miR-21-3p/IL-22 Axes Are Major Drivers of Psoriasis Pathogenesis by Modulating Keratinocytes Proliferation-Survival Balance and Inflammatory Response
Abstract
:1. Introduction
2. Materials and Methods
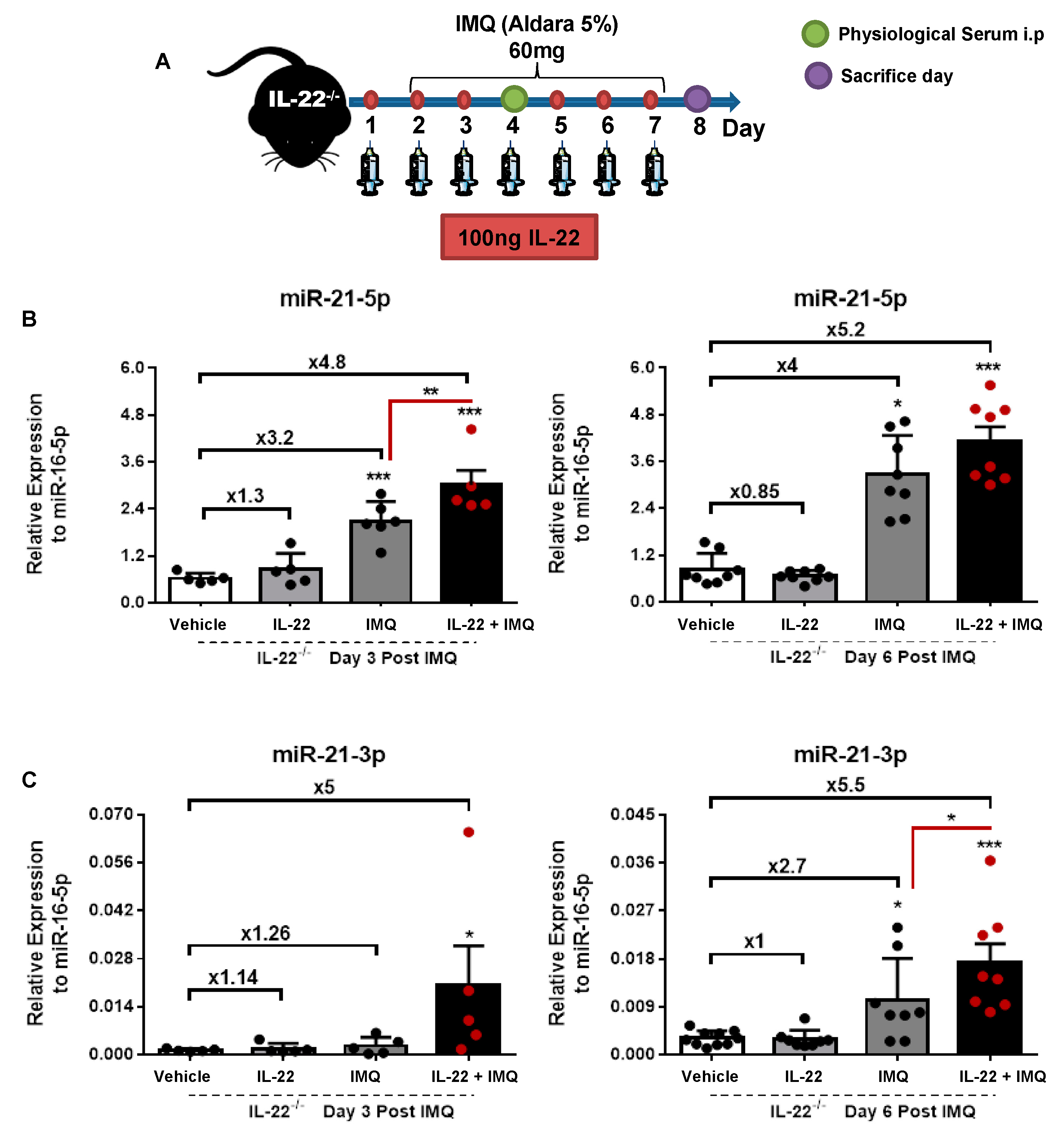
2.1. Mice and In Vivo Treatments
2.2. Scoring of Skin Inflammation Severity
2.3. Patients and Biopsies
2.4. Cell Handling
2.5. Cell Stimulation
2.6. Cell Transfection
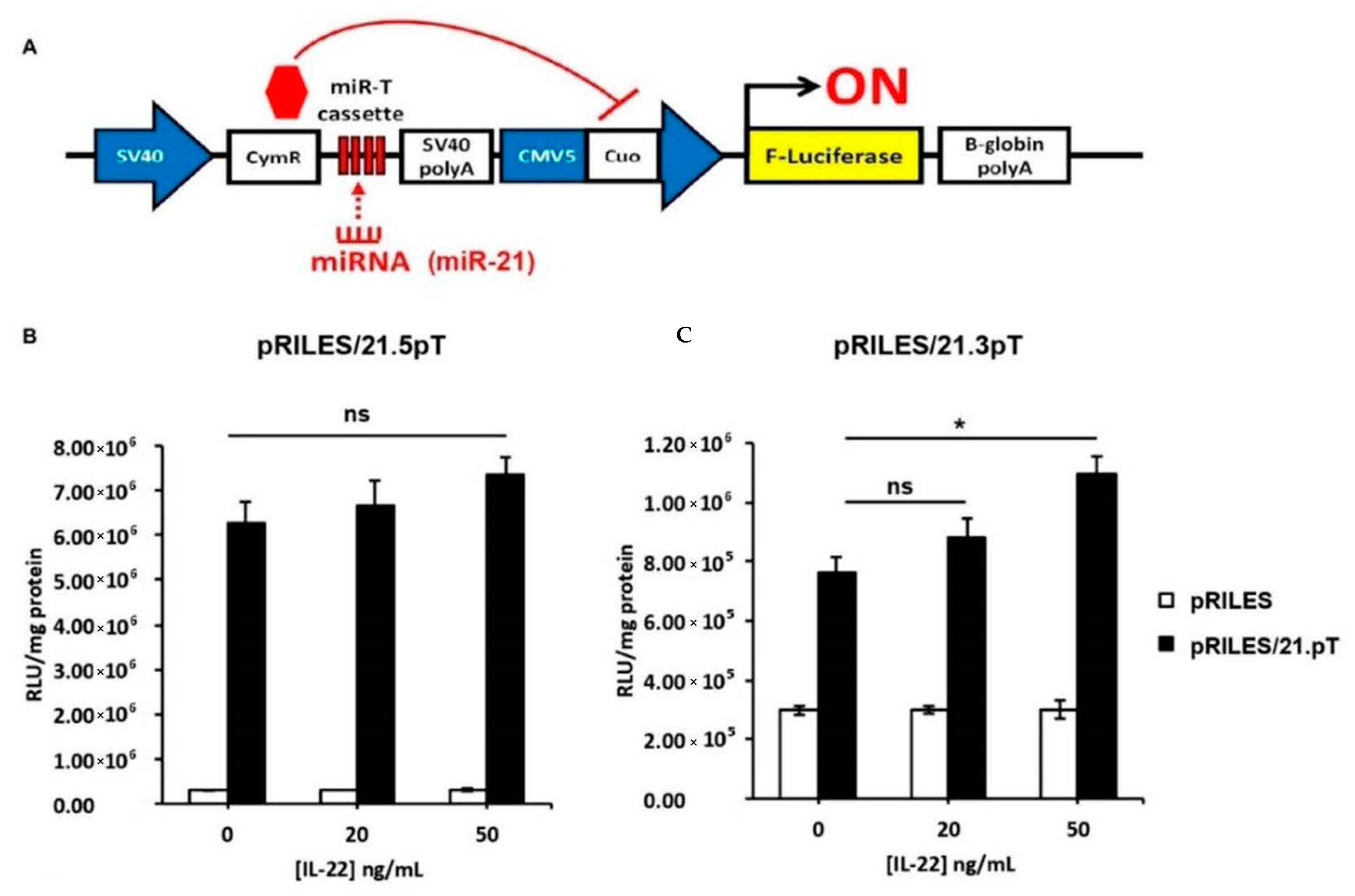
2.7. Luciferase Reporter Assays
2.8. RNA Extraction, Reverse Transcription, and Real-Time PCR
2.9. Enzyme-Linked Immunosorbent Assay
2.10. Phospho-STAT3 (Y705) Assay
2.11. Immunofluorescence Staining
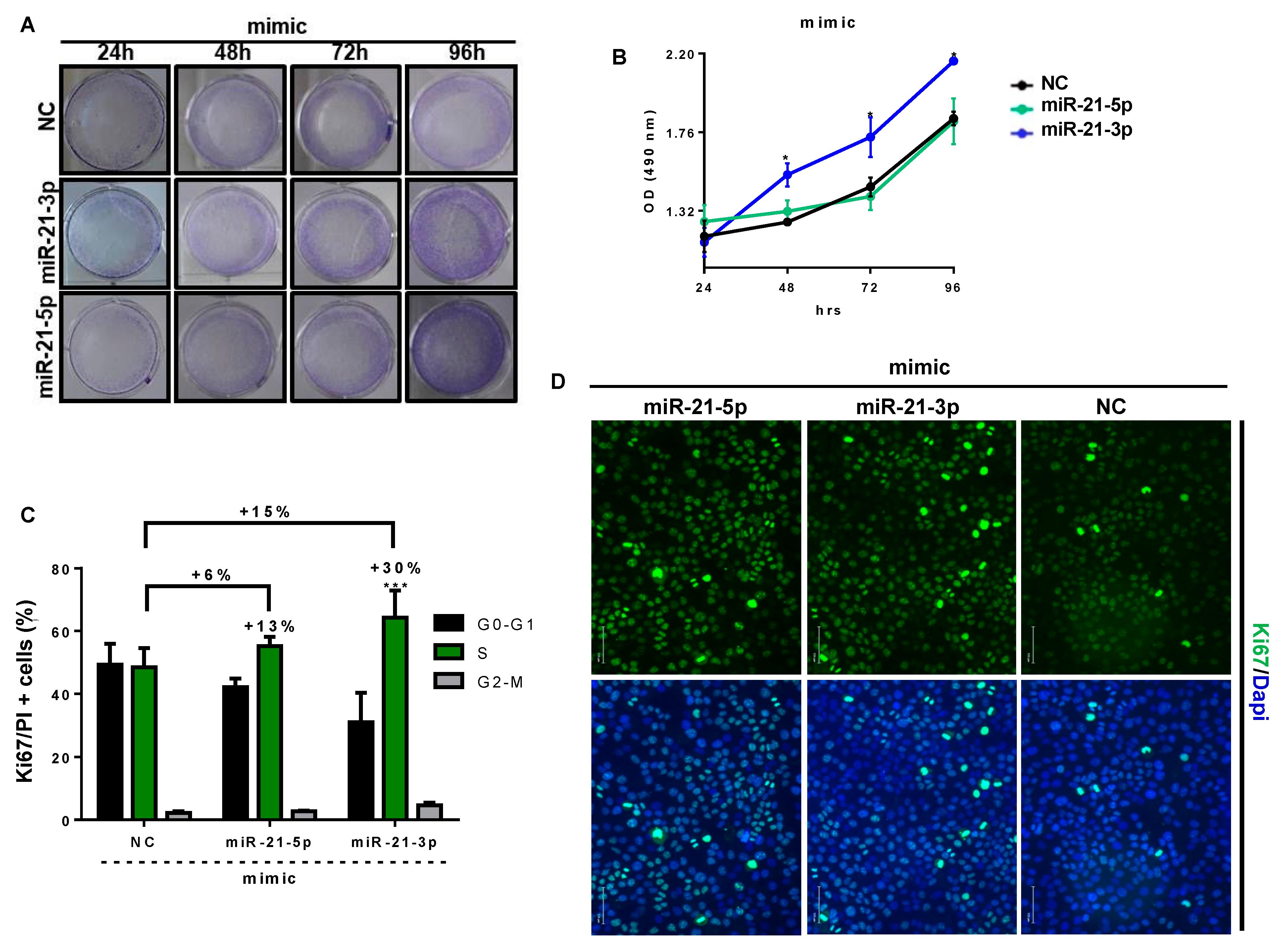
2.12. Crystal Violet Staining
2.13. Cell Metabolic Activity
2.14. Flow Cytometric Analysis for Ki67 and PI Staining
2.15. Monitoring miRNA Activity with RILES
2.16. RNA-Seq from HaCaT Cells
2.17. Statistical Analysis
3. Results
3.1. miR-21-5p/3p Expression in IMQ-Induced Psoriasis in WT versus IL-22−/− Mice
3.2. Validation of miR-21-3p Dependency on IL-22 In Vitro, in Psoriatic Human Biopsies, and In Vivo
3.3. IL-22 Induces miR-21-3p via STAT3 and NF-κB Pathways
3.4. IL-22-Induced miR-21-3p in KCs Is Functional
3.5. Global Changes in the Transcriptome of HaCaT Overexpressing miR-21-3p
3.6. miR-21-3p Promotes Keratinocyte Hyperproliferation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Springate, D.A.; Parisi, R.; Kontopantelis, E.; Reeves, D.; Griffiths, C.E.M.; Ashcroft, D.M. Incidence, prevalence and mortality of patients with psoriasis: A U.K. population-based cohort study. Br. J. Dermatol. 2017, 176, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Wolk, K.; Haugen, H.S.; Xu, W.; Witte, E.; Waggie, K.; Anderson, M.; Vom Baur, E.; Witte, K.; Warszawska, K.; Philipp, S.; et al. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-γ are not. J. Mol. Med. 2009, 87, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, S.; Eyerich, K.; Pennino, D.; Carbone, T.; Nasorri, F.; Pallotta, S.; Cianfarani, F.; Odorisio, T.; Traidl-Hoffmann, C.; Behrendt, H.; et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Investig. 2009, 119, 3573–3585. [Google Scholar] [CrossRef] [Green Version]
- Wolk, K.; Witte, E.; Wallace, E.; Döcke, W.D.; Kunz, S.; Asadullah, K.; Volk, H.D.; Sterry, W.; Sabat, R. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: A potential role in psoriasis. Eur. J. Immunol. 2006, 36, 1309–1323. [Google Scholar] [CrossRef]
- Boniface, K.; Bernard, F.-X.; Garcia, M.; Gurney, A.L.; Lecron, J.-C.; Morel, F. IL-22 Inhibits Epidermal Differentiation and Induces Proinflammatory Gene Expression and Migration of Human Keratinocytes. J. Immunol. 2005, 174, 3695–3702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [Green Version]
