Irradiation Suppresses IFNγ-Mediated PD-L1 and MCL1 Expression in EGFR-Positive Lung Cancer to Augment CD8+ T Cells Cytotoxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Healthy Volunteers
2.2. Cell Culture
2.3. Cell Viability
2.4. Flow Cytometry
2.5. Western Blots
2.6. Quantitative Polymerase Chain Reaction (qPCR)
2.7. Gene Knockdown
2.8. Enzyme-Linked Immunosorbent Assay (ELISA) for Measurement of IFNγ
2.9. RNAseq Profiling and Bioinformatics Analysis
2.10. Isolation of Peripheral Blood Mononuclear Cells (PBMCs) and CD8+ T Cells
2.11. Chromatin Immunoprecipitation (ChIP)
2.12. Statistical Analysis
3. Results
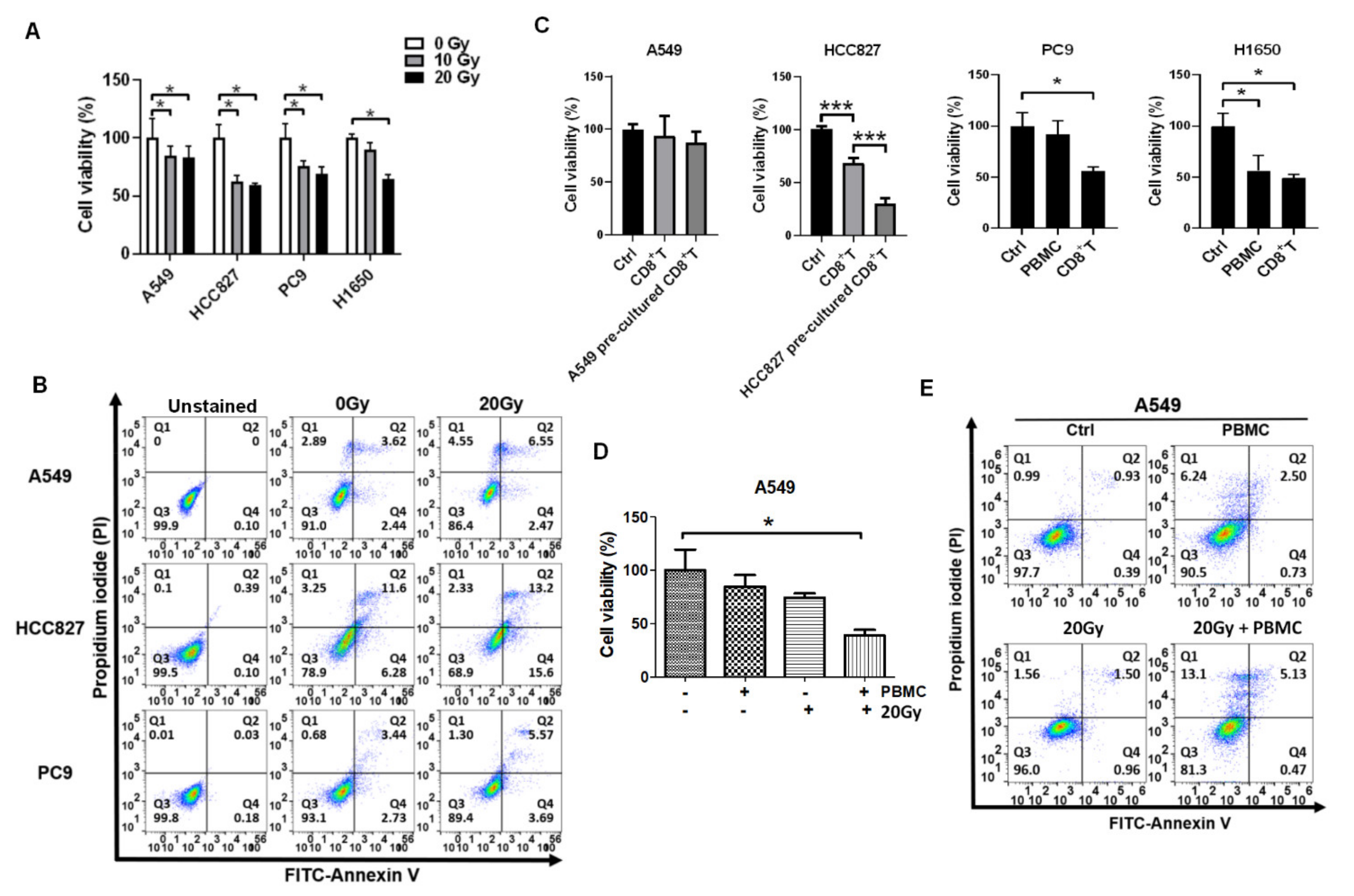
3.1. Irradiation and PBMCs Synergistically Inhibited Tumor Cell Viability and Induced Apoptosis in Lung Cancer Cells
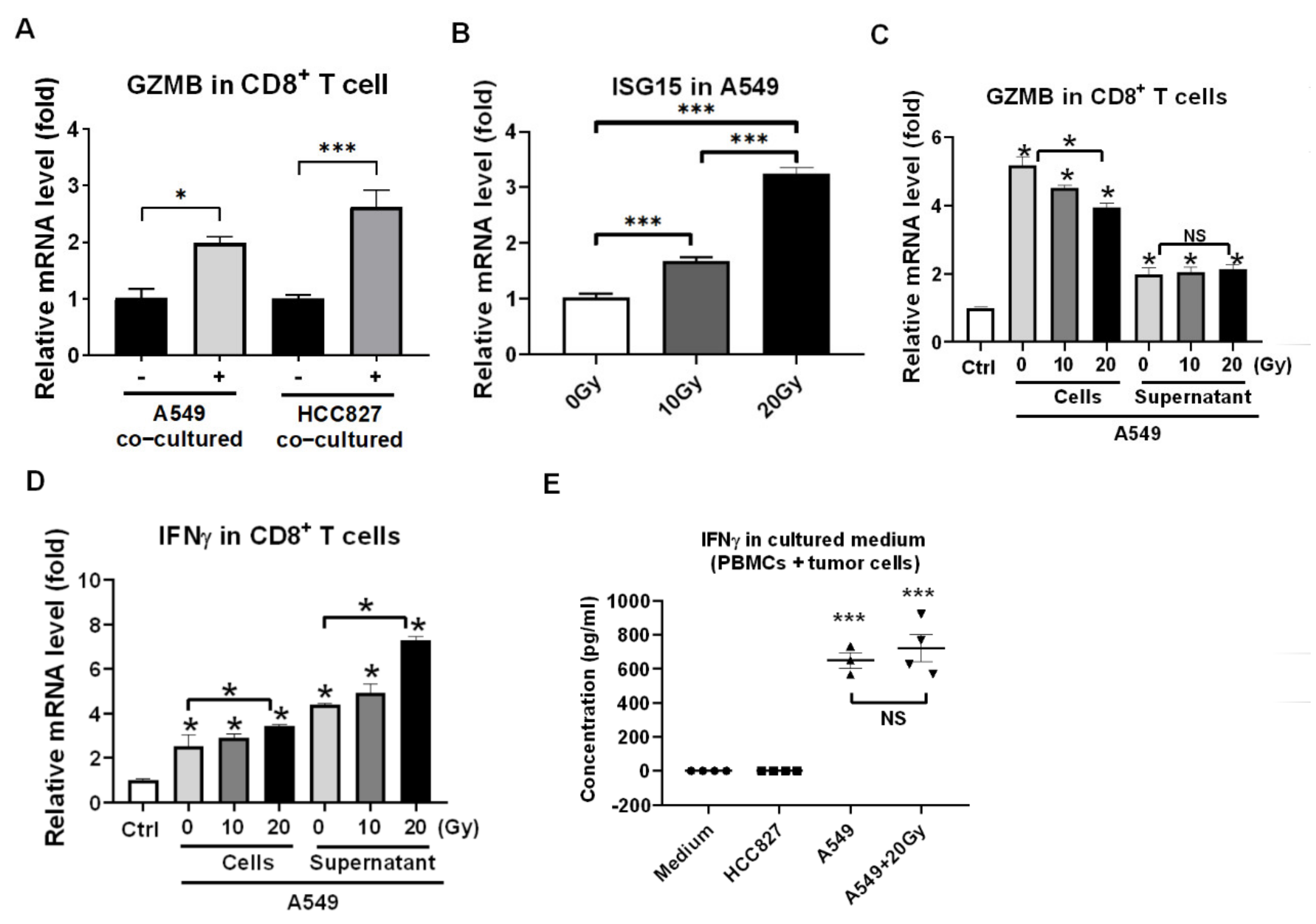
3.2. Reactivation of Healthy CD8+ T Cells in Encountering HCC827 and A549 In Vitro
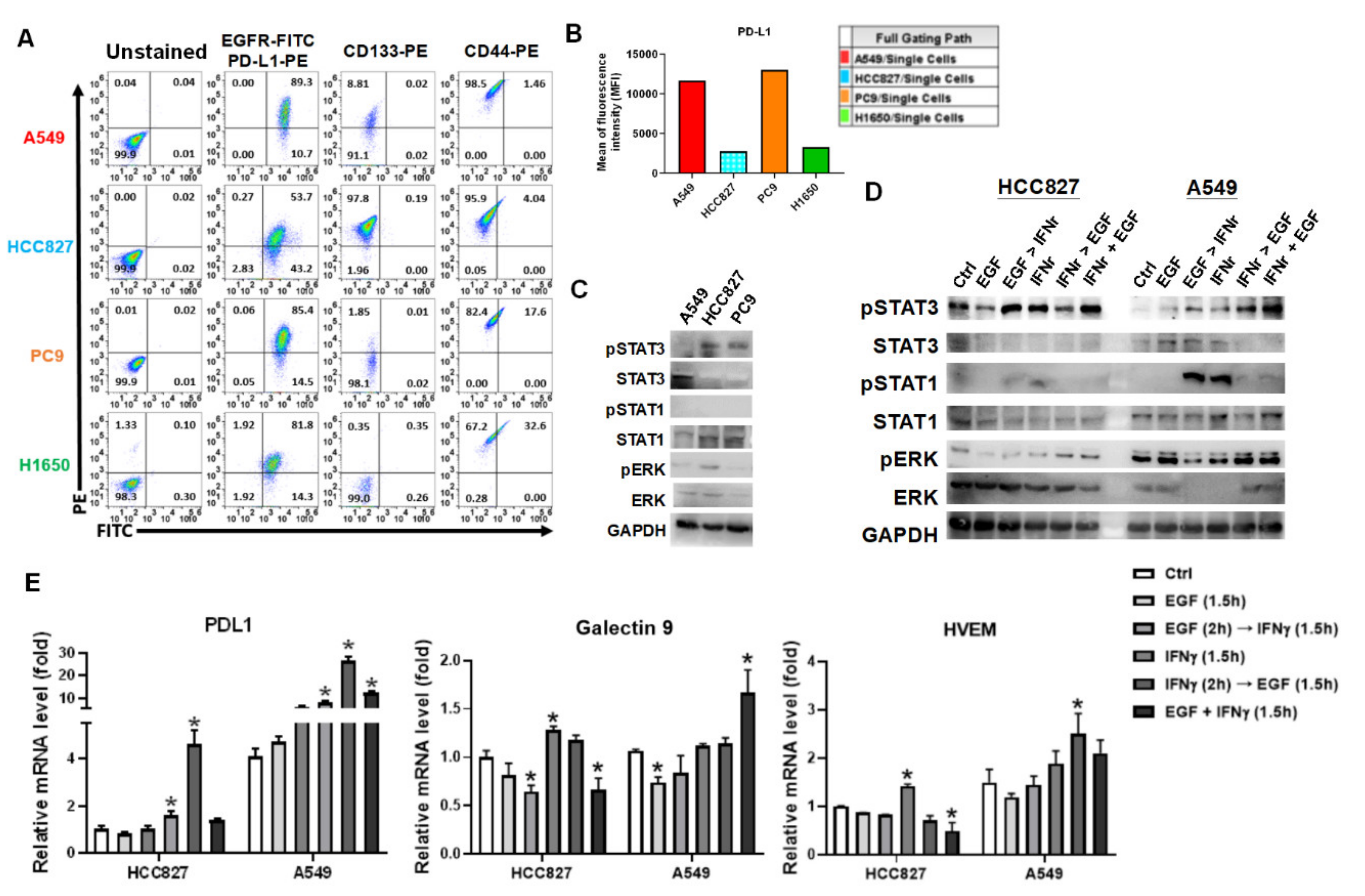
3.3. IFNγ Dominantly Phosphorylated STAT3 in the Premise of Phosphorylated EGFR
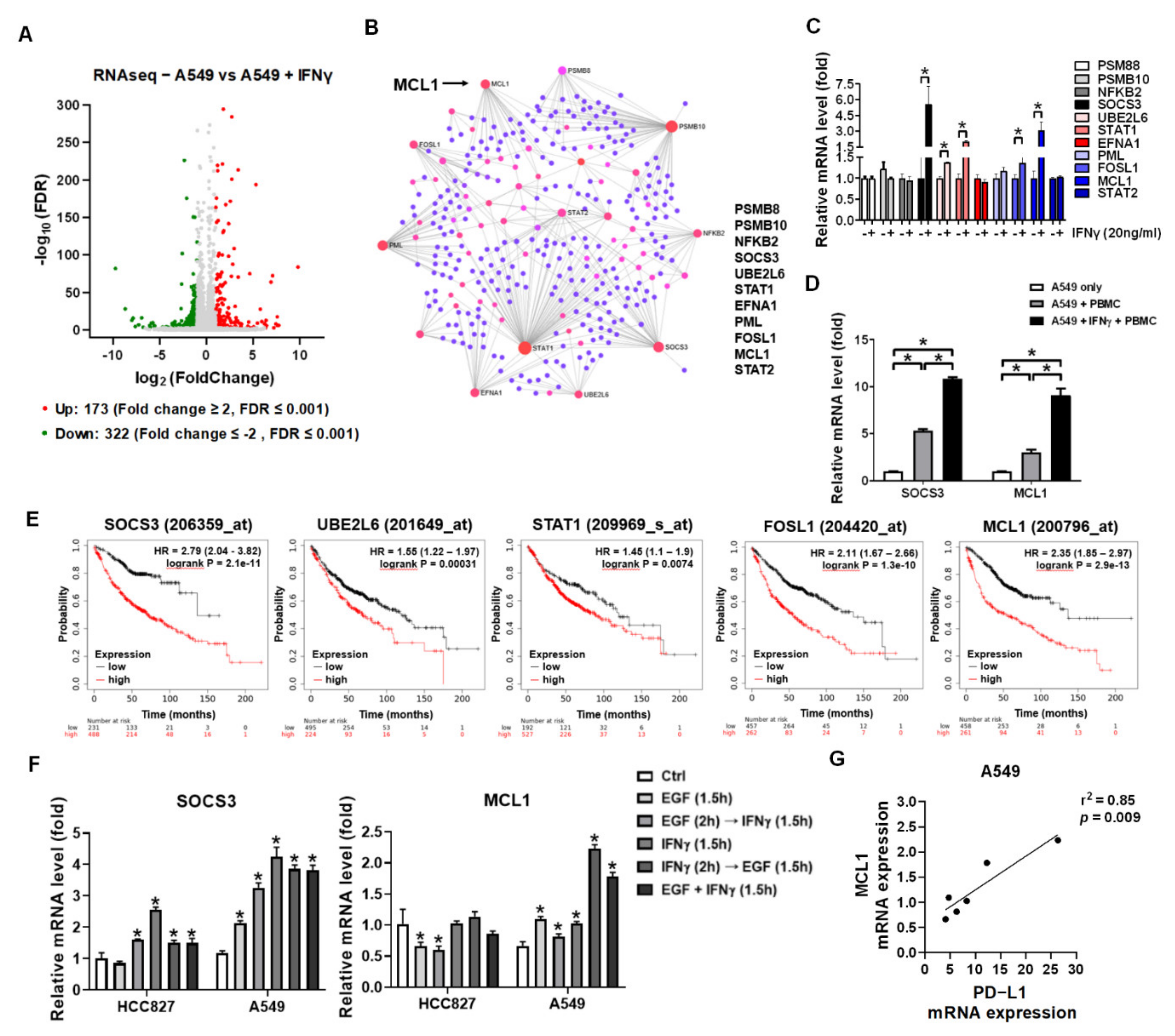
3.4. IFNγ Increased MCL1 Expression in the Premise of Phosphorylated EGFR
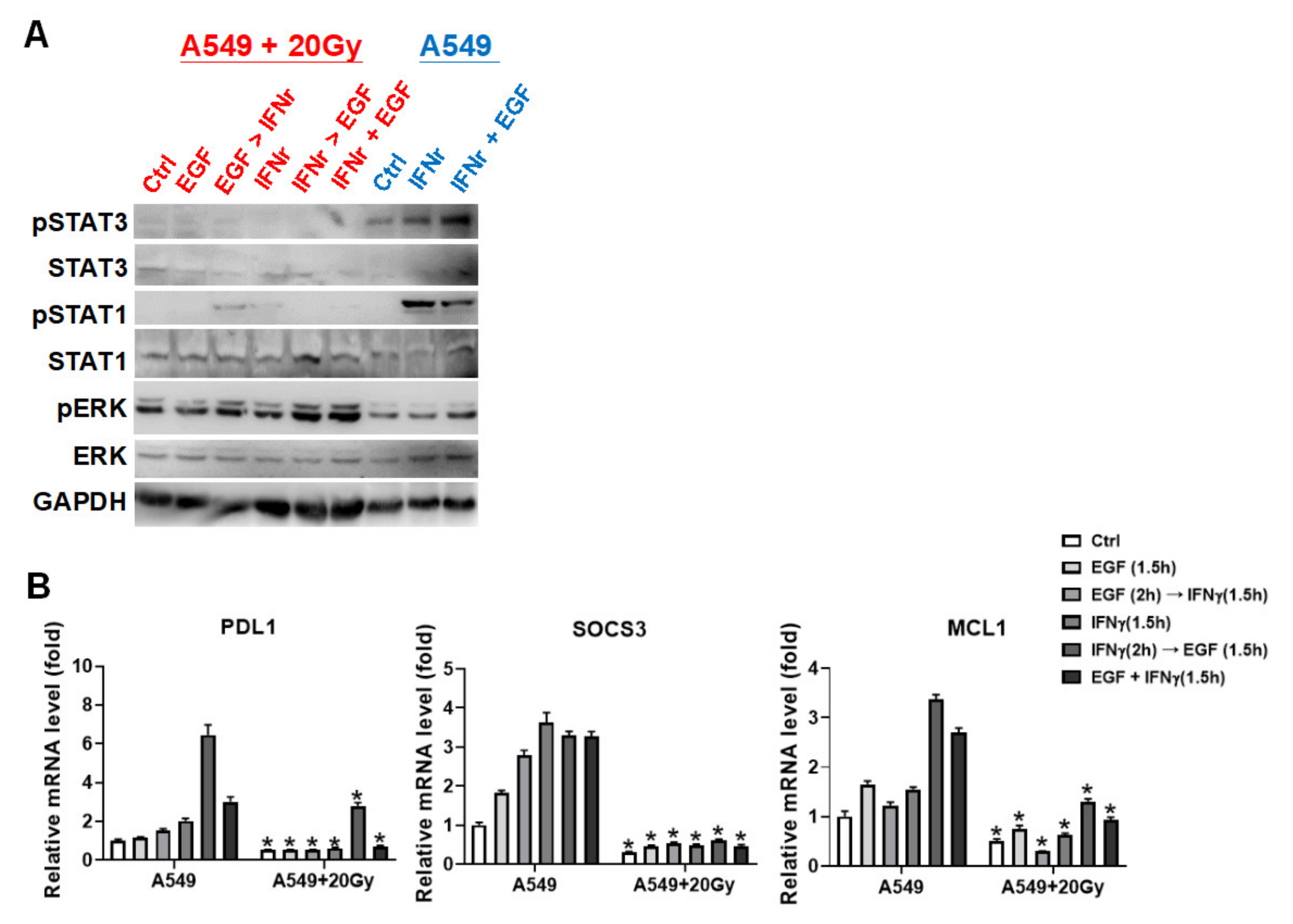
3.5. Irradiation Specifically Blocked IFNγ-Mediated Phosphorylations on STAT1 and STAT3 in A549 Cells
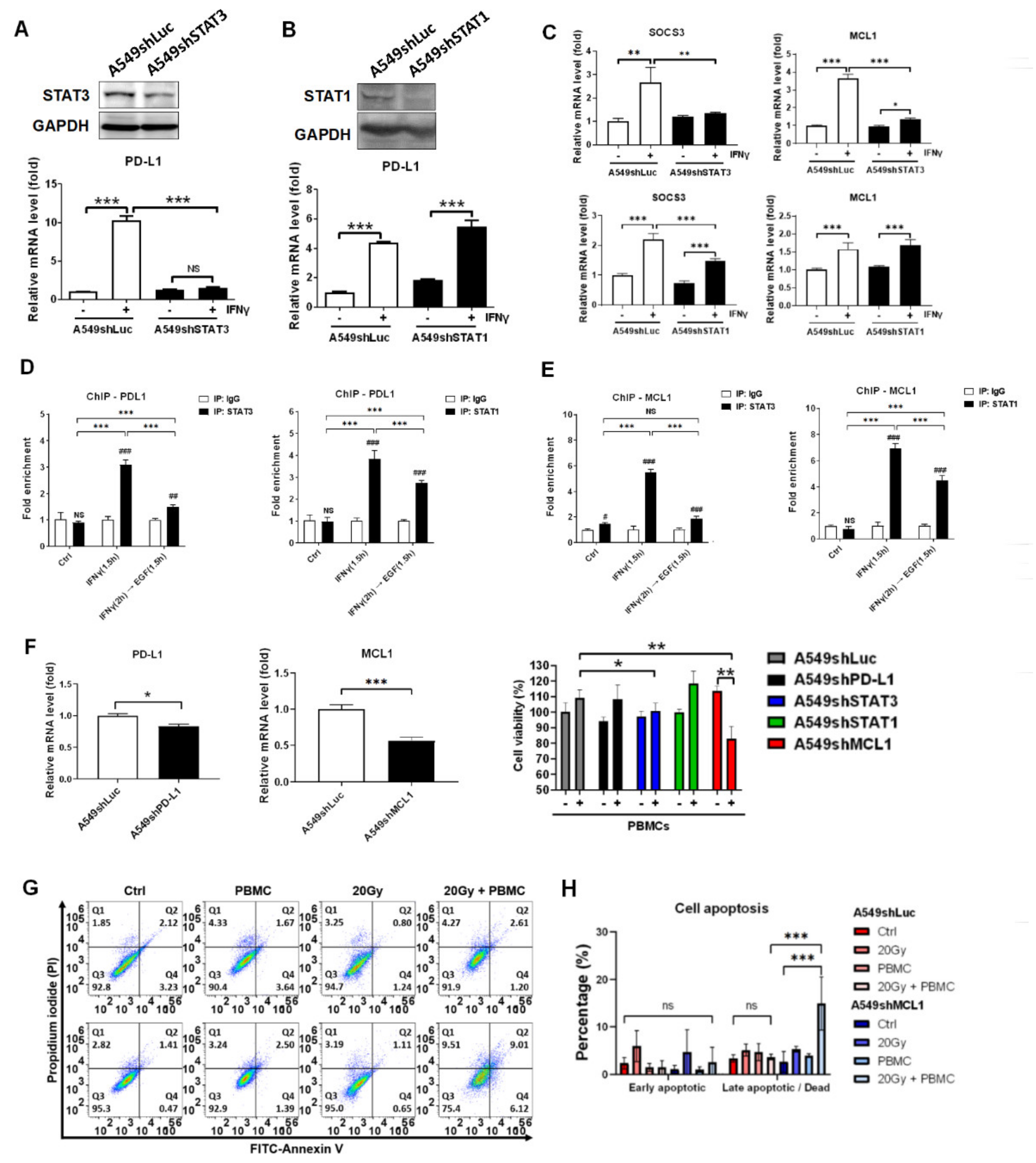
3.6. STAT3 Dominantly Determined IFNγ-Mediated Gene Expression and Knockdown of STAT3-Mediated MCL1 Augmented PBMCs against A549 Cells
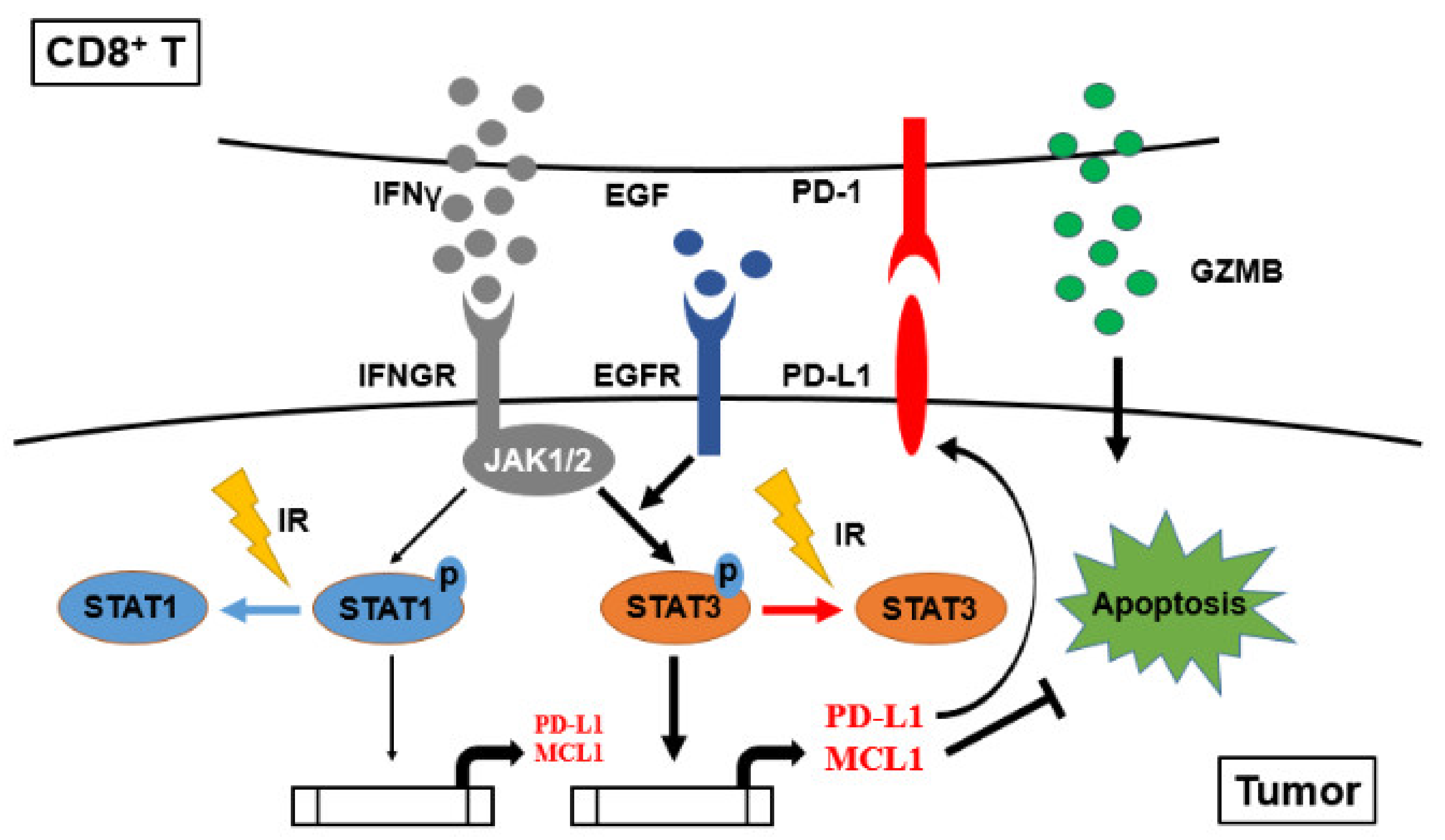
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Schiller, J.H.; Gazdar, A.F. Lung cancer in never smokers—A different disease. Nat. Rev. Cancer 2007, 7, 778–790. [Google Scholar] [CrossRef]
- Kong, F.M.; Zhao, J.; Wang, J.; Faivre-Finn, C. Radiation dose effect in locally advanced non-small cell lung cancer. J. Thorac. Dis. 2014, 6, 336–347. [Google Scholar] [CrossRef]
- Bironzo, P.; Di Maio, M. A review of guidelines for lung cancer. J. Thorac. Dis. 2018, 10, S1556–S1563. [Google Scholar] [CrossRef] [Green Version]
