Characterization of Conserved and Promiscuous Human Rhinovirus CD4 T Cell Epitopes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Subjects
2.2. PBMCs Isolation and HLA Typing
2.3. Peptide Synthesis
2.4. CD4 T Cell Epitope Prediction
2.5. IFNγ-ELISPOT Assays
2.6. Intracellular Cytokine Staining
2.7. Quantitative Binding Affinity Assays
2.8. Other Procedures
3. Results
3.1. Computational Selection of Conserved RV Peptides with Potential CD4 T Cell Epitopes
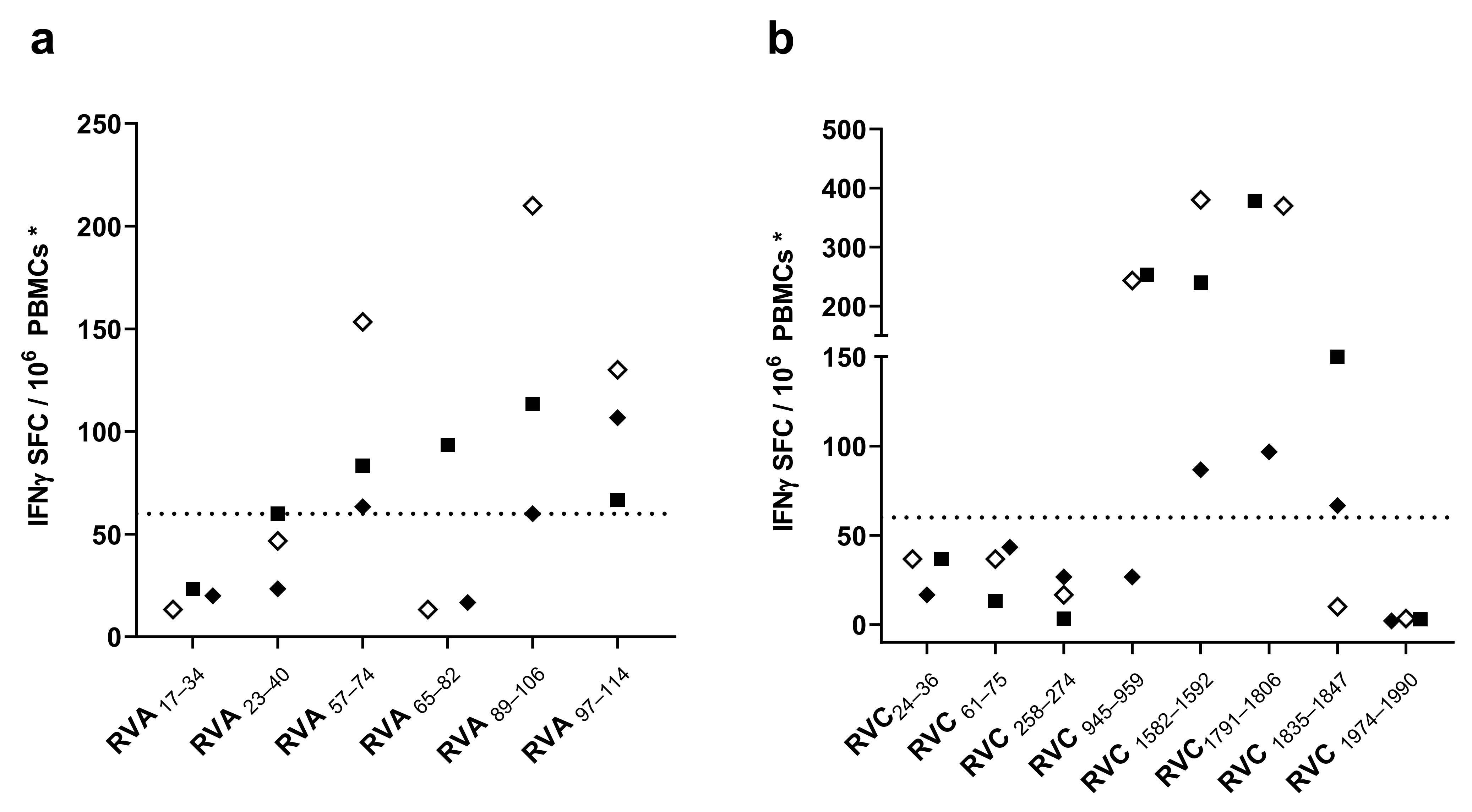
3.2. Screening of CD4 T Cell Epitope Candidates by IFNγ-ELISPOT Assays
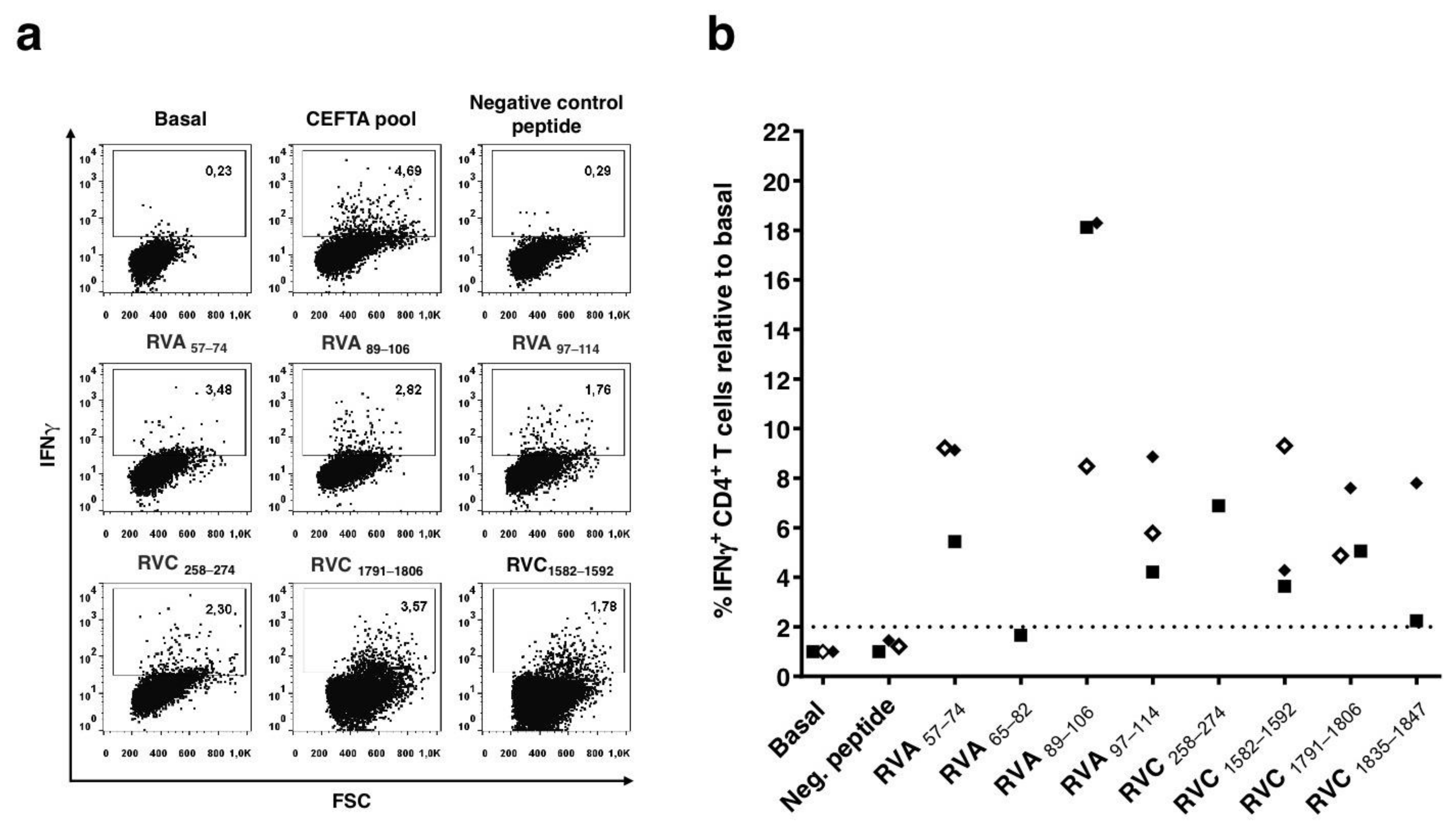
3.3. Characterization of CD4 T Cell Epitopes
3.4. Population Coverage of RV-Specific T Cell Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jacobs, S.E.; Lamson, D.M.; St George, K.; Walsh, T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.J.; Arnold, J.C.; Fairchok, M.P.; Danaher, P.J.; McDonough, E.A.; Blair, P.J.; Garcia, J.; Halsey, E.S.; Schofield, C.; Ottolini, M.; et al. Epidemiologic, clinical, and virologic characteristics of human rhinovirus infection among otherwise healthy children and adults: Rhinovirus among adults and children. J. Clin. Virol. 2015, 64, 74–82. [Google Scholar] [CrossRef]
- Miller, E.K.; Edwards, K.M.; Weinberg, G.A.; Iwane, M.K.; Griffin, M.R.; Hall, C.B.; Zhu, Y.; Szilagyi, P.G.; Morin, L.L.; Heil, L.H.; et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 2009, 123, 98–104.e101. [Google Scholar] [CrossRef]
- Bizzintino, J.; Lee, W.M.; Laing, I.A.; Vang, F.; Pappas, T.; Zhang, G.; Martin, A.C.; Khoo, S.K.; Cox, D.W.; Geelhoed, G.C.; et al. Association between human rhinovirus C and severity of acute asthma in children. Eur. Respir. J. 2011, 37, 1037–1042. [Google Scholar] [CrossRef]
- Vandini, S.; Biagi, C.; Fischer, M.; Lanari, M. Impact of Rhinovirus Infections in Children. Viruses 2019, 11, 521. [Google Scholar] [CrossRef] [Green Version]
- Hope, J.L.; Bradley, L.M. Lessons in antiviral immunity. Science 2021, 371, 464–465. [Google Scholar] [CrossRef]
- Makris, S.; Johnston, S. Recent advances in understanding rhinovirus immunity. F1000Reserch 2018, 7, 15337. [Google Scholar] [CrossRef]