- Van Belle, A.B.; de Heusch, M.; Lemaire, M.M.; Hendrickx, E.; Warnier, G.; Dunussi-Joannopoulos, K.; Fouser, L.A.; Renauld, J.-C.; Dumoutier, L. IL-22 Is Required for Imiquimod-Induced Psoriasiform Skin Inflammation in Mice. J. Immunol. 2012, 188, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Pasquali, L.; Srivastava, A.; Meisgen, F.; Das Mahapatra, K.; Xia, P.; Xu Landén, N.; Pivarcsi, A.; Sonkoly, E. The keratinocyte transcriptome in psoriasis: Pathways related to immune responses, cell cycle and keratinization. Acta Derm. Venereol. 2019, 99, 196–205. [Google Scholar] [CrossRef] [Green Version]
- Sonkoly, E.; Wei, T.; Janson, P.C.J.; Sääf, A.; Lundeberg, L.; Tengvall-Linder, M.; Norstedt, G.; Alenius, H.; Homey, B.; Scheynius, A.; et al. MicroRNAs: Novel Regulators Involved in the Pathogenesis of Psoriasis? PLoS ONE 2007, 2, e610. [Google Scholar] [CrossRef] [Green Version]
- Zibert, J.R.; Løvendorf, M.B.; Litman, T.; Olsen, J.; Kaczkowski, B.; Skov, L. MicroRNAs and potential target interactions in psoriasis. J. Dermatol. Sci. 2010, 58, 177–185. [Google Scholar] [CrossRef]
- Lerman, G.; Avivi, C.; Mardoukh, C.; Barzilai, A.; Tessone, A.; Gradus, B.; Pavlotsky, F.; Barshack, I.; Polak-Charcon, S.; Orenstein, A.; et al. MiRNA expression in psoriatic skin: Reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS ONE 2011, 6, e20916. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Darido, C.; Georgy, S.R.; Wilanowski, T.; Dworkin, S.; Auden, A.; Zhao, Q.; Rank, G.; Srivastava, S.; Finlay, M.J.; Papenfuss, A.T.; et al. Targeting of the Tumor Suppressor GRHL3 by a miR-21-Dependent Proto-Oncogenic Network Results in PTEN Loss and Tumorigenesis. Cancer Cell 2011, 20, 635–648. [Google Scholar] [CrossRef] [Green Version]
- Ren, W.; Qiang, C.; Gao, L.; Li, S.-M.; Zhang, L.-M.; Wang, X.-L.; Dong, J.-W.; Chen, C.; Liu, C.-Y.; Zhi, K.-Q. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers 2014, 19, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, Y.; Lv, P.; Li, L. The role of miR-21 in proliferation and invasion capacity of human tongue squamous cell carcinoma in vitro. Int. J. Clin. Exp. Pathol. 2015, 8, 4555–4563. [Google Scholar] [PubMed]
- Wang, T.; Feng, Y.; Sun, H.; Zhang, L.; Hao, L.; Shi, C.; Wang, J.; Li, R.; Ran, X.; Su, Y.; et al. MiR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am. J. Pathol. 2012, 181, 1911–1920. [Google Scholar] [CrossRef]
