Sphingolipids in Hematopoiesis: Exploring Their Role in Lineage Commitment
Abstract
1. Introduction
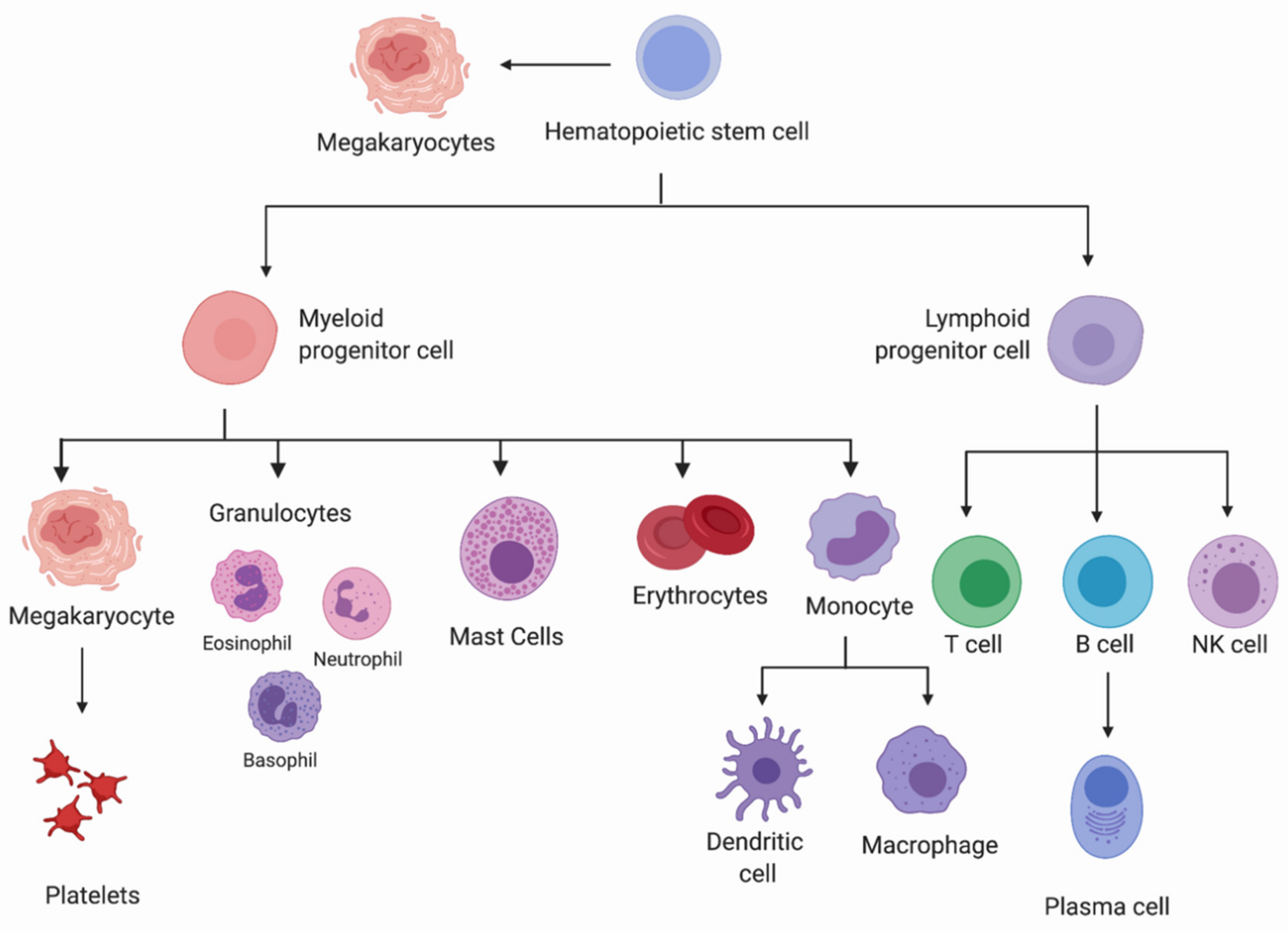
1.1. Hematopoietic Lineage Commitment
1.2. An Overview of Sphingolipid Metabolism
2. Sphingolipids in Hematopoiesis
2.1. Sphingolipids and Hematopoietic Stem Cells
2.2. Sphingolipids in Erythroid Differentiation and Erythrocytes
2.3. Sphingolipids in Thrombopoiesis and Megakaryocytes
2.4. Sphingolipids in Myeloid Differentiation
2.5. Lymphocytes: T-Cells, B-Cells, Natural Killer Cells
3. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chao, M.P.; Seita, J.; Weissman, I.L. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, D. The Hemopoietic Colony Stimulating Factors; Elsevier: Amsterdam, The Netherlands, 1984. [Google Scholar]
- Loughran, S.J.; Haas, S.; Wilkinson, A.C.; Klein, A.M.; Brand, M. Lineage commitment of hematopoietic stem cells and progenitors: Insights from recent single cell and lineage tracing technologies. Exp. Hematol. 2020, 88, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Laurenti, E.; Gottgens, B. From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553, 418–426. [Google Scholar] [CrossRef]
- Takayama, N.; Murison, A.; Takayanagi, S.I.; Arlidge, C.; Zhou, S.; Garcia-Prat, L.; Chan-Seng-Yue, M.; Zandi, S.; Gan, O.I.; Boutzen, H.; et al. The Transition from Quiescent to Activated States in Human Hematopoietic Stem Cells Is Governed by Dynamic 3D Genome Reorganization. Cell Stem Cell 2021, 28, 488–501.e410. [Google Scholar] [CrossRef]
- Woolthuis, C.M.; Park, C.Y. Hematopoietic stem/progenitor cell commitment to the megakaryocyte lineage. Blood 2016, 127, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- Carrelha, J.; Meng, Y.; Kettyle, L.M.; Luis, T.C.; Norfo, R.; Alcolea, V.; Boukarabila, H.; Grasso, F.; Gambardella, A.; Grover, A.; et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 2018, 554, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Zandi, S.; Takayama, N.; Dobson, S.; Gan, O.I.; Wilson, G.; Kaufmann, K.B.; McLeod, J.; Laurenti, E.; Dunant, C.F.; et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016, 351, aab2116. [Google Scholar] [CrossRef]
- Adolfsson, J.; Borge, O.J.; Bryder, D.; Theilgaard-Monch, K.; Astrand-Grundstrom, I.; Sitnicka, E.; Sasaki, Y.; Jacobsen, S.E. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 2001, 15, 659–669. [Google Scholar] [CrossRef]
- Pernes, G.; Flynn, M.C.; Lancaster, G.I.; Murphy, A.J. Fat for fuel: Lipid metabolism in haematopoiesis. Clin. Transl. Immunol. 2019, 8, e1098. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, S.; Xia, J.; Liu, F. Hematopoietic Hierarchy—An Updated Roadmap. Trends Cell Biol. 2018, 28, 976–986. [Google Scholar] [CrossRef]
- Robb, L. Cytokine receptors and hematopoietic differentiation. Oncogene 2007, 26, 6715–6723. [Google Scholar] [CrossRef] [PubMed]
- Rieger, M.A.; Schroeder, T. Hematopoiesis. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J.E. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef]
- Dharampuriya, P.R.; Scapin, G.; Wong, C.; John Wagner, K.; Cillis, J.L.; Shah, D.I. Tracking the origin, development, and differentiation of hematopoietic stem cells. Curr. Opin. Cell Biol. 2017, 49, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Hofer, T.; Rodewald, H.R. Differentiation-based model of hematopoietic stem cell functions and lineage pathways. Blood 2018, 132, 1106–1113. [Google Scholar] [CrossRef]
- Ohta, H.; Sweeney, E.A.; Masamune, A.; Yatomi, Y.; Hakomori, S.; Igarashi, Y. Induction of apoptosis by sphingosine in human leukemic HL-60 cells: A possible endogenous modulator of apoptotic DNA fragmentation occurring during phorbol ester-induced differentiation. Cancer Res. 1995, 55, 691–697. [Google Scholar]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Futerman, A.H.; Riezman, H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005, 15, 312–318. [Google Scholar] [CrossRef]