- Palmenberg, A.C.; Rathe, J.A.; Liggett, S.B. Analysis of the complete genome sequences of human rhinovirus. J. Allergy Clin. Immunol. 2010, 125, 1190–1199; quiz 1200–1191. [Google Scholar] [CrossRef] [Green Version]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser-Liggett, C.M.; Liggett, S.B. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 2009, 324, 55–59. [Google Scholar] [CrossRef] [Green Version]
- McIntyre, C.L.; Knowles, N.J.; Simmonds, P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J. Gen. Virol. 2013, 94, 1791–1806. [Google Scholar] [CrossRef]
- Barclay, W.S.; Al-Nakib, W.; Higgins, P.G.; Tyrrell, D.A. The time course of the humoral immune response to rhinovirus infection. Epidemiol. Infect. 1989, 103, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Glanville, N.; Johnston, S.L. Challenges in developing a cross-serotype rhinovirus vaccine. Curr. Opin. Virol. 2015, 11, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Nguyen, M.T.; Currier, M.G.; Jenkins, J.B.; Strobert, E.A.; Kajon, A.E.; Madan-Lala, R.; Bochkov, Y.A.; Gern, J.E.; Roy, K.; et al. A polyvalent inactivated rhinovirus vaccine is broadly immunogenic in rhesus macaques. Nat. Commun. 2016, 7, 12838. [Google Scholar] [CrossRef] [PubMed]
- Steinke, J.W.; Liu, L.; Turner, R.B.; Braciale, T.J.; Borish, L. Immune surveillance by rhinovirus-specific circulating CD4+ and CD8+ T lymphocytes. PLoS ONE 2015, 10, e0115271. [Google Scholar] [CrossRef] [PubMed]
- Muehling, L.M.; Mai, D.T.; Kwok, W.W.; Heymann, P.W.; Pomes, A.; Woodfolk, J.A. Circulating Memory CD4+ T Cells Target Conserved Epitopes of Rhinovirus Capsid Proteins and Respond Rapidly to Experimental Infection in Humans. J. Immunol. 2016, 197, 3214–3224. [Google Scholar] [CrossRef] [Green Version]
- James, E.; Gern, E.C.D.; Elizabeth, A.B.; Kelly, R.V.; Bruce, K. Rhinovirus-Specific T Cells Recognize both Shared and Serotype-Restricted Viral Epitopes. J. Infect. Dis. 1997, 175, 1108–1114. [Google Scholar]
