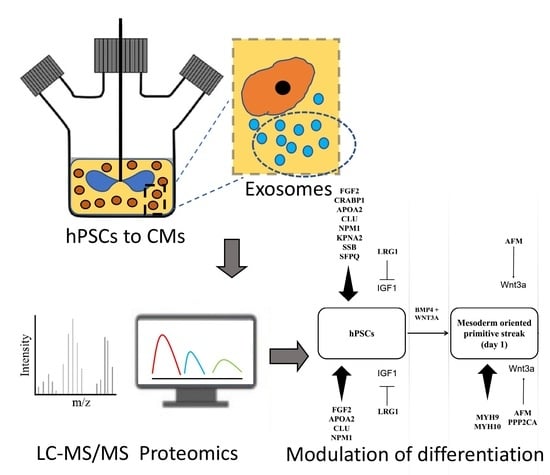
Proteomic Analysis of Exosomes during Cardiogenic Differentiation of Human Pluripotent Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Pluripotent Stem Cell Culture
2.2. Cardiac Differentiation
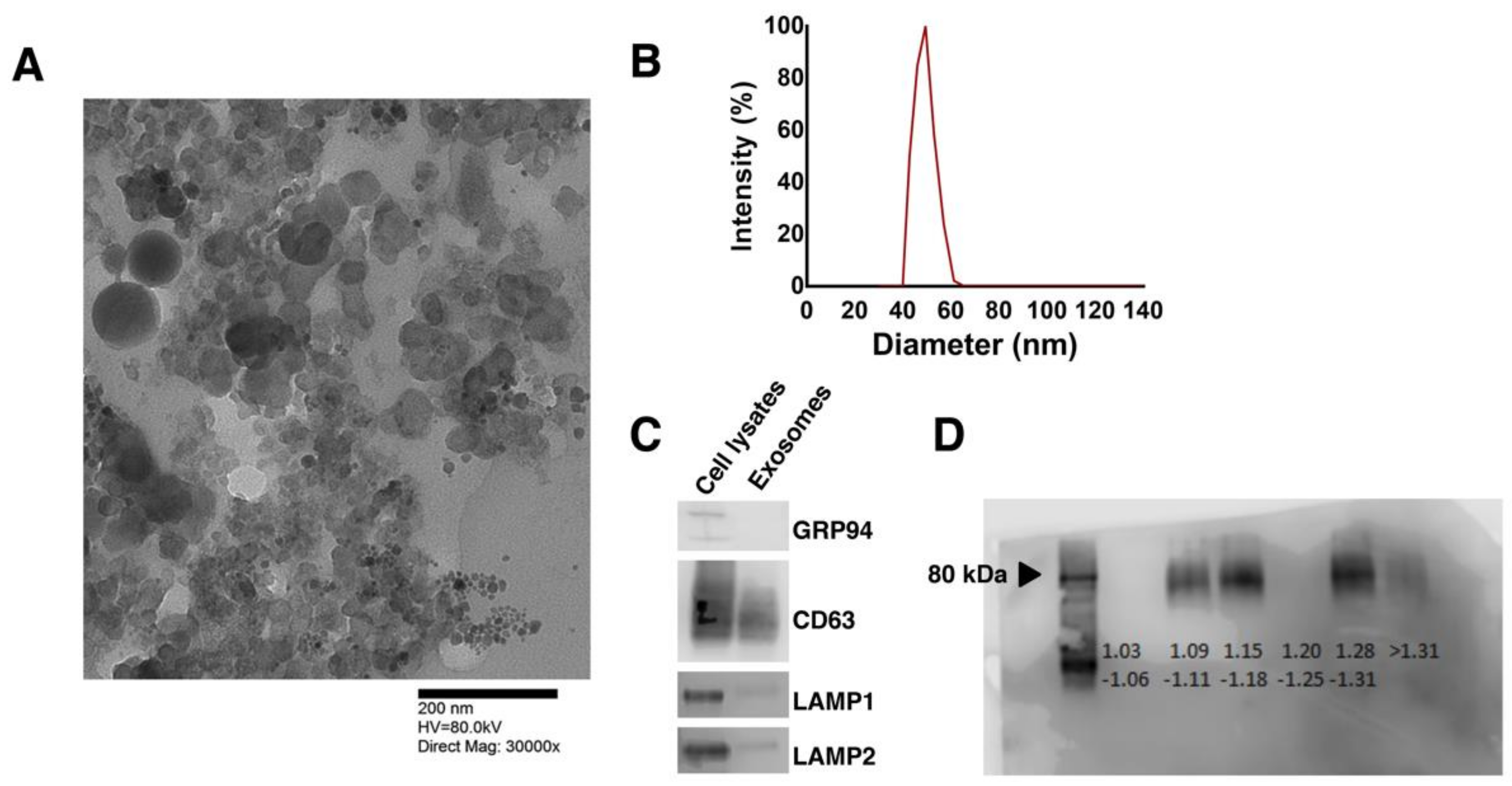
2.3. Exosome Isolation and Characterization
2.4. Size and Morphological Analysis
2.5. RNA Extraction, RT-PCR and Quantitative PCR Analysis
2.6. Western Blot Analysis
2.7. Proteomic Analysis
2.8. Addition of Exosomes to Differentiating Cells
2.9. Flow Cytometry
2.10. Statistical Analysis and Experimental Design
3. Results
3.1. Human Pluripotent Stem Cell-Secreted Exosome Collection and Characterization
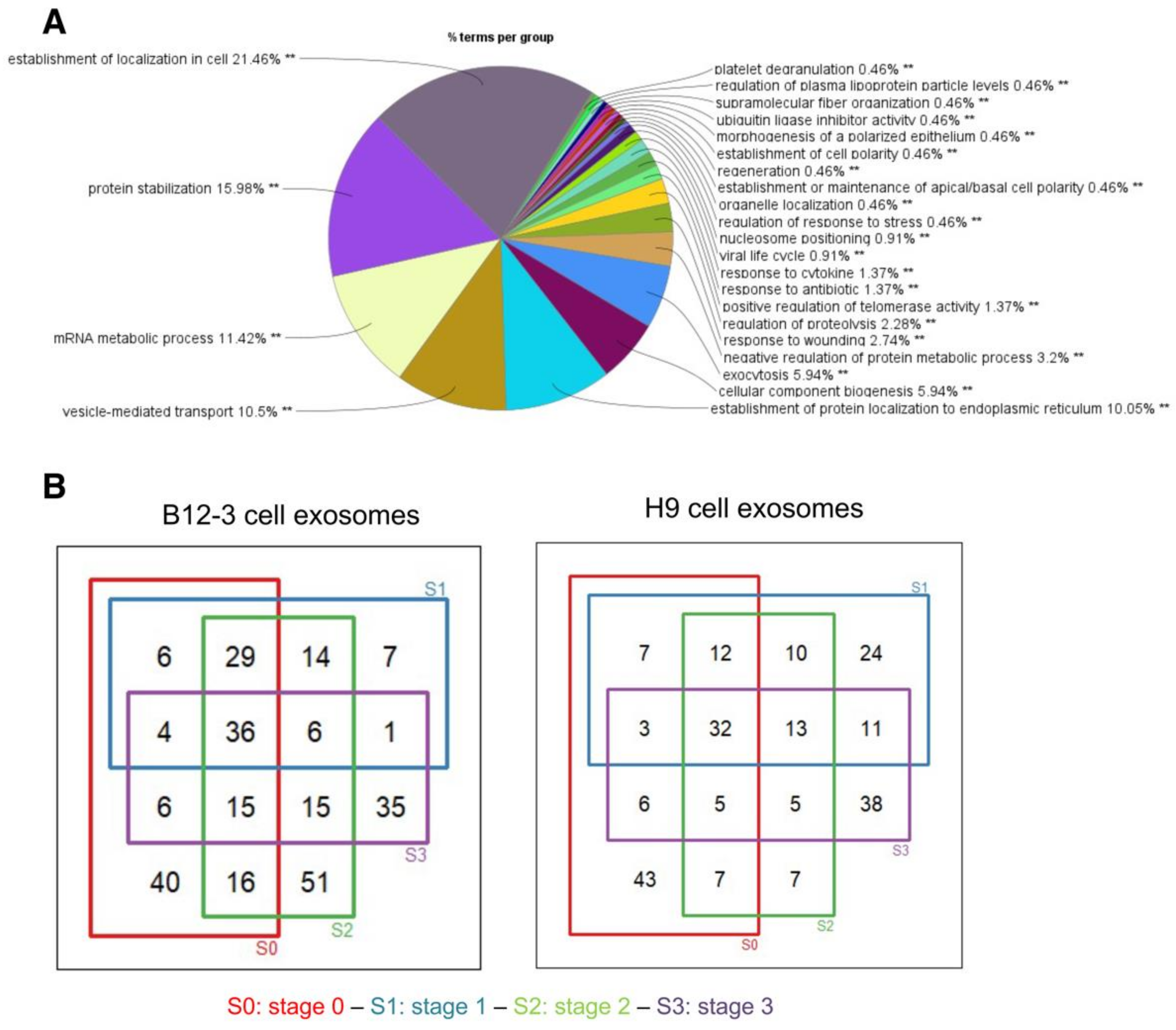
3.2. Proteomic Cargo of Exosomes during Cardiogenic hPSC Differentiation
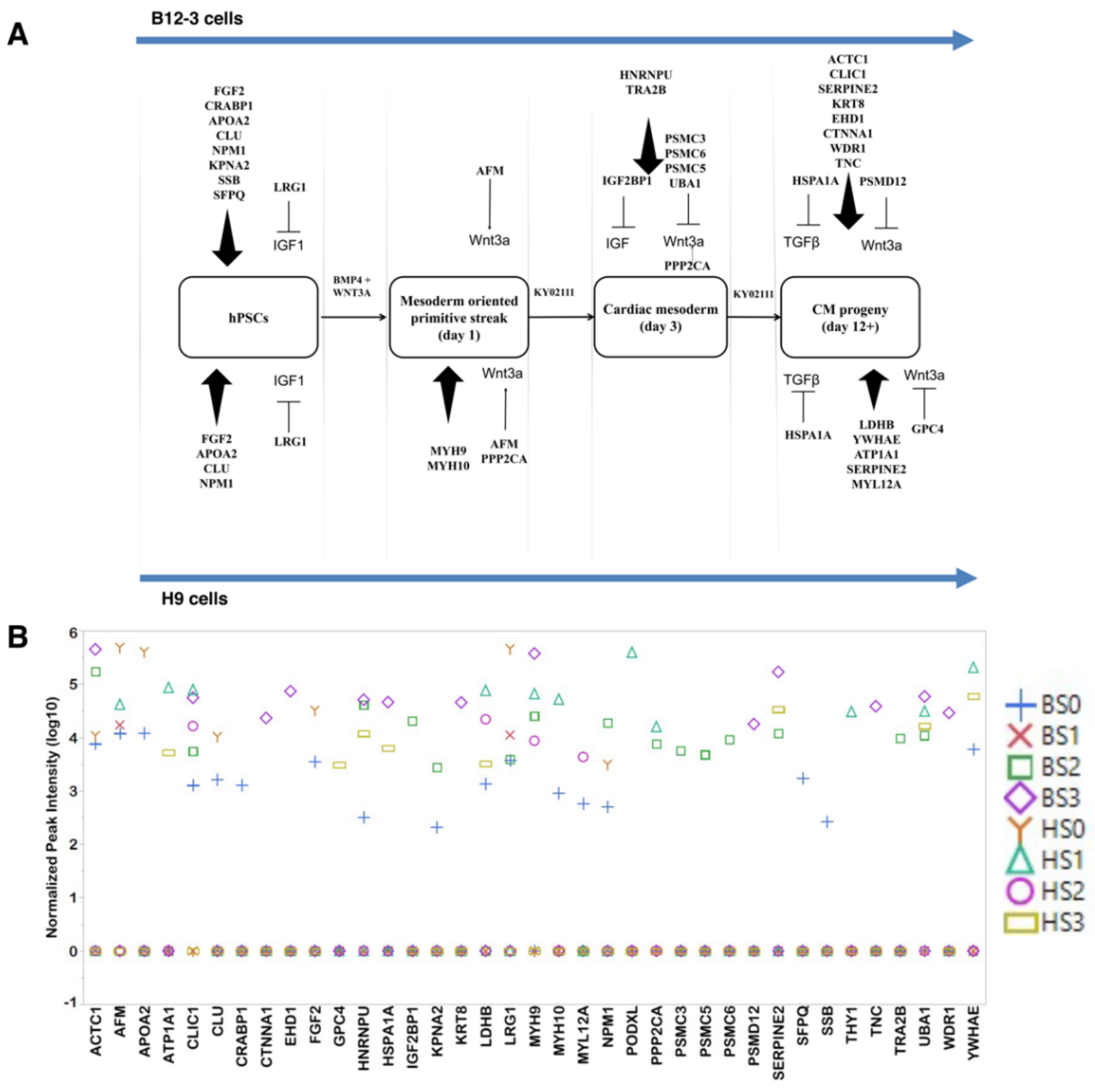
3.3. Exosomes Contain Proteins which Influence hPSC Differentiation
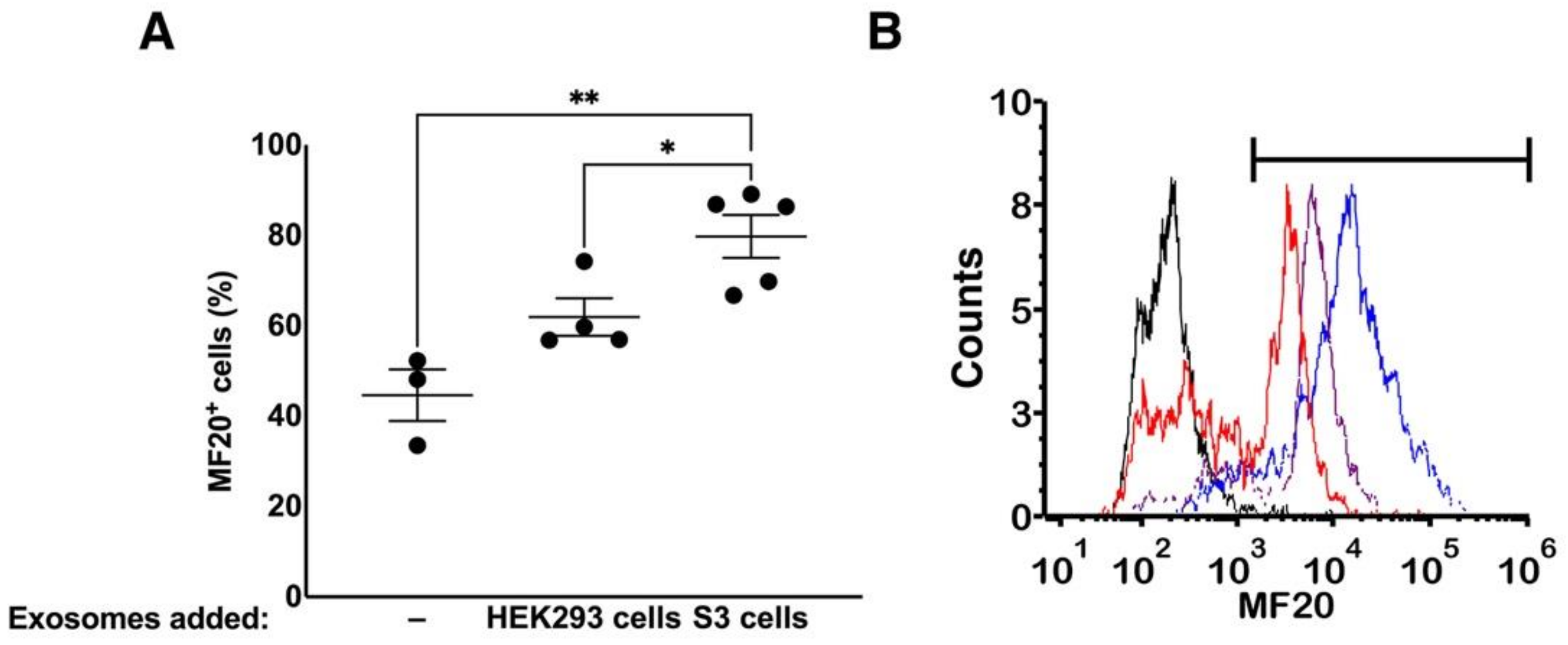
3.4. Effect of Exosomes on Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, M.; Nickoloff, E.; Abramova, T.; Johnson, J.; Verma, S.K.; Krishnamurthy, P.; Mackie, A.R.; Vaughan, E.; Garikipati, V.N.; Benedict, C.; et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ. Res. 2015, 117, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, A.G.-E.; Cheng, K.; Marbán, E. Exosomes as Critical Agents of Cardiac Regeneration Triggered by Cell Therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Yang, J.; Zhao, X.; Zhang, E.; Zeng, Q.; Yu, Y.; Yang, L.; Wu, B.; Yi, G.; Mao, X.; et al. Circulating myocardial microRNAs from infarcted hearts are carried in exosomes and mobilise bone marrow progenitor cells. Nat. Commun. 2019, 10, 959. [Google Scholar] [CrossRef] [PubMed]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Welton, J.L.; Khanna, S.; Giles, P.J.; Brennan, P.; Brewis, I.A.; Staffurth, J.; Mason, M.D.; Clayton, A. Proteomics Analysis of Bladder Cancer Exosomes. Mol. Cell. Proteom. 2010, 9, 1324–1338. [Google Scholar] [CrossRef] [Green Version]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J. 2009, 23, 1541–1557. [Google Scholar] [CrossRef] [Green Version]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Toh, W.S.; Lai, R.C.; Zhang, B.; Lim, S.K. MSC exosome works through a protein-based mechanism of action. Biochem. Soc. Trans. 2018, 46, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Oh, M.; Lee, J.; Kim, Y.J.; Rhee, W.J.; Park, J.H. Exosomes Derived from Human Induced Pluripotent Stem Cells Ameliorate the Aging of Skin Fibroblasts. Int. J. Mol. Sci. 2018, 19, 1715. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.D.; Johansson, H.J.; Graham, C.S.; Vesterlund, M.; Pham, M.T.; Bramlett, C.S.; Montgomery, E.N.; Mellema, M.S.; Bardini, R.L.; Contreras, Z.; et al. Comprehensive Proteomic Analysis of Mesenchymal Stem Cell Exosomes Reveals Modulation of Angiogenesis via Nuclear Factor-KappaB Signaling. Stem Cells 2016, 34, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: Role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 2018, 114, 992–1005. [Google Scholar] [CrossRef] [Green Version]