- Yan, L.; Cao, R.; Liu, Y.B.; Wang, L.Z.; Pan, B.; Lv, X.Y.; Jiao, H.; Zhuang, Q.; Sun, X.J.; Xiao, R. MiR-21-5p Links Epithelial-Mesenchymal Transition Phenotype with Stem-Like Cell Signatures via AKT Signaling in Keloid Keratinocytes. Sci. Rep. 2016, 6, 28281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Zhao, Y.; Fan, R.; Chen, T.; Dong, C. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int. J. Mol. Sci. 2016, 17, 959. [Google Scholar] [CrossRef] [Green Version]
- Meisgen, F.; Xu, N.; Wei, T.; Janson, P.C.; Obad, S.; Broom, O.; Nagy, N.; Kauppinen, S.; Kemény, L.; Ståhle, M.; et al. miR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp. Dermatol. 2012, 21, 312–314. [Google Scholar] [CrossRef]
- Guinea-Viniegra, J.; Jiménez, M.; Schonthaler, H.B.; Navarro, R.; Delgado, Y.; Concha-Garzón, M.J.; Tschachler, E.; Obad, S.; Daudén, E.; Wagner, E.F. Targeting miR-21 to treat psoriasis. Sci. Transl. Med. 2014, 6, 225re1. [Google Scholar] [CrossRef]
- Degueurce, G.; Errico, I.D.; Pich, C.; Ibberson, M.; Schütz, F.; Montagner, A.; Sgandurra, M.; Mury, L.; Jafari, P.; Boda, A.; et al. Identification of a novel PPARb/d/miR-21-3p axis in UV-induced skin inflammation Gwendoline. EMBO Mol. Med. 2016, 8, 919–936. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Park, C.; Young, D.; Sanders, K.M.; Yan, W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007, 35, 5944–5953. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, C.; Ardourel, M.-Y.; Decoville, M.; Breuzard, G.; Midoux, P.; Hartmann, B.; Pichon, C. An optimized extended DNA kappa B site that enhances plasmid DNA nuclear import and gene expression. J. Gene Med. 2009, 11, 401–411. [Google Scholar] [CrossRef]
- Ezzine, S.; Vassaux, G.; Pitard, B.; Barteau, B.; Malinge, J.M.; Midoux, P.; Pichon, C.; Baril, P. RILES, a novel method for temporal analysis of the in vivo regulation of miRNA expression. Nucleic Acids Res. 2013, 41, e192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nestle, F.O.; Di Meglio, P.; Qin, J.-Z.; Nickoloff, B.J. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 2009, 9, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, F.; Mijouin, L.; Pichon, C. Skin Immune Landscape: Inside and Outside the Organism. Mediat. Inflamm. 2017, 2017, 5095293. [Google Scholar] [CrossRef]
- Löffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermüller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6-dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.; Shi, Y.; Tan, G.; Yang, C.H.; Fan, M.; Pfeffer, L.M.; Wu, Z.H. DNA damage induces NF-kB-dependent MicroRNA-21 up-regulation and promotes breast cancer cell invasion. J. Biol. Chem. 2012, 287, 21783–21795. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Fang, S.; Di, Y.; Ying, W.; Tan, Y.; Gu, W. Modulation of NF-kB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PLoS ONE 2015, 10, e0121547. [Google Scholar] [CrossRef]