- Glanville, N.; McLean, G.R.; Guy, B.; Lecouturier, V.; Berry, C.; Girerd, Y.; Gregoire, C.; Walton, R.P.; Pearson, R.M.; Kebadze, T.; et al. Cross-serotype immunity induced by immunization with a conserved rhinovirus capsid protein. PLoS Pathog. 2013, 9, e1003669. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Perosanz, M.; Sanchez-Trincado, J.L.; Fernandez-Arquero, M.; Sidney, J.; Sette, A.; Lafuente, E.M.; Reche, P.A. Human rhinovirus-specific CD8 T cell responses target conserved and unusual epitopes. FASEB J. 2021, 35, e21208. [Google Scholar] [CrossRef]
- Gaido, C.M.; Stone, S.; Chopra, A.; Thomas, W.R.; Le Souef, P.N.; Hales, B.J. Immunodominant T-Cell Epitopes in the VP1 Capsid Protein of Rhinovirus Species A and C. J. Virol. 2016, 90, 10459–10471. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, E.; Isakova-Sivak, I.; Rudenko, L. Overview of human rhinovirus immunogenic epitopes for rational vaccine design. Expert Rev. Vaccines 2019, 18, 877–880. [Google Scholar] [CrossRef] [Green Version]
- Kimura, A.; Sasazuki, T. Eleventh International Histocompatibility Workshop reference protocol for the HLA DNA-typing technique. In HLA 1991: Proceedings of the Eleventh International Histocompatibility Workshop and Conference; Tsuji, K., Aizawa, M., Sasazuki, T., Eds.; Oxford University Press: Oxford, UK, 1991; Volume 1, p. 397. [Google Scholar]
- Reche, P.A.; Glutting, J.-P.; Reinherz, E.L. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 2004, 56, 405–419. [Google Scholar] [CrossRef] [Green Version]
- Jensen, K.K.; Andreatta, M.; Marcatili, P.; Buus, S.; Greenbaum, J.A.; Yan, Z.; Sette, A.; Peters, B.; Nielsen, M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 2018, 154, 394–406. [Google Scholar] [CrossRef]
- Reche, P.A.; Keskin, D.B.; Hussey, R.E.; Ancuta, P.; Gabuzda, D.; Reinherz, E.L. Elicitation from virus-naive individuals of cytotoxic T lymphocytes directed against conserved HIV-1 epitopes. Med. Immunol. 2006, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Sidney, J.; Southwood, S.; Moore, C.; Oseroff, C.; Pinilla, C.; Grey, H.M.; Sette, A. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. 2013, 18, 18.3.1–18.3.36. [Google Scholar] [CrossRef] [Green Version]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Bui, H.H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef] [Green Version]