- Lyu, L.; Wang, H.; Li, B.; Qin, Q.; Qi, L.; Nagarkatti, M.; Nagarkatti, P.; Janicki, J.S.; Wang, X.L.; Cui, T. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J. Mol. Cell. Cardiol. 2015, 89, 268–279. [Google Scholar] [CrossRef] [Green Version]
- Dougherty, J.A.; Kumar, N.; Noor, M.; Angelos, M.G.; Khan, M.; Chen, C.A.; Khan, M. Extracellular Vesicles Released by Human Induced-Pluripotent Stem Cell-Derived Cardiomyocytes Promote Angiogenesis. Front. Physiol. 2018, 9, 1794. [Google Scholar] [CrossRef] [Green Version]
- Van Vliet, P.; Wu, S.M.; Zaffran, S.; Pucéat, M. Early cardiac development: A view from stem cells to embryos. Cardiovasc. Res. 2012, 96, 352–362. [Google Scholar] [CrossRef] [Green Version]
- Keren-Politansky, A.; Keren, A.; Bengal, E. Neural ectoderm-secreted FGF initiates the expression of Nkx2.5 in cardiac progenitors via a p38 MAPK/CREB pathway. Dev. Biol. 2009, 335, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Stary, M.; Schneider, M.; Sheikh, S.P.; Weitzer, G. Parietal endoderm secreted S100A4 promotes early cardiomyogenesis in embryoid bodies. Biochem. Biophys. Res. Commun. 2006, 343, 555–563. [Google Scholar] [CrossRef]
- Ashok, P.; Parikh, A.; Du, C.; Tzanakakis, E.S. Xenogeneic-Free System for Biomanufacturing of Cardiomyocyte Progeny From Human Pluripotent Stem Cells. Front. Bioeng. Biotechnol. 2020, 8, 571425. [Google Scholar] [CrossRef]
- Ashok, P.; Fan, Y.; Rostami, M.R.; Tzanakakis, E.S. Aggregate and Microcarrier Cultures of Human Pluripotent Stem Cells in Stirred-Suspension Systems. In Bioreactors in Stem Cell Biology; Humana Press: New York, NY, USA, 2015. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Chiou, N.-T.; Ansel, K.M. Improved exosome isolation by sucrose gradient fractionation of ultracentrifuged crude exosome pellets. Protoc. Exch. 2016. [Google Scholar] [CrossRef]
- Fan, Y.; Hsiung, M.; Cheng, C.; Tzanakakis, E.S. Facile engineering of xeno-free microcarriers for the scalable cultivation of human pluripotent stem cells in stirred suspension. Tissue Eng. Part A 2014, 20, 588–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, D.; Kehoe, D.E.; Tzanakakis, E.S. Expression of Reg Family Proteins in Embryonic Stem Cells and its Modulation by Wnt/beta-catenin signaling. Stem Cells Dev. 2010, 19, 1307–1319. [Google Scholar] [CrossRef] [Green Version]