- Spiegel, S.; Merrill, A.H., Jr. Sphingolipid metabolism and cell growth regulation. FASEB J. 1996, 10, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, Y.; Zhang, Z.; Gable, K.; Gupta, S.D.; Somashekarappa, N.; Han, G.; Zhao, H.; Myasnikov, A.G.; Kalathur, R.C.; et al. Structural insights into the regulation of human serine palmitoyltransferase complexes. Nat. Struct. Mol. Biol. 2021, 28, 240–248. [Google Scholar] [CrossRef]
- Li, S.; Xie, T.; Liu, P.; Wang, L.; Gong, X. Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex. Nat. Struct. Mol. Biol. 2021, 28, 249–257. [Google Scholar] [CrossRef]
- Hanada, K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 2003, 1632, 16–30. [Google Scholar] [CrossRef]
- Han, G.; Gupta, S.D.; Gable, K.; Niranjanakumari, S.; Moitra, P.; Eichler, F.; Brown, R.H., Jr.; Harmon, J.M.; Dunn, T.M. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA 2009, 106, 8186–8191. [Google Scholar] [CrossRef] [PubMed]
- Hornemann, T.; Wei, Y.; von Eckardstein, A. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem. J. 2007, 405, 157–164. [Google Scholar] [CrossRef]
- Zhao, L.; Spassieva, S.; Gable, K.; Gupta, S.D.; Shi, L.Y.; Wang, J.; Bielawski, J.; Hicks, W.L.; Krebs, M.P.; Naggert, J.; et al. Elevation of 20-carbon long chain bases due to a mutation in serine palmitoyltransferase small subunit b results in neurodegeneration. Proc. Natl. Acad. Sci. USA 2015, 112, 12962–12967. [Google Scholar] [CrossRef]
- Harmon, J.M.; Bacikova, D.; Gable, K.; Gupta, S.D.; Han, G.; Sengupta, N.; Somashekarappa, N.; Dunn, T.M. Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 2013, 288, 10144–10153. [Google Scholar] [CrossRef]
- Levy, M.; Futerman, A.H. Mammalian ceramide synthases. IUBMB Life 2010, 62, 347–356. [Google Scholar] [CrossRef]
- Kihara, A.; Igarashi, Y. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 2004, 279, 49243–49250. [Google Scholar] [CrossRef]
- Michel, C.; van Echten-Deckert, G.; Rother, J.; Sandhoff, K.; Wang, E.; Merrill, A.H., Jr. Characterization of ceramide synthesis. A dihydroceramide desaturase introduces the 4,5-trans-double bond of sphingosine at the level of dihydroceramide. J. Biol. Chem. 1997, 272, 22432–22437. [Google Scholar] [CrossRef] [PubMed]
- Marggraf, W.D.; Kanfer, J.N. The phosphorylcholine acceptor in the phosphatidylcholine:ceramide cholinephosphotransferase reaction. Is the enzyme a transferase or a hydrolase? Biochim. Biophys. Acta 1984, 793, 346–353. [Google Scholar] [CrossRef]
- Vacaru, A.M.; van den Dikkenberg, J.; Ternes, P.; Holthuis, J.C. Ceramide phosphoethanolamine biosynthesis in Drosophila is mediated by a unique ethanolamine phosphotransferase in the Golgi lumen. J. Biol. Chem. 2013, 288, 11520–11530. [Google Scholar] [CrossRef] [PubMed]
- Malgat, M.; Maurice, A.; Baraud, J. Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain. J. Lipid Res. 1986, 27, 251–260. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Hill, R.A.; Li, Y.T. Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Adv. Cancer Res. 2013, 117, 59–89. [Google Scholar] [CrossRef]
- Stoffel, W. GalCer Synthase (Ceramide Galactosyltransferase, CGT). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Clausen, H., Furukawa, K., Hart, G.W., Kannagi, R., Kawasaki, T., Kinoshita, T., Muramatsu, T., et al., Eds.; Springer: Japan, Tokyo, 2002; pp. 51–57. [Google Scholar]
- Sugiura, M.; Kono, K.; Liu, H.; Shimizugawa, T.; Minekura, H.; Spiegel, S.; Kohama, T. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J. Biol. Chem. 2002, 277, 23294–23300. [Google Scholar] [CrossRef]
- Senkal, C.E.; Salama, M.F.; Snider, A.J.; Allopenna, J.J.; Rana, N.A.; Koller, A.; Hannun, Y.A.; Obeid, L.M. Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 2017, 25, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Ohno, Y.; Kamiyama, N.; Nakamichi, S.; Kihara, A. PNPLA1 is a transacylase essential for the generation of the skin barrier lipid omega-O-acylceramide. Nat. Commun. 2017, 8, 14610. [Google Scholar] [CrossRef]
- El Bawab, S.; Mao, C.; Obeid, L.M.; Hannun, Y.A. Ceramidases in the regulation of ceramide levels and function. Subcell Biochem. 2002, 36, 187–205. [Google Scholar] [CrossRef]