- Nagalakshmi, M.L.; Rascle, A.; Zurawski, S.; Menon, S.; De Waal Malefyt, R. Interleukin-22 activates STAT3 and induces IL-10 by colon epithelial cells. Int. Immunopharmacol. 2004, 4, 679–691. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Karin, M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010, 21, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizzul, P.F.; Aphale, A.; Malaviya, R.; Sun, Y.; Masud, S.; Dombrovskiy, V.; Gottlieb, A.B. Differential expression of phosphorylated NF-κB/RelA in normal and psoriatic epidermis and downregulation of NF-κB in response to treatment with etanercept. J. Investig. Dermatol. 2005, 124, 1275–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inui, S.; Lee, Y.F.; Haake, A.R.; Goldsmith, L.A.; Chang, C. Induction of TR4 orphan receptor by retinoic acid in human HaCaT keratinocytes. J. Investig. Dermatol. 1999, 112, 426–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inui, S.; Lee, Y.F.; Chang, E.; Shyr, C.R.; Chang, C. Differential and bi-directional regulation between TR2/TR4 orphan nuclear receptors and a specific ligand mediated-peroxisome proliferator-activated receptor α in human HaCaT keratinocytes. J. Dermatol. Sci. 2003, 31, 65–71. [Google Scholar] [CrossRef]
- Ishiuchi, T.; Ohishi, H.; Sato, T.; Kamimura, S.; Yorino, M.; Abe, S.; Suzuki, A.; Wakayama, T.; Suyama, M.; Sasaki, H. Zfp281 Shapes the Transcriptome of Trophoblast Stem Cells and Is Essential for Placental Development. Cell Rep. 2019, 27, 1742–1754.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushansky, K. Hunting for hematopoietic transcriptional networks. Proc. Natl. Acad. Sci. USA 2018, 115, 9818–9820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pooter, R.F.; Kee, B.L. E proteins and the regulation of early lymphocyte development. Immunol. Rev. 2010, 238, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ali, M.; Yang, J.; Chan, K.W.; Ben-Mustapha, I.; Mekki, N.; Benabdesselem, C.; Mellouli, F.; Bejaoui, M.; Yang, W.; Aissaoui, L.; et al. Homozygous transcription factor 3 gene (TCF3) mutation is associated with severe hypogammaglobulinemia and B-cell acute lymphoblastic leukemia. J. Allergy Clin. Immunol. 2017, 140, 1191–1194.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimova, D.K.; Dyson, N.J. The E2F transcriptional network: Old acquaintances with new faces. Oncogene 2005, 24, 2810–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wang, C.; Wu, J.; Ha, X.; Deng, Y.; Zhang, X.; Wang, J.; Chen, K.; Feng, J.; Zhu, J.; et al. The effect and mechanism of KLF7 in the TLR4/NF-κB/IL-6 inflammatory signal pathway of adipocytes. Mediat. Inflamm. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtsuji, M.; Katsuoka, F.; Kobayashi, A.; Aburatani, H.; Hayes, J.D.; Yamamoto, M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 2008, 283, 33554–33562. [Google Scholar] [CrossRef] [Green Version]