- Potriquet, J.; Laohaviroj, M.; Bethony, J.M.; Mulvenna, J. A modified FASP protocol for high-throughput preparation of protein samples for mass spectrometry. PLoS ONE 2017, 12, e0175967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manteiga, S.; Lee, K. Monoethylhexyl Phthalate Elicits an Inflammatory Response in Adipocytes Characterized by Alterations in Lipid and Cytokine Pathways. Environ. Health Perspect. 2017, 125, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wei, J.F.; Fan, L.Y.; Wang, S.H.; Zhu, L.; Li, T.P.; Lin, G.; Sun, Y.; Sun, Z.J.; Ding, J.; et al. miR-302 regulates pluripotency, teratoma formation and differentiation in stem cells via an AKT1/OCT4-dependent manner. Cell Death Dis. 2016, 7, e2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef] [Green Version]
- Kempf, H.; Olmer, R.; Kropp, C.; Rückert, M.; Jara-Avaca, M.; Robles-Diaz, D.; Franke, A.; Elliott, D.A.; Wojciechowski, D.; Fischer, M.; et al. Controlling Expansion and Cardiomyogenic Differentiation of Human Pluripotent Stem Cells in Scalable Suspension Culture. Stem Cell Rep. 2014, 3, 1132–1146. [Google Scholar] [CrossRef] [Green Version]
- Halloin, C.; Schwanke, K.; Löbel, W.; Franke, A.; Szepes, M.; Biswanath, S.; Wunderlich, S.; Merkert, S.; Weber, N.; Osten, F.; et al. Continuous WNT Control Enables Advanced hPSC Cardiac Processing and Prognostic Surface Marker Identification in Chemically Defined Suspension Culture. Stem Cell Rep. 2019, 13, 366–379. [Google Scholar] [CrossRef] [Green Version]
- Lafontant, P.J.; Behzad, A.R.; Brown, E.; Landry, P.; Hu, N.; Burns, A.R. Cardiac Myocyte Diversity and a Fibroblast Network in the Junctional Region of the Zebrafish Heart Revealed by Transmission and Serial Block-Face Scanning Electron Microscopy. PLoS ONE 2013, 8, e72388. [Google Scholar] [CrossRef] [Green Version]
- Materna, S.C.; Sinha, T.; Barnes, R.M.; Lammerts van Bueren, K.; Black, B.L. Cardiovascular development and survival require Mef2c function in the myocardial but not the endothelial lineage. Dev. Biol. 2019, 445, 170–177. [Google Scholar] [CrossRef]
- Buschow, S.I.; van Balkom, B.W.M.; Aalberts, M.; Heck, A.J.R.; Wauben, M.; Stoorvogel, W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Gauvreau, M.E.; Côté, M.H.; Bourgeois-Daigneault, M.C.; Rivard, L.D.; Xiu, F.; Brunet, A.; Shaw, A.; Steimle, V.; Thibodeau, J. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic 2009, 10, 1518–1527. [Google Scholar] [CrossRef]
- Mayran, N.; Parton, R.G.; Gruenberg, J. Annexin II regulates multivesicular endosome biogenesis in the degradation pathway of animal cells. EMBO J. 2003, 22, 3242–3253. [Google Scholar] [CrossRef] [Green Version]
- Grewal, T.; Hoque, M.; Conway, J.R.W.; Reverter, M.; Wahba, M.; Beevi, S.S.; Timpson, P.; Enrich, C.; Rentero, C. Annexin A6-A multifunctional scaffold in cell motility. Cell Adhes. Migr. 2017, 11, 288–304. [Google Scholar] [CrossRef] [Green Version]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Wermuth, P.J.; Piera-Velazquez, S.; Jimenez, S.A. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin. Exp. Rheumatol. 2017, 35 (Suppl. 106), 21–30. [Google Scholar]