- Forde, P.M.; Ettinger, D.S. Targeted therapy for non-small-cell lung cancer: Past, present and future. Expert Rev. Anticancer Ther. 2013, 13, 745–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 504–535. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, M.; Wu, Y.L.; Thongprasert, S.; Sunpaweravong, P.; Leong, S.S.; Sriuranpong, V.; Chao, T.Y.; Nakagawa, K.; Chu, D.T.; Saijo, N.; et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J. Clin. Oncol. 2011, 29, 2866–2874. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Lin, C.S.; Tai, W.T.; Hsieh, C.Y.; Shiau, C.W.; Cheng, A.L.; Chen, K.F. Sorafenib enhances radiation-induced apoptosis in hepatocellular carcinoma by inhibiting STAT3. Int. J. Radiat. Oncol. Biol. Phys. 2013, 86, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Shekarian, T.; Valsesia-Wittmann, S.; Caux, C.; Marabelle, A. Paradigm shift in oncology: Targeting the immune system rather than cancer cells. Mutagenesis 2015, 30, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Sui, H.; Ma, N.; Wang, Y.; Li, H.; Liu, X.; Su, Y.; Yang, J. Anti-PD-1/PD-L1 Therapy for Non-Small-Cell Lung Cancer: Toward Personalized Medicine and Combination Strategies. J. Immunol. Res. 2018, 2018, 6984948. [Google Scholar] [CrossRef] [Green Version]
- Ko, E.C.; Raben, D.; Formenti, S.C. The Integration of Radiotherapy with Immunotherapy for the Treatment of Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 5792–5806. [Google Scholar] [CrossRef] [Green Version]
- Cullen, S.P.; Brunet, M.; Martin, S.J. Granzymes in cancer and immunity. Cell Death Differ. 2010, 17, 616–623. [Google Scholar] [CrossRef] [Green Version]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef]
- Anagnostou, V.K.; Brahmer, J.R. Cancer immunotherapy: A future paradigm shift in the treatment of non-small cell lung cancer. Clin. Cancer Res. 2015, 21, 976–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sideras, K.; Biermann, K.; Verheij, J.; Takkenberg, B.R.; Mancham, S.; Hansen, B.E.; Schutz, H.M.; de Man, R.A.; Sprengers, D.; Buschow, S.I.; et al. PD-L1, Galectin-9 and CD8(+) tumor-infiltrating lymphocytes are associated with survival in hepatocellular carcinoma. Oncoimmunology 2017, 6, e1273309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.C.; Lin, H.C.; Tsai, K.J.; Chiang, Y.W.; Lim, K.H.; Chen, C.G.; Su, Y.W.; Peng, C.L.; Ho, A.S.; Huang, L.; et al. Epidermal growth factor induces STAT1 expression to exacerbate the IFNr-mediated PD-L1 axis in epidermal growth factor receptor-positive cancers. Mol. Carcinog. 2018, 57, 1588–1598. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Yamamoto, M.; Ikushima, H.; Ohara, S.; Takeshima, H.; Sakatani, T.; Usui, K. Successful Treatment of Afatinib-Refractory Non-Small Cell Lung Cancer with Uncommon Complex EGFR Mutations Using Pembrolizumab: A Case Report. Case Rep. Oncol. 2019, 12, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wang, R.; Zhang, X.; Ma, Y.; Zhong, L.; Li, K.; Nishiyama, A.; Arai, S.; Yano, S.; Wang, W. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol. Cancer 2019, 18, 165. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.; Patin, E.C.; Pedersen, M.; Wilkins, A.; Dillon, M.T.; Melcher, A.A.; Harrington, K.J. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat. Rev. Cancer 2020, 20, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Formenti, S.C.; Demaria, S. Systemic effects of local radiotherapy. Lancet Oncol. 2009, 10, 718–726. [Google Scholar] [CrossRef] [Green Version]
- Lugade, A.A.; Sorensen, E.W.; Gerber, S.A.; Moran, J.P.; Frelinger, J.G.; Lord, E.M. Radiation-induced IFN-gamma production within the tumor microenvironment influences antitumor immunity. J. Immunol. 2008, 180, 3132–3139. [Google Scholar] [CrossRef] [PubMed]
- Ganss, R.; Ryschich, E.; Klar, E.; Arnold, B.; Hammerling, G.J. Combination of T-cell therapy and trigger of inflammation induces remodeling of the vasculature and tumor eradication. Cancer Res. 2002, 62, 1462–1470. [Google Scholar]
- Matsumura, S.; Wang, B.; Kawashima, N.; Braunstein, S.; Badura, M.; Cameron, T.O.; Babb, J.S.; Schneider, R.J.; Formenti, S.C.; Dustin, M.L.; et al. Radiation-induced CXCL16 release by breast cancer cells attracts effector T cells. J. Immunol. 2008, 181, 3099–3107. [Google Scholar] [CrossRef]
- Hodge, J.W.; Guha, C.; Neefjes, J.; Gulley, J.L. Synergizing radiation therapy and immunotherapy for curing incurable cancers. Opportunities and challenges. Oncology 2008, 22, 1064–1070. [Google Scholar]
- Lu, C.; Guan, J.; Lu, S.; Jin, Q.; Rousseau, B.; Lu, T.; Stephens, D.; Zhang, H.; Zhu, J.; Yang, M.; et al. DNA Sensing in Mismatch Repair-Deficient Tumor Cells Is Essential for Anti-tumor Immunity. Cancer Cell 2021, 39, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.K.; Postow, M.A.; Salama, J.K. Irradiation and immunotherapy: From concept to the clinic. Cancer 2016, 122, 1659–1671. [Google Scholar] [CrossRef] [Green Version]