- Maceyka, M.; Milstien, S.; Spiegel, S. Sphingosine kinases, sphingosine-1-phosphate and sphingolipidomics. Prostaglandins Lipid Mediat. 2005, 77, 15–22. [Google Scholar] [CrossRef]
- Serra, M.; Saba, J.D. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv. Enzyme Regul. 2010, 50, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.; Stevens, R.C.; Hanson, M.; Roberts, E.; Oldstone, M.B. Sphingosine-1-phosphate and its receptors: Structure, signaling, and influence. Annu. Rev. Biochem 2013, 82, 637–662. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, T.; Hla, T. Structural and functional characteristics of S1P receptors. J. Cell Biochem. 2004, 92, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.C.; Han, M.H. Sphingosine-1-Phosphate (S1P) and S1P Signaling Pathway: Therapeutic Targets in Autoimmunity and Inflammation. Drugs 2016, 76, 1067–1079. [Google Scholar] [CrossRef]
- Olesch, C.; Ringel, C.; Brune, B.; Weigert, A. Beyond Immune Cell Migration: The Emerging Role of the Sphingosine-1-phosphate Receptor S1PR4 as a Modulator of Innate Immune Cell Activation. Mediat. Inflamm. 2017, 2017, 6059203. [Google Scholar] [CrossRef] [PubMed]
- Graler, M.H.; Bernhardt, G.; Lipp, M. A lymphoid tissue-specific receptor, EDG6, with potential immune modulatory functions mediated by extracellular lysophospholipids. Curr. Top. Microbiol. Immunol. 1999, 246, 131–136. [Google Scholar] [CrossRef]
- Graler, M.H.; Bernhardt, G.; Lipp, M. EDG6, a novel G-protein-coupled receptor related to receptors for bioactive lysophospholipids, is specifically expressed in lymphoid tissue. Genomics 1998, 53, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Evangelisti, C.; Evangelisti, C.; Buontempo, F.; Lonetti, A.; Orsini, E.; Chiarini, F.; Barata, J.T.; Pyne, S.; Pyne, N.J.; Martelli, A.M. Therapeutic potential of targeting sphingosine kinases and sphingosine 1-phosphate in hematological malignancies. Leukemia 2016, 30, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Kitatani, K.; Taniguchi, M.; Okazaki, T. Role of Sphingolipids and Metabolizing Enzymes in Hematological Malignancies. Mol. Cells 2015, 38, 482–495. [Google Scholar] [CrossRef]
- Moorthi, S.; Luberto, C. Role of Sphingolipids in Hematological Malignancies: Myeloproliferative Disorders. In Bioactive Sphingolipids in Cancer Biology and Therapy; Moorthi, S.; Luberto, C. Springer International Publishing: Cham, Switzerland, 2015; pp. 53–79. [Google Scholar]
- Sawai, H.; Taniguchi, M.; Okazaki, T. Role of Sphingolipids in Hematological Malignancies: Lymphoproliferative Disorders. In Bioactive Sphingolipids in Cancer Biology and Therapy; Hannun, Y.A., Luberto, C., Mao, C., Obeid, L.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 23–51. [Google Scholar]
- Maguer-Satta, V.; Oostendorp, R.; Reid, D.; Eaves, C.J. Evidence that ceramide mediates the ability of tumor necrosis factor to modulate primitive human hematopoietic cell fates. Blood 2000, 96, 4118–4123. [Google Scholar] [CrossRef]
- Xie, S.Z.; Garcia-Prat, L.; Voisin, V.; Ferrari, R.; Gan, O.I.; Wagenblast, E.; Kaufmann, K.B.; Zeng, A.G.X.; Takayanagi, S.I.; Patel, I.; et al. Sphingolipid Modulation Activates Proteostasis Programs to Govern Human Hematopoietic Stem Cell Self-Renewal. Cell Stem Cell 2019, 25, 639–653.e637. [Google Scholar] [CrossRef] [PubMed]
- Golan, K.; Vagima, Y.; Ludin, A.; Itkin, T.; Cohen-Gur, S.; Kalinkovich, A.; Kollet, O.; Kim, C.; Schajnovitz, A.; Ovadya, Y.; et al. S1P promotes murine progenitor cell egress and mobilization via S1P1-mediated ROS signaling and SDF-1 release. Blood 2012, 119, 2478–2488. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Lee, H.; Wysoczynski, M.; Wan, W.; Marlicz, W.; Laughlin, M.J.; Kucia, M.; Janowska-Wieczorek, A.; Ratajczak, J. Novel insight into stem cell mobilization-plasma sphingosine-1-phosphate is a major chemoattractant that directs the egress of hematopoietic stem progenitor cells from the bone marrow and its level in peripheral blood increases during mobilization due to activation of complement cascade/membrane attack complex. Leukemia 2010, 24, 976–985. [Google Scholar] [CrossRef]