- Wallimann, T.; Hemmer, W. Creatine kinase in non-muscle tissues and cells. Mol. Cell. Biochem. 1994, 133, 193–220. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.K.; Han, Y.M.; Kim, J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008, 40, 1043–1054. [Google Scholar] [CrossRef]
- Rangrez, A.Y.; Kilian, L.; Stiebeling, K.; Dittmann, S.; Yadav, P.; Schulze-Bahr, E.; Frey, N.; Frank, D. Data on the role of cardiac α-actin (ACTC1) gene mutations on SRF-signaling. Data Brief 2020, 28, 105071. [Google Scholar] [CrossRef]
- Bouton, M.-C.; Boulaftali, Y.; Richard, B.; Arocas, V.; Michel, J.-B.; Jandrot-Perrus, M. Emerging role of serpinE2/protease nexin-1 in hemostasis and vascular biology. Blood 2012, 119, 2452–2457. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Collins, K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 2007, 28, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Beetz, N.; O’Keeffe, S.; Tapia, J.C.; Macpherson, L.; Chen, W.V.; Bassel-Duby, R.; Olson, E.N.; Maniatis, T. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc. Natl. Acad. Sci. USA 2015, 112, E3020–E3029. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Zhao, B.; Sun, L.; Bhuripanyo, K.; Wang, Y.; Bi, Y.; Davuluri, R.V.; Duong, D.M.; Nanavati, D.; Yin, J.; et al. Orthogonal ubiquitin transfer identifies ubiquitination substrates under differential control by the two ubiquitin activating enzymes. Nat. Commun. 2017, 8, 14286. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, P.; Macaya, I.; Bazzocco, S.; Mazzolini, R.; Andretta, E.; Dopeso, H.; Mateo-Lozano, S.; Bilić, J.; Cartón-García, F.; Nieto, R.; et al. RHOA inactivation enhances Wnt signalling and promotes colorectal cancer. Nat. Commun. 2014, 5, 5458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlund, S.; Landström, M.; Heldin, C.-H.; Aspenström, P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol. Biol. Cell 2002, 13, 902–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbediwy, A.; Zihni, C.; Terry, S.J.; Clark, P.; Matter, K.; Balda, M.S. Epithelial junction formation requires confinement of Cdc42 activity by a novel SH3BP1 complex. J. Cell Biol. 2012, 198, 677–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galan, A.; Diaz-Gimeno, P.; Poo, M.E.; Valbuena, D.; Sanchez, E.; Ruiz, V.; Dopazo, J.; Montaner, D.; Conesa, A.; Simon, C. Defining the genomic signature of totipotency and pluripotency during early human development. PLoS ONE 2013, 8, e62135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, X.; Miura, T.; Luo, Y.; Bhattacharya, B.; Condie, B.; Chen, J.; Ginis, I.; Lyons, I.; Mejido, J.; Puri, R.K.; et al. Properties of Pluripotent Human Embryonic Stem Cells BG01 and BG02. Stem Cells 2004, 22, 292–312. [Google Scholar] [CrossRef]