- Dovedi, S.J.; Adlard, A.L.; Lipowska-Bhalla, G.; McKenna, C.; Jones, S.; Cheadle, E.J.; Stratford, I.J.; Poon, E.; Morrow, M.; Stewart, R.; et al. Acquired resistance to fractionated radiotherapy can be overcome by concurrent PD-L1 blockade. Cancer Res. 2014, 74, 5458–5468. [Google Scholar] [CrossRef] [Green Version]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Obeid, M.; Ortiz, C.; Criollo, A.; Mignot, G.; Maiuri, M.C.; Ullrich, E.; Saulnier, P.; et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007, 13, 1050–1059. [Google Scholar] [CrossRef]
- Gong, X.; Li, X.; Jiang, T.; Xie, H.; Zhu, Z.; Zhou, F.; Zhou, C. Combined Radiotherapy and Anti-PD-L1 Antibody Synergistically Enhances Antitumor Effect in Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Herter-Sprie, G.S.; Koyama, S.; Korideck, H.; Hai, J.; Deng, J.; Li, Y.Y.; Buczkowski, K.A.; Grant, A.K.; Ullas, S.; Rhee, K.; et al. Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer. JCI Insight 2016, 1, e87415. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Song, M.; Ping, Y.; Zhang, K.; Yang, L.; Li, F.; Zhang, C.; Cheng, S.; Yue, D.; Maimela, N.R.; Qu, J.; et al. Low-Dose IFNgamma Induces Tumor Cell Stemness in Tumor Microenvironment of Non-Small Cell Lung Cancer. Cancer Res. 2019, 79, 3737–3748. [Google Scholar] [CrossRef] [PubMed]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-gamma in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Aragane, Y.; Kulms, D.; Luger, T.A.; Schwarz, T. Down-regulation of interferon gamma-activated STAT1 by UV light. Proc. Natl. Acad. Sci. USA 1997, 94, 11490–11495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.C.; Lin, H.C.; Chiang, Y.W.; Chang, J.; Sie, Z.L.; Yang, B.L.; Lim, K.H.; Peng, C.L.; Ho, A.S.; Chang, Y.F. Nicotine exhausts CD8(+) T cells against tumor cells through increasing miR-629-5p to repress IL2RB-mediated granzyme B expression. Cancer Immunol. Immunother. 2021, 70, 1351–1364. [Google Scholar] [CrossRef]
- Cheng, C.C.; Chang, J.; Huang, S.C.; Lin, H.C.; Ho, A.S.; Lim, K.H.; Chang, C.C.; Huang, L.; Chang, Y.C.; Chang, Y.F.; et al. YM155 as an inhibitor of cancer stemness simultaneously inhibits autophosphorylation of epidermal growth factor receptor and G9a-mediated stemness in lung cancer cells. PLoS ONE 2017, 12, e0182149. [Google Scholar] [CrossRef]