- Juarez, J.G.; Harun, N.; Thien, M.; Welschinger, R.; Baraz, R.; Pena, A.D.; Pitson, S.M.; Rettig, M.; DiPersio, J.F.; Bradstock, K.F.; et al. Sphingosine-1-phosphate facilitates trafficking of hematopoietic stem cells and their mobilization by CXCR4 antagonists in mice. Blood 2012, 119, 707–716. [Google Scholar] [CrossRef]
- Brinkmann, V.; Davis, M.D.; Heise, C.E.; Albert, R.; Cottens, S.; Hof, R.; Bruns, C.; Prieschl, E.; Baumruker, T.; Hiestand, P.; et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002, 277, 21453–21457. [Google Scholar] [CrossRef]
- Graler, M.H.; Goetzl, E.J. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G-protein-coupled receptors. FASEB J. 2004, 18, 551–553. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kohara, H.; Noda, M.; Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006, 25, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Han, Y.C.; Zou, Y.R. CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 2008, 205, 777–783. [Google Scholar] [CrossRef]
- Rosu-Myles, M.; Gallacher, L.; Murdoch, B.; Hess, D.A.; Keeney, M.; Kelvin, D.; Dale, L.; Ferguson, S.S.; Wu, D.; Fellows, F.; et al. The human hematopoietic stem cell compartment is heterogeneous for CXCR4 expression. Proc. Natl. Acad. Sci. USA 2000, 97, 14626–14631. [Google Scholar] [CrossRef] [PubMed]
- Massberg, S.; Schaerli, P.; Knezevic-Maramica, I.; Kollnberger, M.; Tubo, N.; Moseman, E.A.; Huff, I.V.; Junt, T.; Wagers, A.J.; Mazo, I.B.; et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell 2007, 131, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, J.; Lee, J.F.; Gartung, A.; Jawadi, H.; Zhang, W.; Lominadze, D.; Lee, M.J. 3-amino-4-(3-hexylphenylamino)-4-oxobutyl phosphonic acid (W146), a Selective Antagonist of Sphingosine-1-phospahte Receptor Subtype 1, Enhances AMD3100-stimulated Mobilization of Hematopoietic Stem Progenitor Cells in Animals. J. Biochem. Pharmacol. Res. 2013, 1, 197–203. [Google Scholar] [PubMed]
- Gregory, C.J.; Eaves, A.C. Human marrow cells capable of erythropoietic differentiation in vitro: Definition of three erythroid colony responses. Blood 1977, 49, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Koury, M.J.; Bondurant, M.C. Control of red cell production: The roles of programmed cell death (apoptosis) and erythropoietin. Transfusion 1990, 30, 673–674. [Google Scholar] [CrossRef]
- Koury, M.J.; Bondurant, M.C. The molecular mechanism of erythropoietin action. Eur. J. Biochem. 1992, 210, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Palis, J. Primitive and definitive erythropoiesis in mammals. Front. Physiol. 2014, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Moras, M.; Lefevre, S.D.; Ostuni, M.A. From Erythroblasts to Mature Red Blood Cells: Organelle Clearance in Mammals. Front. Physiol. 2017, 8, 1076. [Google Scholar] [CrossRef]
- Clayton, R.B.; Cooper, J.M.; Curstedt, T.; Sjovall, J.; Borsook, H.; Chin, J.; Schwarz, A. Stimulation of erythroblast maturation in vitro by sphingolipids. J. Lipid Res. 1974, 15, 557–562. [Google Scholar] [CrossRef]
- Scaro, J.L.; Miranda, C.; Martin, B.M.; Carrera, M. Effects of sphingolipids on erythroblastic maturation in the mouse. Experientia 1982, 38, 401–403. [Google Scholar] [CrossRef]
- Dallalio, G.; North, M.; Worden, B.D.; Means, R.T., Jr. Inhibition of human erythroid colony formation by ceramide. Exp. Hematol. 1999, 27, 1133–1138. [Google Scholar] [CrossRef]
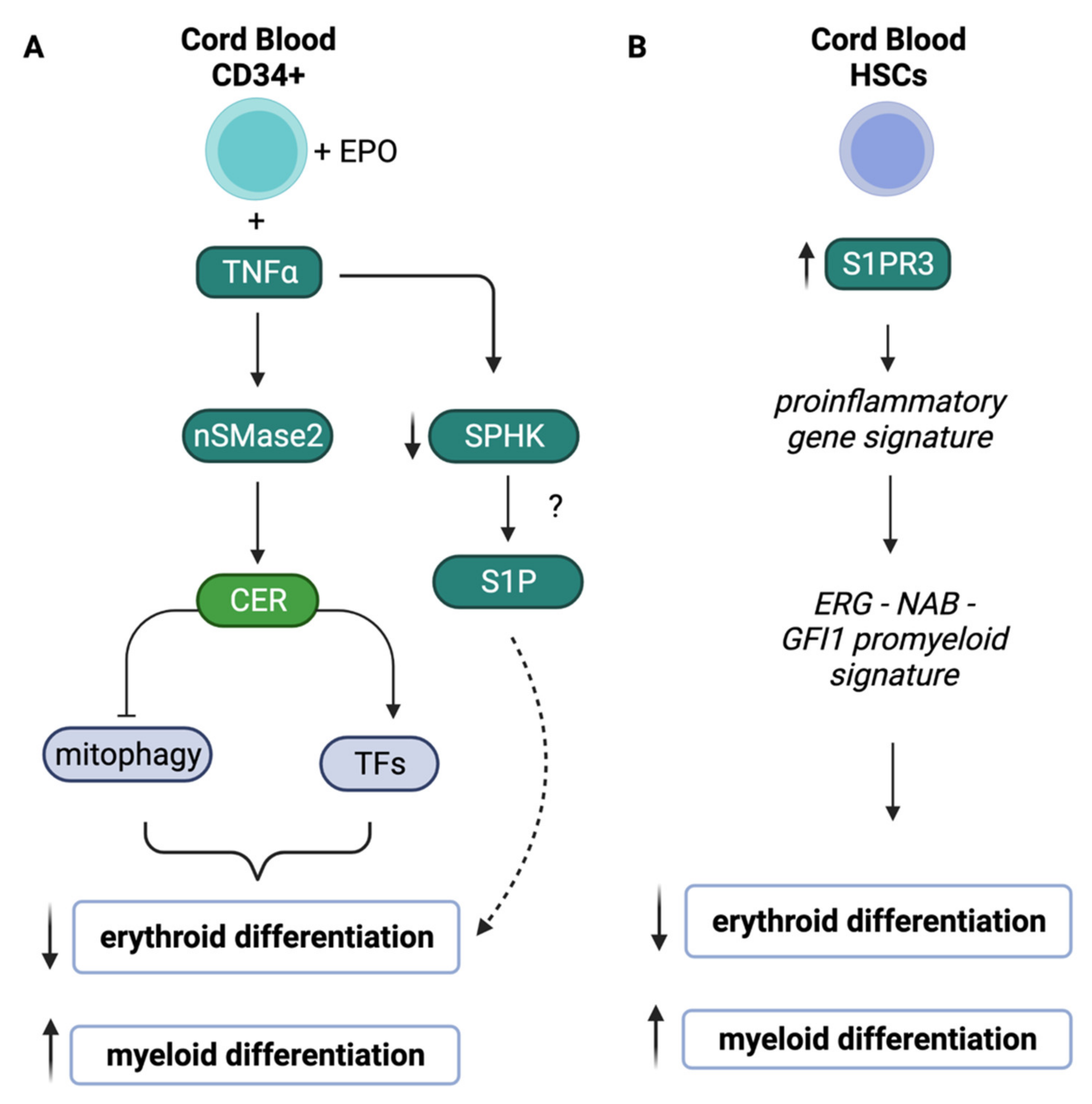
- Orsini, M.; Chateauvieux, S.; Rhim, J.; Gaigneaux, A.; Cheillan, D.; Christov, C.; Dicato, M.; Morceau, F.; Diederich, M. Sphingolipid-mediated inflammatory signaling leading to autophagy inhibition converts erythropoiesis to myelopoiesis in human hematopoietic stem/progenitor cells. Cell Death Differ. 2019, 26, 1796–1812. [Google Scholar] [CrossRef]