- Diez-Rivero, C.M.; Reche, P.A. CD8 T cell epitope distribution in viruses reveals patterns of protein biosynthesis. PLoS ONE 2012, 7, e43674. [Google Scholar] [CrossRef]
- Sidney, J.; Steen, A.; Moore, C.; Ngo, S.; Chung, J.; Peters, B.; Sette, A. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J. Immunol. 2010, 185, 4189–4198. [Google Scholar] [CrossRef] [Green Version]
- Sidney, J.; Steen, A.; Moore, C.; Ngo, S.; Chung, J.; Peters, B.; Sette, A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J. Immunol. 2010, 184, 2492–2503. [Google Scholar] [CrossRef] [PubMed]
- Southwood, S.; Sidney, J.; Kondo, A.; del Guercio, M.F.; Appella, E.; Hoffman, S.; Kubo, R.T.; Chesnut, R.W.; Grey, H.M.; Sette, A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998, 160, 3363–3373. [Google Scholar]
- Calvo-Calle, J.M.; Strug, I.; Nastke, M.D.; Baker, S.P.; Stern, L.J. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog 2007, 3, 1511–1529. [Google Scholar] [CrossRef]
- Sette, A.; Rappuoli, R. Reverse vaccinology: Developing vaccines in the era of genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, P.E. Recent advances in antigen processing and presentation. Nat. Immunol. 2007, 8, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Fendrick, A.M.; Monto, A.S.; Nightengale, B.; Sarnes, M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 2003, 163, 487–494. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Unger, S.D.; Zhang, N.; Taka, S.; Michel, S.; Akdağ, N.; Lan, F.; Helfer, M.; Hudemann, C.; Eickmann, M.; et al. Development and characterization of DNAzyme candidates demonstrating significant efficiency against human rhinoviruses. J. Allergy Clin. Immunol. 2019, 143, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Potaczek, D.P.; Garn, H.; Unger, S.D.; Renz, H. Antisense molecules: A new class of drugs. J. Allergy Clin. Immunol. 2016, 137, 1334–1346. [Google Scholar] [CrossRef] [Green Version]
