [83] | MOR1 [84] | XMAP215 [85] | GI: 1976646509 |
| EB1 [86] | EB1 [87] | YEB1/Bim1p [88] | Mal3 [89] | EB1c [90] | EB1 | EB1 |
| Moe1 [91] | eIF-3 subunit 7 | - | Moe1 [92] | eIF-3 subunit 7 | eIF-3 subunit 7 | eIF-3D |
| CP248/CP250 [64,93] | C-Nap1/Cep250 [94] | - | - | - | - | - |
| CenA/DdCrp [95] | Centrin-3 [96] | Cdc31p [97] | Cdc31 [98] | Centrin [99] | Centrin [100] | Centrin [101] |
| CP103 [64] | - | - | - | - | - | - |
| Corona-associated | ||||||
| Dynein DHC [102,103] | DHC [104] | Dyn1p [105] | Dhc1 [106] | - | DHC [107] | DHC [108] |
| Dynactin (including p50, p62, Arp1/Centractin) (own unpubl [109]) | Dynactin [110] | Dynactin [111] | Dynactin [106] | - | Dynactin | Dynactin [112] |
| Lis1 [103] | Lis1/PAFAH1B1 [113] | Pac1p [114] | - | - | SMU1 | GI: 1678234918 |
| Centrosome-associated (no sublocation determined) | ||||||
| AurK [115] | AuroraA/B/C [116] | Ipl1p [117] | Ark1 [118] | ATAUR1/2/3 | TcAUK1/2/3 [119] | Pfark-1/2/3 [120] |
| Plk [64] | Plk1 [121] | Cdc5p [121] | Plo1 [122] | - | TbPLK [123] | - [123] |
| Sun1 [124,125] | Sun1/Matefin [126] | Mps3p [127] | Sad1 [128] | Sun1 [129] | GI: 686631607 | GI: 221061315 |
| Kif9 [130] | Kif9 [131] | - | - | - | - | - |
| Kif12 [132] | Kif12 [133] | - | - | Kinesin-12 | - | - |
| Nup53 (Meyer in prep) | Nup35 [134] | Nup53p [135] | Nup40 [136] | NUP35 [137] | TcCL_ESM01172 | GI: 325183342 |
| phr2AB [138] | PPP2R5D [139] | Cdc55p [140] | Pab1 [141] | AtB beta [142] | NCBI: XP_829543 | phr2AB |
| HSBP1 [143] | HSBP1 [144] | - | - | AtHSBP [145] | - | HSBP [146] |
| NdrA [147] | NDR1 [148] | Cbk1p [149] | Orb6 [149] | AGC§ [150] | PK50/PK53 § [151] | AGC/AKT § |
| NdrC [152] | LATS2 | Dbf2p [149] | Sid2 [149] | AGC§ [150] | PK50/PK53 § [151] | AGC/AKT § [153] |
| SepA [154] | - | Cdc15p [155] | Cdc7 [156] | - | - | - |
| Spg1 [154] | - | Tem1p [157] | Sid3 [158] | AtSGP1 [159] | - | - |
| SvkA/Hrk-Svk [160] | MST1/2 [161] | Kic1p [162] | Sid1 [158] | SIK1 [163] | GI: 1919796340 | - $ [164] |
| NumA1 [165] | BRCT domain proteins & | BRCT domain proteins & | BRCT domain proteins & | BRCT domain proteins & | BRCT domain proteins & | BRCT domain proteins & |
| fttB [52] | 14-3-3 | |||||
| Clathrin light chain (clcA) [166] | CLC [167] | Clc1p [168] | Clc1 [169] | CLC2 [170] | TcClc [171] | GI: 124809181 |
| AbpF [172] | - | - | - | - | - | - |
| AdcA [173,174] | b-arrestin 2 [175] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gräf, R.; Grafe, M.; Meyer, I.; Mitic, K.; Pitzen, V. The Dictyostelium Centrosome. Cells 2021, 10, 2657. https://doi.org/10.3390/cells10102657
Gräf R, Grafe M, Meyer I, Mitic K, Pitzen V. The Dictyostelium Centrosome. Cells. 2021; 10(10):2657. https://doi.org/10.3390/cells10102657
Chicago/Turabian StyleGräf, Ralph, Marianne Grafe, Irene Meyer, Kristina Mitic, and Valentin Pitzen. 2021. "The Dictyostelium Centrosome" Cells 10, no. 10: 2657. https://doi.org/10.3390/cells10102657
APA StyleGräf, R., Grafe, M., Meyer, I., Mitic, K., & Pitzen, V. (2021). The Dictyostelium Centrosome. Cells, 10(10), 2657. https://doi.org/10.3390/cells10102657