- Cheng, C.C.; Yang, B.L.; Chen, W.C.; Ho, A.S.; Sie, Z.L.; Lin, H.C.; Chang, C.C. STAT3 Mediated miR-30a-5p Inhibition Enhances Proliferation and Inhibits Apoptosis in Colorectal Cancer Cells. Int. J. Mol. Sci. 2020, 21, 7315. [Google Scholar] [CrossRef]
- Lu, Y.; Fukuyama, S.; Yoshida, R.; Kobayashi, T.; Saeki, K.; Shiraishi, H.; Yoshimura, A.; Takaesu, G. Loss of SOCS3 gene expression converts STAT3 function from anti-apoptotic to pro-apoptotic. J. Biol. Chem. 2006, 281, 36683–36690. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.W.; Lam, C.; Edwards, S.W. Mcl-1; the molecular regulation of protein function. FEBS Lett. 2010, 584, 2981–2989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef]
- Kerr, W.G.; Chisholm, J.D. The Next Generation of Immunotherapy for Cancer: Small Molecules Could Make Big Waves. J. Immunol. 2019, 202, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.; Cui, Y.; Liu, Q.; Chen, S. CAR-T cell therapy for lung cancer: A promising but challenging future. J. Thorac. Dis. 2020, 12, 4516–4521. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Teixeira, L.; Sousa, J.; McNab, F.W.; Torrado, E.; Cardoso, F.; Machado, H.; Castro, F.; Cardoso, V.; Gaifem, J.; Wu, X.; et al. Type I IFN Inhibits Alternative Macrophage Activation during Mycobacterium tuberculosis Infection and Leads to Enhanced Protection in the Absence of IFN-gamma Signaling. J. Immunol. 2016, 197, 4714–4726. [Google Scholar] [CrossRef] [Green Version]
- Leopold Wager, C.M.; Wormley, F.L., Jr. Classical versus alternative macrophage activation: The Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol. 2014, 7, 1023–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjorkstrom, N.K.; Gonzalez, V.D.; Malmberg, K.J.; Falconer, K.; Alaeus, A.; Nowak, G.; Jorns, C.; Ericzon, B.G.; Weiland, O.; Sandberg, J.K.; et al. Elevated numbers of Fc gamma RIIIA+ (CD16+) effector CD8 T cells with NK cell-like function in chronic hepatitis C virus infection. J. Immunol. 2008, 181, 4219–4228. [Google Scholar] [CrossRef] [Green Version]
- Martos, S.N.; Campbell, M.R.; Lozoya, O.A.; Wang, X.; Bennett, B.D.; Thompson, I.J.B.; Wan, M.; Pittman, G.S.; Bell, D.A. Single-cell analyses identify dysfunctional CD16(+) CD8 T cells in smokers. Cell Rep. Med. 2020, 1, 100054. [Google Scholar] [CrossRef]