- Gaido, C.M.; Granland, C.; Laing, I.A.; Souef, P.N.L.; Thomas, W.R.; Currie, A.J.; Hales, B.J. T-cell responses against rhinovirus species A and C in asthmatic and healthy children. Immun. Inflamm. Dis. 2018, 6, 143–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stone, C.A., Jr.; Miller, E.K. Understanding the Association of Human Rhinovirus with Asthma. Clin. Vaccine Immunol. 2016, 23, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef] [Green Version]
- Reche, P.A.; Reinherz, E.L. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 2003, 331, 623–641. [Google Scholar] [CrossRef] [Green Version]


| Virus | Peptide | Sequence | Protein 1 | Position 2 | Predicted HLA II Binding Profile 3 | |
|---|---|---|---|---|---|---|
| HLA-DRB1 | HLA-DQA1/B1 | |||||
| RV A | RVA17–34 | NSVSNGSSLNYFNINYFK | VP4 | 17–34 | DRB1*12:01 | DQA1*01:01/B1*05:01 |
| RVA23–40 | LNYFNINYFKDAASSGAS | VP4 | 23–40 | DRB1*04:01 DRB1*04:04 DRB1*08:02 DRB1*12:01 | DQA1*01:01/B1*05:01 DQA1*01:01/B1*05:02 DQA1*01:04/B1*05:03 | |
| RVA57–74 | VKDVLEKGIPTLQSPTVE | VP4 | 57–74 | DRB1*11:01 | - | |
| RVA65–82 | IPTLQSPTVEACGYSDRI | VP4 | 65–82 | - | DQA1*05:01/B1*03:02 | |
| RVA89–106 | DSTITSQDVANAVVGYGV | VP4 | 89–106 | - | DQA1*01:02/B1*06:02 DQA1*02:01/B1*02:02 DQA1*03:01/B1*03:01 DQA1*05:01/B1*03:01 | |
| RVA97–114 | VANAVVGYGVWPHYLTPE | VP4 | 97–114 | DRB1*04:04 | DQA1*01:01/B1*05:01 DQA1*03:01/B1*03:01 DQA1*05:01/B1*03:01 DQA1*05:01/B1*04:02 DQA1*06:01/B1*04:02 | |
| RV C | RVC24–36 | VVKYFNINYYKDA | VP4 | 24–36 | DRB1*12:01 DRB1*15:01 | DQA1*01:01/B1*05:01 DQA1*01:02/B1*05:02 |
| RVC61–75 | LTNPALMSPSVEACG | VP4 | 61–75 | - | DQA1*01:03/B1*06:03 DQA1*02:01/B1*03:03 DQA1*05:01/B1*03:02 DQA1*05:01/B1*03:03 | |
| RVC258–274 | INLRTNNSSTIVVPYIN | VP2 | 258–274 | DRB1*13:02 | DQA1*01:02/B1*05:01 DQA1*01:02/B1*06:02 DQA1*01:03/B1*06:03 DQA1*02:01/B1*03:03 DQA1*05:01/B1*03:03 | |
| RVC945–959 | YEIQESEYYPKHIQY | 2A | 945–959 | - | DQA1*01:04/B1*05:03 | |
| RVC1582–1592 | KEKFRDIRRFIP | 3A | 1582–1592 | DRB1*08:02 DRB1*11:01 | - | |
| RVC1791–1806 | GLEPLDLNTSAGFPYV | 3D | 1791–1806 | DRB1*07:01 DRB1*09:01 DRB1*13:02 | - | |
| RVC1835–1847 | DLPYVTYLKDELR | 3D | 1835–1847 | - | DQA1*02:01/B1*02:02 DQA1*05:01/B1*02:01 | |
| RVC1974–1990 | GTSVFNTMINNIILRTL | 3D | 1974–1990 | DRB1*01:01 DRB1*01:03 DRB1*04:01 DRB1*04:03 DRB1*04:04 DRB1*04:05 DRB1*07:01 DRB1*13:02 | DQA1*01:02/B1*05:01 | |