- Kiyama, T.; Chen, C.K.; Wang, S.W.; Pan, P.; Ju, Z.; Wang, J.; Takada, S.; Klein, W.H.; Mao, C.A. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis 06 Biological Sciences 0601 Biochemistry and Cell Biology 11 Medical and Health Sciences 1109 Neurosciences. Mol. Neurodegener. 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Stacey, D.W. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 2003, 15, 158–163. [Google Scholar] [CrossRef]
- Li, X.; Li, J.; Lu, F.; Cao, Y.; Xing, J.; Li, J.; Hou, R.; Yin, G. Role of SPRED1 in keratinocyte proliferation in psoriasis. J. Dermatol. 2020, 47, 735–742. [Google Scholar] [CrossRef]
- Mah, S.M.; Buske, C.; Humphries, R.K.; Kuchenbauer, F. miRNA*: A Passenger Stranded in RNA-Induced Silencing Complex? Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 141–148. [Google Scholar] [CrossRef]
- Báez-Vega, P.M.; Echevarría Vargas, I.M.; Valiyeva, F.; Encarnación-Rosado, J.; Roman, A.; Flores, J.; Marcos-Martínez, M.J.; Vivas-Mejía, P.E. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget 2016, 7, 36321–36337. [Google Scholar] [CrossRef]
- Guo, L.; Lu, Z. The fate of miRNA* strand through evolutionary analysis: Implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE 2010, 5, e11387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijer, H.A.; Smith, E.M.; Bushell, M. Regulation of miRNA strand selection: Follow the leader? Biochem. Soc. Trans. 2014, 42, 1135–1140. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Romay, M.C.; Che, N.; Becker, S.N.; Pouldar, D.; Hagopian, R.; Xiao, X.; Lusis, A.J.; Berliner, J.A.; Civelek, M. Regulation of NF-κB signaling by oxidized glycerophospholipid and IL-1β induced miRs-21-3p and -27a-5p in human aortic endothelial cells. J. Lipid Res. 2015, 56, 38–50. [Google Scholar] [CrossRef] [Green Version]
- Lerman, G.; Sharon, M.; Leibowitz-Amit, R.; Sidi, Y.; Avni, D. The crosstalk between IL-22 signaling and miR-197 in human keratinocytes. PLoS ONE 2014, 9, e107467. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, F.; Pichon, C. Evidence on the direct correlation between miR-31 and IL-22 axis in IMQ induced psoriasis. Exp. Dermatol. 2019, 28, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Xu, Z.; Lou, F.; Zhang, L.; Ke, F.; Bai, J.; Liu, Z.; Liu, J.; Wang, H.; Zhu, H.; et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 2015, 6, 7652. [Google Scholar] [CrossRef] [Green Version]