- Warren, H.S.; Rana, P.M.; Rieger, D.T.; Hewitt, K.A.; Dahlstrom, J.E.; Kent, A.L. CD8 T cells expressing killer Ig-like receptors and NKG2A are present in cord blood and express a more naive phenotype than their counterparts in adult blood. J. Leukoc. Biol. 2006, 79, 1252–1259. [Google Scholar] [CrossRef]
- Barbarin, A.; Cayssials, E.; Jacomet, F.; Nunez, N.G.; Basbous, S.; Lefevre, L.; Abdallah, M.; Piccirilli, N.; Morin, B.; Lavoue, V.; et al. Phenotype of NK-Like CD8(+) T Cells with Innate Features in Humans and Their Relevance in Cancer Diseases. Front. Immunol. 2017, 8, 316. [Google Scholar] [CrossRef] [Green Version]
- Intlekofer, A.M.; Takemoto, N.; Wherry, E.J.; Longworth, S.A.; Northrup, J.T.; Palanivel, V.R.; Mullen, A.C.; Gasink, C.R.; Kaech, S.M.; Miller, J.D.; et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005, 6, 1236–1244. [Google Scholar] [CrossRef]
- Chinnaiyan, P.; Huang, S.; Vallabhaneni, G.; Armstrong, E.; Varambally, S.; Tomlins, S.A.; Chinnaiyan, A.M.; Harari, P.M. Mechanisms of enhanced radiation response following epidermal growth factor receptor signaling inhibition by erlotinib (Tarceva). Cancer Res. 2005, 65, 3328–3335. [Google Scholar] [CrossRef] [Green Version]
- Moschini, I.; Dell’Anna, C.; Losardo, P.L.; Bordi, P.; D’Abbiero, N.; Tiseo, M. Radiotherapy of non-small-cell lung cancer in the era of EGFR gene mutations and EGF receptor tyrosine kinase inhibitors. Future Oncol. 2015, 11, 2329–2342. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Qiu, C.; Yang, N. STAT3 Contributes to Radioresistance in Cancer. Front. Oncol. 2020, 10, 1120. [Google Scholar] [CrossRef]
- You, S.; Li, R.; Park, D.; Xie, M.; Sica, G.L.; Cao, Y.; Xiao, Z.Q.; Deng, X. Disruption of STAT3 by niclosamide reverses radioresistance of human lung cancer. Mol. Cancer Ther. 2014, 13, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Kida, H.; Ihara, S.; Kumanogoh, A. Involvement of STAT3 in immune evasion during lung tumorigenesis. Oncoimmunology 2013, 2, e22653. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int. Rev. Immunol. 2009, 28, 239–260. [Google Scholar] [CrossRef] [PubMed]