| Peptide | Sequence | Protein | Confirmed HLA II Binding Profile 1 | PPC 2 | |
|---|---|---|---|---|---|
| HLA-DRB1 | HLA-DQA1/B1 | ||||
| RVA57–74 | VKDVLEKGIPTLQSPTVE | VP4 | - | DQA1*01:02/B1*06:02 | 34.55 |
| RVA89–106 | DSTITSQDVANAVVGYGV | VP4 | DRB1*04:04 DRB1*07:01 | DQA1*01:02/B1*06:02 DQA1*02:01/B1*02:02 DQA1*03:01/B1*03:01 DQA1*05:01/B1*02:01 DQA1*05:01/B1*03:01 | 95.09 |
| RVA97–114 | VANAVVGYGVWPHYLTPE | VP4 | DRB1*08:02 | DQA1*01:01/B1*05:01 DQA1*05:01/B1*02:01 DQA1*05:01/B1*03:01 | 81.58 |
| RVC258–274 | INLRTNNSSTIVVPYIN | VP2 | DRB1*07:01 DRB1*09:01 DRB1*13:02 | DQA1*01:02/B1*06:02 DQA1*02:01/B1*02:02 | 67.68 |
| RVC1582–1592 | KEKFRDIRRFIP | 3A | DRB1*08:02 DRB1*11:01 DRB1*11:04 | - | 17.37 |
| RVC1791–1806 | GLEPLDLNTSAGFPYV | 3D | DRB1*09:01 DRB1*13:02 | DQA1*02:01/B1*02:02 | 36.28 |
| RVC1835–1847 | DLPYVTYLKDELR | 3D | DRB1*04:04 | DQA1*05:01/B1*02:01 | 52.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez-Perosanz, M.; Fiyouzi, T.; Fernandez-Arquero, M.; Sidney, J.; Sette, A.; Reinherz, E.L.; Lafuente, E.M.; Reche, P.A. Characterization of Conserved and Promiscuous Human Rhinovirus CD4 T Cell Epitopes. Cells 2021, 10, 2294. https://doi.org/10.3390/cells10092294
Gomez-Perosanz M, Fiyouzi T, Fernandez-Arquero M, Sidney J, Sette A, Reinherz EL, Lafuente EM, Reche PA. Characterization of Conserved and Promiscuous Human Rhinovirus CD4 T Cell Epitopes. Cells. 2021; 10(9):2294. https://doi.org/10.3390/cells10092294
Chicago/Turabian StyleGomez-Perosanz, Marta, Tara Fiyouzi, Miguel Fernandez-Arquero, John Sidney, Alessandro Sette, Ellis L. Reinherz, Esther M. Lafuente, and Pedro A. Reche. 2021. "Characterization of Conserved and Promiscuous Human Rhinovirus CD4 T Cell Epitopes" Cells 10, no. 9: 2294. https://doi.org/10.3390/cells10092294
APA StyleGomez-Perosanz, M., Fiyouzi, T., Fernandez-Arquero, M., Sidney, J., Sette, A., Reinherz, E. L., Lafuente, E. M., & Reche, P. A. (2021). Characterization of Conserved and Promiscuous Human Rhinovirus CD4 T Cell Epitopes. Cells, 10(9), 2294. https://doi.org/10.3390/cells10092294