- Sano, S.; Chan, K.S.; Carbajal, S.; Clifford, J.; Peavey, M.; Kiguchi, K.; Itami, S.; Nickoloff, B.J.; DiGiovanni, J. Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat. Med. 2005, 11, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.A.; Jetten, A.M. Control of growth regulatory and differentiation-specific genes in human epidermal keratinocytes by interferon γ: Antagonism by retinoic acid and transforming growth factor β1. J. Biol. Chem. 1994, 269, 2016–2022. [Google Scholar] [CrossRef]
- Shoeib, M.; El-Shafey, E.; Sonbol, A.; Radwan Lashin, S. Assessment of serum interferon-γ in psoriasis. Menoufia Med. J. 2015, 28, 488. [Google Scholar] [CrossRef]
- Sun, S.C. The noncanonical NF-κB pathway. Immunol. Rev. 2012, 246, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Zhou, X.F.; Yu, J.; Cheng, X.; Sun, S.C. Regulation of Th17 cell differentiation and EAE induction by MAP3K NIK. Blood 2009, 113, 6603–6610. [Google Scholar] [CrossRef] [PubMed]
- Nast, A.; Spuls, P.; Nijsten, T. Treatment of Psoriasis. Evid. Based Dermatol. Third Ed. 2014, 201, 175–199. [Google Scholar] [CrossRef]
- Hou, Y.; Zhu, L.; Tian, H.; Sun, H.X.; Wang, R.; Zhang, L.; Zhao, Y. IL-23-induced macrophage polarization and its pathological roles in mice with imiquimod-induced psoriasis. Protein Cell 2018, 9, 1027–1038. [Google Scholar] [CrossRef]
- Bai, F.; Li, G.G.; Liu, Q.; Niu, X.; Li, R.; Ma, H. Short-Term Efficacy and Safety of IL-17, IL-12/23, and IL-23 Inhibitors Brodalumab, Secukinumab, Ixekizumab, Ustekinumab, Guselkumab, Tildrakizumab, and Risankizumab for the Treatment of Moderate to Severe Plaque Psoriasis: A Systematic Review and Network Meta-Analysis of Randomized Controlled Trials. J. Immunol. Res. 2019, 2019, 2546161. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Oak, A.S.W.; Elewski, B.E. Use of IL-23 Inhibitors for the Treatment of Plaque Psoriasis and Psoriatic Arthritis: A Comprehensive Review. Am. J. Clin. Dermatol. 2021, 22, 173–192. [Google Scholar] [CrossRef]
- Reich, K.; Papp, K.A.; Blauvelt, A.; Langley, R.G.; Armstrong, A.; Warren, R.B.; Gordon, K.B.; Merola, J.F.; Okubo, Y.; Madden, C.; et al. Bimekizumab versus ustekinumab for the treatment of moderate to severe plaque psoriasis (BE VIVID): Efficacy and safety from a 52-week, multicentre, double-blind, active comparator and placebo controlled phase 3 trial. Lancet 2021, 397, 487–498. [Google Scholar] [CrossRef]