- Pietra, G.; Romagnani, C.; Manzini, C.; Moretta, L.; Mingari, M.C. The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. J. Biomed. Biotechnol. 2010, 2010, 907092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Li, F.S.; Chen, X.H.; Liu, Q.W.; Feng, J.B.; Liu, Q.J.; Su, X. Radiation induces phosphorylation of STAT3 in a dose- and time-dependent manner. Asian Pac. J. Cancer Prev. 2014, 15, 6161–6164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Corte, C.M.D.; Sen, T.; Gay, C.M.; Ramkumar, K.; Diao, L.; Cardnell, R.J.; Rodriguez, B.L.; Stewart, C.A.; Papadimitrakopoulou, V.A.; Gibson, L.; et al. STING Pathway Expression Identifies NSCLC With an Immune-Responsive Phenotype. J. Thorac. Oncol. 2020, 15, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Urban, S.L.; Berg, L.J.; Welsh, R.M. Type 1 interferon licenses naive CD8 T cells to mediate anti-viral cytotoxicity. Virology 2016, 493, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Tremblay, M.L.; Digiovanni, J. Protein tyrosine phosphatases, TC-PTP, SHP1, and SHP2, cooperate in rapid dephosphorylation of Stat3 in keratinocytes following UVB irradiation. PLoS ONE 2010, 5, e10290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.T.; Su, J.C.; Liu, C.Y.; Shiau, C.W.; Chen, K.F. Alteration of SHP-1/p-STAT3 Signaling: A Potential Target for Anticancer Therapy. Int. J. Mol. Sci. 2017, 18, 1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene | Direction | Primer Sequence |
|---|---|---|
| GAPDH | Forward | GAGTCAACGGATTTGGTCGT |
| Reverse | TTGATTTTGGAGGGATCTCG | |
| GZMB | Forward | ACTGCAGCTGGAGAGAAAGG |
| Reverse | TTCGCACTTTCGATCTTCCT | |
| CD274 (PD-L1) | Forward | GTACCTTGGCTTTGCCACAT |
| Reverse | CCAACACCACAAGGAGGAGT | |
| TNFRSF14 (HVEM) | Forward | CCACTGGGTATGGTGGTTTC |
| Reverse | TCACCTTCTGCCTCCTGTCT | |
| LGALS9 (Galectin-9) | Forward | CCTTTGACCTCTGCTTCCTG |
| Reverse | AAACAGACAGGCTGGGAGAA | |
| STAT1 | Forward | CCGTTTTCATGACCTCCTGT |
| Reverse | TGAATATTCCCCGACTGAGC | |
| STAT2 | Forward | GAGGCCTCAACTCAGACCAG |
| Reverse | GCGTCCATCATTCCAGAGAT | |
| STAT3 | Forward | TTTCACTTGGGTGGAGAAGG |
| Reverse | GCTACCTGGGTCAGCTTCAG | |
| PSM88 | Forward | CACGGGTAGTGGGAACACTT |
| Reverse | TCACCCAACCATCTTCCTTC | |
| PSMB10 | Forward | AATGTGGACGCATGTGTGAT |
| Reverse | TCCAGGGTTAGTGGCTTCAC | |
| NFKB2 | Forward | GAACAGCCTTGCATCTAGCC |