- Brandenberger, R.; Khrebtukova, I.; Thies, R.S.; Miura, T.; Jingli, C.; Puri, R.; Vasicek, T.; Lebkowski, J.; Rao, M. MPSS profiling of human embryonic stem cells. BMC Dev. Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Adjaye, J.; Huntriss, J.; Herwig, R.; BenKahla, A.; Brink, T.C.; Wierling, C.; Hultschig, C.; Groth, D.; Yaspo, M.-L.; Picton, H.M.; et al. Primary Differentiation in the Human Blastocyst: Comparative Molecular Portraits of Inner Cell Mass and Trophectoderm Cells. Stem Cells 2005, 23, 1514–1525. [Google Scholar] [CrossRef]
- Li, S.S.; Liu, Y.H.; Tseng, C.N.; Chung, T.L.; Lee, T.Y.; Singh, S. Characterization and gene expression profiling of five new human embryonic stem cell lines derived in Taiwan. Stem Cells Dev. 2006, 15, 532–555. [Google Scholar] [CrossRef]
- Corsini, N.S.; Peer, A.M.; Moeseneder, P.; Roiuk, M.; Burkard, T.R.; Theussl, H.C.; Moll, I.; Knoblich, J.A. Coordinated Control of mRNA and rRNA Processing Controls Embryonic Stem Cell Pluripotency and Differentiation. Cell Stem Cell 2018, 22, 543–558.e512. [Google Scholar] [CrossRef] [Green Version]
- Chia, N.-Y.; Chan, Y.-S.; Feng, B.; Lu, X.; Orlov, Y.L.; Moreau, D.; Kumar, P.; Yang, L.; Jiang, J.; Lau, M.-S.; et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 2010, 468, 316–320. [Google Scholar] [CrossRef]
- Li, X.; Sun, L.; Jin, Y. Identification of karyopherin-alpha 2 as an Oct4 associated protein. J. Genet. Genom. 2008, 35, 723–728. [Google Scholar] [CrossRef]
- Johansson, H.; Simonsson, S. Core transcription factors, Oct4, Sox2 and Nanog, individually form complexes with nucleophosmin (Npm1) to control embryonic stem (ES) cell fate determination. Aging 2010, 2, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Tian, Y.; Liang, Y.; Li, Q. Circ_0008035 contributes to cell proliferation and inhibits apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1 axis. Cancer Cell Int. 2020, 20, 84. [Google Scholar] [CrossRef] [Green Version]
- Ratnadiwakara, M.; Archer, S.K.; Dent, C.I.; Ruiz De Los Mozos, I.; Beilharz, T.H.; Knaupp, A.S.; Nefzger, C.M.; Polo, J.M.; Anko, M.-L. SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. eLife 2018, 7, e37419. [Google Scholar] [CrossRef]
- Rowe, H.M.; Kapopoulou, A.; Corsinotti, A.; Fasching, L.; Macfarlan, T.S.; Tarabay, Y.; Viville, S.; Jakobsson, J.; Pfaff, S.L.; Trono, D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013, 23, 452–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleksiewicz, U.; Gładych, M.; Raman, A.T.; Heyn, H.; Mereu, E.; Chlebanowska, P.; Andrzejewska, A.; Sozańska, B.; Samant, N.; Fąk, K.; et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem Cell Rep. 2017, 9, 2065–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. H1.0 Linker Histone as an Epigenetic Regulator of Cell Proliferation and Differentiation. Genes 2018, 9, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihara, E.; Hirai, H.; Yamamoto, H.; Tamura-Kawakami, K.; Matano, M.; Kikuchi, A.; Sato, T.; Takagi, J. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. eLife 2016, 5, e11621. [Google Scholar] [CrossRef]