- Yang, C.; Hashimoto, M.; Lin, Q.X.X.; Tan, D.Q.; Suda, T. Sphingosine-1-phosphate signaling modulates terminal erythroid differentiation through the regulation of mitophagy. Exp. Hematol. 2019, 72, 47-5.e41. [Google Scholar] [CrossRef]
- Sandoval, H.; Thiagarajan, P.; Dasgupta, S.K.; Schumacher, A.; Prchal, J.T.; Chen, M.; Wang, J. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008, 454, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xu, W.; Xu, L.; Kong, Q.; Fang, J. Mitophagy is increased during erythroid differentiation in beta-thalassemia. Int. J. Hematol. 2017, 105, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yang, P.; Proia, R.L.; Hla, T. Erythrocyte-derived sphingosine 1-phosphate is essential for vascular development. J. Clin. Investig. 2014, 124, 4823–4828. [Google Scholar] [CrossRef] [PubMed]
- Rusten, L.S.; Jacobsen, S.E. Tumor necrosis factor (TNF)-alpha directly inhibits human erythropoiesis in vitro: Role of p55 and p75 TNF receptors. Blood 1995, 85, 989–996. [Google Scholar] [CrossRef]
- Jacobs-Helber, S.M.; Roh, K.H.; Bailey, D.; Dessypris, E.N.; Ryan, J.J.; Chen, J.; Wickrema, A.; Barber, D.L.; Dent, P.; Sawyer, S.T. Tumor necrosis factor-alpha expressed constitutively in erythroid cells or induced by erythropoietin has negative and stimulatory roles in normal erythropoiesis and erythroleukemia. Blood 2003, 101, 524–531. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Wu, K.; Xiao, X.; Liao, J.; Hu, Q.; Chen, H.; Liu, J.; An, X. Autophagy as a regulatory component of erythropoiesis. Int. J. Mol. Sci. 2015, 16, 4083–4094. [Google Scholar] [CrossRef]
- Pappu, R.; Schwab, S.R.; Cornelissen, I.; Pereira, J.P.; Regard, J.B.; Xu, Y.; Camerer, E.; Zheng, Y.W.; Huang, Y.; Cyster, J.G.; et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science 2007, 316, 295–298. [Google Scholar] [CrossRef]
- Schwab, S.R.; Pereira, J.P.; Matloubian, M.; Xu, Y.; Huang, Y.; Cyster, J.G. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science 2005, 309, 1735–1739. [Google Scholar] [CrossRef]
- Obinata, H.; Hla, T. Sphingosine 1-phosphate and inflammation. Int. Immunol. 2019, 31, 617–625. [Google Scholar] [CrossRef]
- Matloubian, M.; Lo, C.G.; Cinamon, G.; Lesneski, M.J.; Xu, Y.; Brinkmann, V.; Allende, M.L.; Proia, R.L.; Cyster, J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004, 427, 355–360. [Google Scholar] [CrossRef]
- Ito, K.; Anada, Y.; Tani, M.; Ikeda, M.; Sano, T.; Kihara, A.; Igarashi, Y. Lack of sphingosine 1-phosphate-degrading enzymes in erythrocytes. Biochem. Biophys. Res. Commun. 2007, 357, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.Q.; Vu, T.M.; Tukijan, F.; Muralidharan, S.; Foo, J.C.; Chin, J.F.L.; Hasan, Z.; Torta, F.; Nguyen, L.N. Erythrocytes efficiently utilize exogenous sphingosines for S1P synthesis and export via Mfsd2b. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Li, F.; Xu, R.; Low, B.E.; Lin, C.L.; Garcia-Barros, M.; Schrandt, J.; Mileva, I.; Snider, A.; Luo, C.K.; Jiang, X.C.; et al. Alkaline ceramidase 2 is essential for the homeostasis of plasma sphingoid bases and their phosphates. FASEB J. 2018, 32, 3058–3069. [Google Scholar] [CrossRef]
- Vu, T.M.; Ishizu, A.N.; Foo, J.C.; Toh, X.R.; Zhang, F.; Whee, D.M.; Torta, F.; Cazenave-Gassiot, A.; Matsumura, T.; Kim, S.; et al. Mfsd2b is essential for the sphingosine-1-phosphate export in erythrocytes and platelets. Nature 2017, 550, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Kawasaki-Nishi, S.; Otsuka, M.; Hisano, Y.; Yamaguchi, A.; Nishi, T. MFSD2B is a sphingosine 1-phosphate transporter in erythroid cells. Sci. Rep. 2018, 8, 4969. [Google Scholar] [CrossRef]
- Kurano, M.; Nishikawa, M.; Kuma, H.; Jona, M.; Yatomi, Y. Involvement of Band3 in the efflux of sphingosine 1-phosphate from erythrocytes. PLoS ONE 2017, 12, e0177543. [Google Scholar] [CrossRef]
- Hanel, P.; Andreani, P.; Graler, M.H. Erythrocytes store and release sphingosine 1-phosphate in blood. FASEB J. 2007, 21, 1202–1209. [Google Scholar] [CrossRef]