| Reverse | TCCGAGTCGCTATCAGAGGT | |
| SOCS3 | Forward | GCCACCTACTGAACCCTCCT |
| Reverse | ACGGTCTTCCGACAGAGATG | |
| UBE2L6 | Forward | CAACCTCCCTACCACCTGAA |
| Reverse | GCAAGGCTTCCAGTTCTCAC | |
| EFNA1 | Forward | GGTGACTGTCAGTGGCAAAA |
| Reverse | AGTGGAAGGAGCAGCACAGT | |
| PML | Forward | ACACAACGTGAGCTTCATGG |
| Reverse | AAGTGGGGTGGAGACTCCTT | |
| FOSL1 | Forward | AGCTGCAGAAGCAGAAGGAG |
| Reverse | GGAGTTAGGGAGGGTGTGGT | |
| MCL1 | Forward | AGAAAGCTGCATCGAACCAT |
| Reverse | CCAGCTCCTACTCCAGCAAC | |
| ISG15 | Forward | TGTCGGTGTCAGAGCTGAAG |
| Reverse | GCCCTTGTTATTCCTCACCA | |
| ChIP_STAT1/3_MCL1 | Forward | AAAAGGGCTCACAAATCAGGT |
| Reverse | GTCTTCGGAGGCTCTGAGTG | |
| ChIP_STAT1/3_PD-L1 | Forward | ACTAGCATGGCTGAGACAGTGA |
| Reverse | CATACCTAGTAGAACCTGCCCTGT |
| Gene | Clone ID | Targeted Sequence |
|---|---|---|
| Luciferase | TRCN0000072249 | GCGGTTGCCAAGAGGTTCCAT |
| STAT1 | TRCN0000004266 | CGACAGTATGATGAACACAGT |
| STAT3 | TRCN0000020842 | CACAATCTACGAAGAATCAA. |
| MCL1 | TRCN0000005515 | GCAGAAAGTATCACAGACGTT |
| PDL1 | TRCN0000056914 | CGAATTACTGTGAAAGTCAAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-I.; Chang, Y.-F.; Sie, Z.-L.; Ho, A.-S.; Chang, J.-S.; Peng, C.-L.; Cheng, C.-C. Irradiation Suppresses IFNγ-Mediated PD-L1 and MCL1 Expression in EGFR-Positive Lung Cancer to Augment CD8+ T Cells Cytotoxicity. Cells 2021, 10, 2515. https://doi.org/10.3390/cells10102515
Wang C-I, Chang Y-F, Sie Z-L, Ho A-S, Chang J-S, Peng C-L, Cheng C-C. Irradiation Suppresses IFNγ-Mediated PD-L1 and MCL1 Expression in EGFR-Positive Lung Cancer to Augment CD8+ T Cells Cytotoxicity. Cells. 2021; 10(10):2515. https://doi.org/10.3390/cells10102515
Chicago/Turabian StyleWang, Chun-I., Yi-Fang Chang, Zong-Lin Sie, Ai-Sheng Ho, Jung-Shan Chang, Cheng-Liang Peng, and Chun-Chia Cheng. 2021. "Irradiation Suppresses IFNγ-Mediated PD-L1 and MCL1 Expression in EGFR-Positive Lung Cancer to Augment CD8+ T Cells Cytotoxicity" Cells 10, no. 10: 2515. https://doi.org/10.3390/cells10102515
APA StyleWang, C.-I., Chang, Y.-F., Sie, Z.-L., Ho, A.-S., Chang, J.-S., Peng, C.-L., & Cheng, C.-C. (2021). Irradiation Suppresses IFNγ-Mediated PD-L1 and MCL1 Expression in EGFR-Positive Lung Cancer to Augment CD8+ T Cells Cytotoxicity. Cells, 10(10), 2515. https://doi.org/10.3390/cells10102515