- Ikeda, S.; Kishida, M.; Matsuura, Y.; Usui, H.; Kikuchi, A. GSK-3β-dependent phosphorylation of adenomatous polyposis coli gene product can be modulated by β-catenin and protein phosphatase 2A complexed with Axin. Oncogene 2000, 19, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Hsu, W.; Zeng, L.; Costantini, F. Identification of a Domain of Axin That Binds to the Serine/Threonine Protein Phosphatase 2A and a Self-binding Domain. J. Biol. Chem. 1999, 274, 3439–3445. [Google Scholar] [CrossRef] [Green Version]
- Rozbicki, E.; Chuai, M.; Karjalainen, A.I.; Song, F.; Sang, H.M.; Martin, R.; Knölker, H.-J.; MacDonald, M.P.; Weijer, C.J. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat. Cell Biol. 2015, 17, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.S.K.; Hagen, H.R.; Swanson, S.A.; Stewart, R.; Boll, K.A.; Aho, J.; Thomson, J.A.; Kyba, M. Development of Bipotent Cardiac/Skeletal Myogenic Progenitors from MESP1+ Mesoderm. Stem Cell Rep. 2016, 6, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Dichmann, D.S.; Walentek, P.; Harland, R.M. The Alternative Splicing Regulator Tra2b Is Required for Somitogenesis and Regulates Splicing of an Inhibitory Wnt11b Isoform. Cell Rep. 2015, 10, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Sahara, M.; Santoro, F.; Sohlmér, J.; Zhou, C.; Witman, N.; Leung, C.Y.; Mononen, M.; Bylund, K.; Gruber, P.; Chien, K.R. Population and Single-Cell Analysis of Human Cardiogenesis Reveals Unique LGR5 Ventricular Progenitors in Embryonic Outflow Tract. Dev. Cell 2019, 48, 475–490.e477. [Google Scholar] [CrossRef] [Green Version]
- Gregoire, S.; Li, G.; Sturzu, A.C.; Schwartz, R.J.; Wu, S.M. YY1 Expression Is Sufficient for the Maintenance of Cardiac Progenitor Cell State. Stem Cells 2017, 35, 1913–1923. [Google Scholar] [CrossRef] [Green Version]
- Timberlake, A.T.; Furey, C.G.; Choi, J.; Nelson-Williams, C.; Yale Center for Genome, A.; Loring, E.; Galm, A.; Kahle, K.T.; Steinbacher, D.M.; Larysz, D.; et al. De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proc. Natl. Acad. Sci. USA 2017, 114, E7341–E7347. [Google Scholar] [CrossRef] [Green Version]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef] [Green Version]
- Wishart, T.M.; Mutsaers, C.A.; Riessland, M.; Reimer, M.M.; Hunter, G.; Hannam, M.L.; Eaton, S.L.; Fuller, H.R.; Roche, S.L.; Somers, E.; et al. Dysregulation of ubiquitin homeostasis and β-catenin signaling promote spinal muscular atrophy. J. Clin. Investig. 2014, 124, 1821–1834. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.; Christiansen, J.; Lykke-Andersen, J.; Johnsen, A.H.; Wewer, U.M.; Nielsen, F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell Biol. 1999, 19, 1262–1270. [Google Scholar] [CrossRef] [Green Version]