- Bode, C.; Sensken, S.C.; Peest, U.; Beutel, G.; Thol, F.; Levkau, B.; Li, Z.; Bittman, R.; Huang, T.; Tolle, M.; et al. Erythrocytes serve as a reservoir for cellular and extracellular sphingosine 1-phosphate. J. Cell Biochem. 2010, 109, 1232–1243. [Google Scholar] [CrossRef]
- Sutter, I.; Park, R.; Othman, A.; Rohrer, L.; Hornemann, T.; Stoffel, M.; Devuyst, O.; von Eckardstein, A. Apolipoprotein M modulates erythrocyte efflux and tubular reabsorption of sphingosine-1-phosphate. J. Lipid Res. 2014, 55, 1730–1737. [Google Scholar] [CrossRef]
- Sun, K.; Zhang, Y.; D’Alessandro, A.; Nemkov, T.; Song, A.; Wu, H.; Liu, H.; Adebiyi, M.; Huang, A.; Wen, Y.E.; et al. Sphingosine-1-phosphate promotes erythrocyte glycolysis and oxygen release for adaptation to high-altitude hypoxia. Nat. Commun. 2016, 7, 12086. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Peng, Z.; Brown, B.C.; Reisz, J.A.; Xu, P.; Zhou, Z.; Song, A.; Zhang, Y.; Bogdanov, M.V.; et al. Erythrocyte Metabolic Reprogramming by Sphingosine 1-Phosphate in Chronic Kidney Disease and Therapies. Circ. Res. 2020, 127, 360–375. [Google Scholar] [CrossRef]
- Akunov, A.; Sydykov, A.; Toktash, T.; Doolotova, A.; Sarybaev, A. Hemoglobin Changes After Long-Term Intermittent Work at High Altitude. Front. Physiol. 2018, 9, 1552. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.J.; Lin, Y.C.; Lin, C.Y.; Pishesha, N.; Lewis, C.A.; Freinkman, E.; Farquharson, C.; Millan, J.L.; Lodish, H. Enhanced phosphocholine metabolism is essential for terminal erythropoiesis. Blood 2018, 131, 2955–2966. [Google Scholar] [CrossRef]
- Leonard, C.; Conrard, L.; Guthmann, M.; Pollet, H.; Carquin, M.; Vermylen, C.; Gailly, P.; Van Der Smissen, P.; Mingeot-Leclercq, M.P.; Tyteca, D. Contribution of plasma membrane lipid domains to red blood cell (re)shaping. Sci. Rep. 2017, 7, 4264. [Google Scholar] [CrossRef]
- Conrard, L.; Stommen, A.; Cloos, A.S.; Steinkuhler, J.; Dimova, R.; Pollet, H.; Tyteca, D. Spatial Relationship and Functional Relevance of Three Lipid Domain Populations at the Erythrocyte Surface. Cell Physiol. Biochem. 2018, 51, 1544–1565. [Google Scholar] [CrossRef]
- Patel, S.R.; Hartwig, J.H.; Italiano, J.E., Jr. The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Investig. 2005, 115, 3348–3354. [Google Scholar] [CrossRef] [PubMed]
- Machlus, K.R.; Italiano, J.E., Jr. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 2013, 201, 785–796. [Google Scholar] [CrossRef]
- Zhang, L.; Orban, M.; Lorenz, M.; Barocke, V.; Braun, D.; Urtz, N.; Schulz, C.; von Bruhl, M.L.; Tirniceriu, A.; Gaertner, F.; et al. A novel role of sphingosine 1-phosphate receptor S1pr1 in mouse thrombopoiesis. J. Exp. Med. 2012, 209, 2165–2181. [Google Scholar] [CrossRef]
- Bunting, S.; Widmer, R.; Lipari, T.; Rangell, L.; Steinmetz, H.; Carver-Moore, K.; Moore, M.W.; Keller, G.A.; de Sauvage, F.J. Normal platelets and megakaryocytes are produced in vivo in the absence of thrombopoietin. Blood 1997, 90, 3423–3429. [Google Scholar] [CrossRef] [PubMed]
- Hla, T.; Maciag, T. An abundant transcript induced in differentiating human endothelial cells encodes a polypeptide with structural similarities to G-protein-coupled receptors. J. Biol. Chem. 1990, 265, 9308–9313. [Google Scholar] [CrossRef]
- Okazaki, H.; Ishizaka, N.; Sakurai, T.; Kurokawa, K.; Goto, K.; Kumada, M.; Takuwa, Y. Molecular cloning of a novel putative G protein-coupled receptor expressed in the cardiovascular system. Biochem. Biophys. Res. Commun. 1993, 190, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Van Brocklyn, J.R.; Thangada, S.; Liu, C.H.; Hand, A.R.; Menzeleev, R.; Spiegel, S.; Hla, T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science 1998, 279, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, F.; Tokuda, M.; Hatase, O.; Brenner, S. Molecular cloning of the novel human G protein-coupled receptor (GPCR) gene mapped on chromosome 9. Biochem. Biophys. Res. Commun. 1996, 227, 608–614. [Google Scholar] [CrossRef]
- Im, D.S.; Heise, C.E.; Ancellin, N.; O’Dowd, B.F.; Shei, G.J.; Heavens, R.P.; Rigby, M.R.; Hla, T.; Mandala, S.; McAllister, G.; et al. Characterization of a novel sphingosine 1-phosphate receptor, Edg-8. J. Biol. Chem. 2000, 275, 14281–14286. [Google Scholar] [CrossRef]