- Gordon, K.B.; Foley, P.; Krueger, J.G.; Pinter, A.; Reich, K.; Vender, R.; Vanvoorden, V.; Madden, C.; White, K.; Cioffi, C.; et al. Bimekizumab efficacy and safety in moderate to severe plaque psoriasis (BE READY): A multicentre, double-blind, placebo-controlled, randomised withdrawal phase 3 trial. Lancet 2021, 397, 475–486. [Google Scholar] [CrossRef]
- Warren, R.B.; Gottlieb, A.B.; Merola, J.F.; Garcia, L.; Cioffi, C.; Peterson, L.; Pelligra, C.; Ciaravino, V. Psychometric Validation of the Psoriasis Symptoms and Impacts Measure (P-SIM), a Novel Patient-Reported Outcome Instrument for Patients with Plaque Psoriasis, Using Data from the BE VIVID and BE READY Phase 3 Trials. Dermatol. Ther. 2021. [Google Scholar] [CrossRef]
- Reich, K.; Warren, R.B.; Lebwohl, M.; Gooderham, M.; Strober, B.; Langley, R.G.; Paul, C.; De Cuyper, D.; Vanvoorden, V.; Madden, C.; et al. Bimekizumab versus Secukinumab in Plaque Psoriasis. N. Engl. J. Med. 2021, 385, 142–152. [Google Scholar] [CrossRef]





| Primers | Assay ID | Catalog Number |
|---|---|---|
| miR-21-5p | Hsa-miR-21-5p | 477,975 |
| miR-21-3p | Hsa-miR-21-3p | 477,973 |
| miR-21-3p | Rno-miR-21-3p | Rno480,993 |
| miR-16-5p | Hsa-miR-16-5p | 477,860 |
| miR-16-5p | Rno-miR-16-5p | Rno481,312 |
| Pasquali et al. PP-H | This Work | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Term | Overlap | p-Value | Adjusted p-Value | Z-Score | Enrichment | Count | Pop Hits | p-Value | Benjiamini | FDR % | Enrichment |
| DNA-dependent DNA replication (GO:0006261) | 40/118 | 1.56 × 10−15 | 1.21 × 10−11 | −3.19 | GO-BP-UP | 5 | 18 | 0.014144 | 0.28673763 | 22.8965119 | DAVID-UP |
| DNA replication (GO:0006260) | 37/125 | 2.01 × 10−12 | 3.9 × 10−9 | −3.16 | GO-BP-UP | 41 | 155 | 3.95 × 10−17 | 2.68 × 10−14 | 7.21 × 10−14 | DAVID-UP |
| Double-strand break repair (GO:0006302) | 42/164 | 1.32 × 10−11 | 2.04 × 10−8 | −3.26 | GO-BP-UP | 14 | 66 | 4.39 × 10−5 | 0.00275283 | 0.08007114 | DAVID-UP |
| G2/M transition of mitotic cell cycle (GO:0000086) | 35/130 | 1.49 × 10−10 | 1.44 × 10−7 | −3.13 | GO-BP-UP | 34 | 137 | 2.22 × 10−13 | 8.35 × 10−11 | 4.04 × 10−10 | DAVID-UP |
| Viral DNA repair (GO:0046787) | 38/160 | 1.28 × 10−9 | 8.27 × 10−7 | −3.31 | GO-BP-UP | ||||||
| DNA repair (GO:0006281) | 54/279 | 1.7 × 10−9 | 1.01 × 10−6 | −3.99 | GO-BP-UP | 39 | 235 | 1.23 × 10−7 | 3.2 × 10−7 | 2.24 × 10−6 | DAVID-UP |
| Keratinocyte development (GO: 0003334) | 16/59 | 1.29 × 10−5 | 8.56 × 10−4 | −2.32 | GO-BP-UP | ||||||
| Keratinocyte differentiation (GO:0030216) | 19/80 | 1.68 × 10−5 | 1.03 × 10−3 | −2.56 | GO-BP-UP | ||||||
| NIK/NF-κB signaling (GO:0038061) | 16/64 | 3.9 × 10−5 | 2.19 × 10−3 | −2.28 | GO-BP-UP | 18 | 66 | 5.1 × 10−8 | 8.65 × 10−6 | 9.31 × 10−5 | DAVID-UP |