- Strate, I.; Tessadori, F.; Bakkers, J. Glypican4 promotes cardiac specification and differentiation by attenuating canonical Wnt and Bmp signaling. Development 2015, 142, 1767–1776. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Xu, X.; Duan, X.; Guo, J.; Wang, Y.; Ren, F.; He, D.; Chang, Z. Hsp70 and Hsp90 oppositely regulate TGF-β signaling through CHIP/Stub1. Biochem. Biophys. Res. Commun. 2014, 446, 387–392. [Google Scholar] [CrossRef]
- Kodama, M.; Furutani, K.; Kimura, R.; Ando, T.; Sakamoto, K.; Nagamori, S.; Ashihara, T.; Kurachi, Y.; Sekino, Y.; Furukawa, T.; et al. Systematic expression analysis of genes related to generation of action potentials in human iPS cell-derived cardiomyocytes. J. Pharmacol. Sci. 2019, 140, 325–330. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome Uptake through Clathrin-mediated Endocytosis and Macropinocytosis and Mediating miR-21 Delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [Green Version]
- Kagan, A.; Melman, Y.F.; Krumerman, A.; McDonald, T.V. 14-3-3 amplifies and prolongs adrenergic stimulation of HERG K+ channel activity. EMBO J. 2002, 21, 1889–1898. [Google Scholar] [CrossRef] [Green Version]
- Linde, C.I.; Di Leva, F.; Domi, T.; Tosatto, S.C.E.; Brini, M.; Carafoli, E. Inhibitory interaction of the 14-3-3 proteins with ubiquitous (PMCA1) and tissue-specific (PMCA3) isoforms of the plasma membrane Ca2+ pump. Cell Calcium 2008, 43, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Janssens, B.; Goossens, S.; Staes, K.; Gilbert, B.; van Hengel, J.; Colpaert, C.; Bruyneel, E.; Mareel, M.; van Roy, F. αT-Catenin: A novel tissue-specific β-catenin-binding protein mediating strong cell-cell adhesion. J. Cell Sci. 2001, 114, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, E.; Vite, A.; Yi, R.; Gomez, L.; Goossens, S.; van Roy, F.; Radice, G.L. Alpha-catenins control cardiomyocyte proliferation by regulating Yap activity. Circ. Res. 2015, 116, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Shi, Y.; Xia, M.; Liu, Z.; Zhang, R.; Luo, H.; Zhang, T.; Yang, Z.; Yuan, B. WDR1-regulated actin dynamics is required for outflow tract and right ventricle development. Dev. Biol. 2018, 438, 124–137. [Google Scholar] [CrossRef]
- Yuan, B.; Wan, P.; Chu, D.; Nie, J.; Cao, Y.; Luo, W.; Lu, S.; Chen, J.; Yang, Z. A cardiomyocyte-specific Wdr1 knockout demonstrates essential functional roles for actin disassembly during myocardial growth and maintenance in mice. Am. J. Pathol. 2014, 184, 1967–1980. [Google Scholar] [CrossRef]