- Spiegel, S. Sphingosine 1-phosphate: A ligand for the EDG-1 family of G-protein-coupled receptors. Ann. N. Y. Acad. Sci. 2000, 905, 54–60. [Google Scholar] [CrossRef]
- Golfier, S.; Kondo, S.; Schulze, T.; Takeuchi, T.; Vassileva, G.; Achtman, A.H.; Graler, M.H.; Abbondanzo, S.J.; Wiekowski, M.; Kremmer, E.; et al. Shaping of terminal megakaryocyte differentiation and proplatelet development by sphingosine-1-phosphate receptor S1P4. FASEB J. 2010, 24, 4701–4710. [Google Scholar] [CrossRef]
- Niazi, H.; Zoghdani, N.; Couty, L.; Leuci, A.; Nitzsche, A.; Allende, M.L.; Mariko, B.; Ishaq, R.; Aslan, Y.; Becker, P.H.; et al. Murine platelet production is suppressed by S1P release in the hematopoietic niche, not facilitated by blood S1P sensing. Blood Adv. 2019, 3, 1702–1713. [Google Scholar] [CrossRef]
- Zhang, L.; Urtz, N.; Gaertner, F.; Legate, K.R.; Petzold, T.; Lorenz, M.; Mazharian, A.; Watson, S.P.; Massberg, S. Sphingosine kinase 2 (Sphk2) regulates platelet biogenesis by providing intracellular sphingosine 1-phosphate (S1P). Blood 2013, 122, 791–802. [Google Scholar] [CrossRef]
- Takeichi, T.; Torrelo, A.; Lee, J.Y.W.; Ohno, Y.; Lozano, M.L.; Kihara, A.; Liu, L.; Yasuda, Y.; Ishikawa, J.; Murase, T.; et al. Biallelic Mutations in KDSR Disrupt Ceramide Synthesis and Result in a Spectrum of Keratinization Disorders Associated with Thrombocytopenia. J. Investig. Dermatol. 2017, 137, 2344–2353. [Google Scholar] [CrossRef]
- Bariana, T.K.; Labarque, V.; Heremans, J.; Thys, C.; De Reys, M.; Greene, D.; Jenkins, B.; Grassi, L.; Seyres, D.; Burden, F.; et al. Sphingolipid dysregulation due to lack of functional KDSR impairs proplatelet formation causing thrombocytopenia. Haematologica 2019, 104, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.Z.; Kaufmann, K.B.; Wang, W.; Chan-Seng-Yue, M.; Gan, O.I.; Laurenti, E.; Garcia-Prat, L.; Takayanagi, S.I.; Ng, S.W.K.; Xu, C.; et al. Sphingosine-1-phosphate receptor 3 potentiates inflammatory programs in normal and leukemia stem cells to promote differentiation. Blood Cancer Discov. 2021, 2, 32–53. [Google Scholar] [CrossRef]
- Yatomi, Y.; Ruan, F.; Hakomori, S.; Igarashi, Y. Sphingosine-1-phosphate: A platelet-activating sphingolipid released from agonist-stimulated human platelets. Blood 1995, 86, 193–202. [Google Scholar] [CrossRef]
- Murate, T.; Banno, Y.; T-Koizumi, K.; Watanabe, K.; Mori, N.; Wada, A.; Igarashi, Y.; Takagi, A.; Kojima, T.; Asano, H.; et al. Cell type-specific localization of sphingosine kinase 1a in human tissues. J. Histochem. Cytochem. 2001, 49, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Urtz, N.; Gaertner, F.; von Bruehl, M.L.; Chandraratne, S.; Rahimi, F.; Zhang, L.; Orban, M.; Barocke, V.; Beil, J.; Schubert, I.; et al. Sphingosine 1-Phosphate Produced by Sphingosine Kinase 2 Intrinsically Controls Platelet Aggregation In Vitro and In Vivo. Circ. Res. 2015, 117, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Yatomi, Y.; Yamamura, S.; Hisano, N.; Nakahara, K.; Igarashi, Y.; Ozaki, Y. Sphingosine 1-phosphate breakdown in platelets. J. Biochem. 2004, 136, 495–502. [Google Scholar] [CrossRef]
- Tani, M.; Sano, T.; Ito, M.; Igarashi, Y. Mechanisms of sphingosine and sphingosine 1-phosphate generation in human platelets. J. Lipid Res. 2005, 46, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, K.; Lee, Y.M.; Michaud, J.; Thangada, S.; Ai, Y.; Bonkovsky, H.L.; Parikh, N.S.; Habrukowich, C.; Hla, T. Vascular endothelium as a contributor of plasma sphingosine 1-phosphate. Circ. Res. 2008, 102, 669–676. [Google Scholar] [CrossRef]
- Gazit, S.L.; Mariko, B.; Therond, P.; Decouture, B.; Xiong, Y.; Couty, L.; Bonnin, P.; Baudrie, V.; Le Gall, S.M.; Dizier, B.; et al. Platelet and Erythrocyte Sources of S1P Are Redundant for Vascular Development and Homeostasis, but Both Rendered Essential After Plasma S1P Depletion in Anaphylactic Shock. Circ. Res. 2016, 119, e110–e126. [Google Scholar] [CrossRef]