| Type I interferon signaling pathway (GO:0060337) | 27/148 | 5.68 × 10−5 | 3 × 10−3 | −2.95 | GO-BP-UP | ||||||
| Limb spinous cell differentiation (GO:0060890) | 18/59 | 1.88 × 10−12 | 6.87 × 10−10 | −2.66 | GO-BP-Down | ||||||
| Limb granular cell differentiation (GO:0060891) | 18/59 | 1.88 × 10−12 | 6.87 × 10−10 | −2.65 | GO-BP-Down | ||||||
| Keratinocyte development (GO:0003334) | 18/59 | 1.88 × 10−12 | 6.87 × 10−10 | −2.63 | GO-BP-Down | ||||||
| Keratinocyte differentiation (GO:0030216) | 20/80 | 6.78 × 10−12 | 1.2 × 10−9 | −2.8 | GO-BP-Down | ||||||
| Skin epidermis development (GO:0098773) | 18/94 | 7.82 × 10−9 | 1.18 × 10−6 | −2.84 | GO-BP-Down | ||||||
| Epidermis development (GO:0008584) | 14/68 | 1.39 × 10−7 | 2.05 × 10−5 | −2.49 | GO-BP-Down | ||||||
| Epidermis morphogenesis (GO:0048730) | 14/75 | 5 × 10−7 | 7.18 × 10−5 | −2.54 | GO-BP-Down | ||||||
| Epidermis cell differentiation (GO:0009913) | 17/124 | 2.91 × 10−6 | 4.08 × 10−4 | −2.98 | GO-BP-Down | ||||||
| Peptide cross-linking (GO:0018149) | 17/128 | 4.52 × 10−6 | 6.19 × 10−4 | −3.22 | GO-BP-Down | ||||||
| Fat cells differentiation (GO:0045444) | Sep-44 | 2.57 × 10−5 | 3.36 × 10−3 | −2.64 | GO-BP-Down | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdallah, F.; Henriet, E.; Suet, A.; Arar, A.; Clemençon, R.; Malinge, J.-M.; Lecellier, G.; Baril, P.; Pichon, C. miR-21-3p/IL-22 Axes Are Major Drivers of Psoriasis Pathogenesis by Modulating Keratinocytes Proliferation-Survival Balance and Inflammatory Response. Cells 2021, 10, 2547. https://doi.org/10.3390/cells10102547
Abdallah F, Henriet E, Suet A, Arar A, Clemençon R, Malinge J-M, Lecellier G, Baril P, Pichon C. miR-21-3p/IL-22 Axes Are Major Drivers of Psoriasis Pathogenesis by Modulating Keratinocytes Proliferation-Survival Balance and Inflammatory Response. Cells. 2021; 10(10):2547. https://doi.org/10.3390/cells10102547
Chicago/Turabian StyleAbdallah, Florence, Elodie Henriet, Amandine Suet, Ali Arar, Rudy Clemençon, Jean-Marc Malinge, Gaël Lecellier, Patrick Baril, and Chantal Pichon. 2021. "miR-21-3p/IL-22 Axes Are Major Drivers of Psoriasis Pathogenesis by Modulating Keratinocytes Proliferation-Survival Balance and Inflammatory Response" Cells 10, no. 10: 2547. https://doi.org/10.3390/cells10102547
APA StyleAbdallah, F., Henriet, E., Suet, A., Arar, A., Clemençon, R., Malinge, J.-M., Lecellier, G., Baril, P., & Pichon, C. (2021). miR-21-3p/IL-22 Axes Are Major Drivers of Psoriasis Pathogenesis by Modulating Keratinocytes Proliferation-Survival Balance and Inflammatory Response. Cells, 10(10), 2547. https://doi.org/10.3390/cells10102547