- Curran, J.; Makara, M.A.; Mohler, P.J. Endosome-based protein trafficking and Ca2+ homeostasis in the heart. Front. Physiol. 2015, 6, 34. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.D., Jr.; Swanson, K.E.; Alvarez, M.G.; Krishnan, S.; Earley, J.U.; Band, H.; Pytel, P.; McNally, E.M.; Demonbreun, A.R. EHD1 mediates vesicle trafficking required for normal muscle growth and transverse tubule development. Dev. Biol. 2014, 387, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Martins-Marques, T.; Catarino, S.; Gonçalves, A.; Miranda-Silva, D.; Gonçalves, L.; Antunes, P.; Coutinho, G.; Moreira, A.L.; Pires, I.F.; Girão, H. EHD1 Modulates Cx43 Gap Junction Remodeling Associated With Cardiac Diseases. Circ. Res. 2020, 126, e97–e113. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukuda, A.; Hisatake, K. Mechanisms of the Metabolic Shift during Somatic Cell Reprogramming. Int. J. Mol. Sci. 2019, 20, 2254. [Google Scholar] [CrossRef] [Green Version]
- Morita, Y.; Tohyama, S. Metabolic Regulation of Cardiac Differentiation and Maturation in Pluripotent Stem Cells: A Lesson from Heart Development. JMA J. 2020, 3, 193–200. [Google Scholar] [CrossRef]
- Arthur, S.A.; Blaydes, J.P.; Houghton, F.D. Glycolysis Regulates Human Embryonic Stem Cell Self-Renewal under Hypoxia through HIF-2α and the Glycolytic Sensors CTBPs. Stem Cell Rep. 2019, 12, 728–742. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Teng, X.; Liu, L.; Mattaini, K.R.; Looper, R.E.; Vander Heiden, M.G.; Rabinowitz, J.D. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate. ACS Chem. Biol. 2015, 10, 510–516. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, T.; Wang, L.; Cai, Y.; Zhong, X.; He, X.; Hu, L.; Tian, S.; Wu, M.; Hui, L.; et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017, 36, 1330–1347. [Google Scholar] [CrossRef] [Green Version]
- Dahan, P.; Lu, V.; Nguyen, R.M.T.; Kennedy, S.A.L.; Teitell, M.A. Metabolism in pluripotency: Both driver and passenger? J. Biol. Chem. 2019, 294, 5420–5429. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, E.F.; Chen, Z.; Stoukides, D.M.; Nair, G.G.; Hebrok, M.; Tzanakakis, E.S. Non-xenogeneic expansion and definitive endoderm differentiation of human pluripotent stem cells in an automated bioreactor. Biotechnol. Bioeng. 2021, 118, 979–991. [Google Scholar] [CrossRef]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashok, P.; Tzanakakis, E.S. Proteomic Analysis of Exosomes during Cardiogenic Differentiation of Human Pluripotent Stem Cells. Cells 2021, 10, 2622. https://doi.org/10.3390/cells10102622
Ashok P, Tzanakakis ES. Proteomic Analysis of Exosomes during Cardiogenic Differentiation of Human Pluripotent Stem Cells. Cells. 2021; 10(10):2622. https://doi.org/10.3390/cells10102622
Chicago/Turabian StyleAshok, Preeti, and Emmanuel S. Tzanakakis. 2021. "Proteomic Analysis of Exosomes during Cardiogenic Differentiation of Human Pluripotent Stem Cells" Cells 10, no. 10: 2622. https://doi.org/10.3390/cells10102622
APA StyleAshok, P., & Tzanakakis, E. S. (2021). Proteomic Analysis of Exosomes during Cardiogenic Differentiation of Human Pluripotent Stem Cells. Cells, 10(10), 2622. https://doi.org/10.3390/cells10102622