- Yatomi, Y.; Yamamura, S.; Ruan, F.; Igarashi, Y. Sphingosine 1-phosphate induces platelet activation through an extracellular action and shares a platelet surface receptor with lysophosphatidic acid. J. Biol. Chem. 1997, 272, 5291–5297. [Google Scholar] [CrossRef]
- Aoki, S.; Osada, M.; Kaneko, M.; Ozaki, Y.; Yatomi, Y. Fluid shear stress enhances the sphingosine 1-phosphate responses in cell-cell interactions between platelets and endothelial cells. Biochem. Biophys. Res. Commun. 2007, 358, 1054–1057. [Google Scholar] [CrossRef]
- Ono, Y.; Kurano, M.; Ohkawa, R.; Yokota, H.; Igarashi, K.; Aoki, J.; Tozuka, M.; Yatomi, Y. Sphingosine 1-phosphate release from platelets during clot formation: Close correlation between platelet count and serum sphingosine 1-phosphate concentration. Lipids Health Dis. 2013, 12, 20. [Google Scholar] [CrossRef]
- Vogt, K.; Mahajan-Thakur, S.; Wolf, R.; Broderdorf, S.; Vogel, C.; Bohm, A.; Ritter, C.A.; Graler, M.; Oswald, S.; Greinacher, A.; et al. Release of Platelet-Derived Sphingosine-1-Phosphate Involves Multidrug Resistance Protein 4 (MRP4/ABCC4) and Is Inhibited by Statins. Thromb. Haemost. 2018, 118, 132–142. [Google Scholar] [CrossRef]
- Decouture, B.; Becker, P.H.; Therond, P.; Gaussem, P.; Bachelot-Loza, C. Evidence that MRP4 is Only Partly Involved in S1P Secretion during Platelet Activation. Thromb. Haemost. 2018, 118, 1116–1118. [Google Scholar] [CrossRef]
- Akashi, K.; Traver, D.; Miyamoto, T.; Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000, 404, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Zon, L.I.; Yamaguchi, Y.; Yee, K.; Albee, E.A.; Kimura, A.; Bennett, J.C.; Orkin, S.H.; Ackerman, S.J. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: Potential role in gene transcription. Blood 1993, 81, 3234–3241. [Google Scholar] [CrossRef] [PubMed]
- Drissen, R.; Thongjuea, S.; Theilgaard-Monch, K.; Nerlov, C. Identification of two distinct pathways of human myelopoiesis. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Parthibane, V.; Acharya, D.; Srideshikan, S.M.; Lin, J.; Myerscough, D.G.; Abimannan, T.; Vijaykrishna, N.; Blankenberg, D.; Bondada, L.; Klarmann, K.D.; et al. Sptlc1 is essential for myeloid differentiation and hematopoietic homeostasis. Blood Adv. 2019, 3, 3635–3649. [Google Scholar] [CrossRef]
- Parthibane, V.; Lin, J.; Acharya, D.; Abimannan, T.; Srideshikan, S.M.; Klarmann, K.; Yang, A.; Soheilian, F.; Nagashima, K.; Fox, S.D.; et al. SSSPTA is essential for serine palmitoyltransferase function during development and hematopoiesis. J. Biol. Chem. 2021, 296, 100491. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, H.; Takaku, F.; Tetsuka, T.; Motoyoshi, K.; Miura, Y.; Saito, M. Characteristic expression of glycosphingolipid profiles in the bipotential cell differentiation of human promyelocytic leukemia cell line HL-60. Blood 1984, 64, 534–541. [Google Scholar] [CrossRef]
- Ryan, J.L.; Yohe, H.C.; Malech, H.L. Changes in membrane gangliosides: Differentiation of human and murine monocytic cells. Yale J. Biol Med. 1985, 58, 125–131. [Google Scholar] [PubMed]
- Delannoy, C.P.; Rombouts, Y.; Groux-Degroote, S.; Holst, S.; Coddeville, B.; Harduin-Lepers, A.; Wuhrer, M.; Elass-Rochard, E.; Guerardel, Y. Glycosylation Changes Triggered by the Differentiation of Monocytic THP-1 Cell Line into Macrophages. J. Proteome Res. 2017, 16, 156–169. [Google Scholar] [CrossRef]
- Momoi, T.; Shinmoto, M.; Kasuya, J.; Senoo, H.; Suzuki, Y. Activation of CMP-N-acetylneuraminic acid:lactosylceramide sialyltransferase during the differentiation of HL-60 cells induced by 12-O-tetradecanoylphorbol-13-acetate. J. Biol. Chem. 1986, 261, 16270–16273. [Google Scholar] [CrossRef]
- Senoo, H.; Momoi, T. The differentiation of HL-60 cells in the synthetic medium induced by GM3 ganglioside. Biosci. Rep. 1985, 5, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, H.; Takaku, F.; Terui, Y.; Miura, Y.; Saito, M. Ganglioside GM3: An acidic membrane component that increases during macrophage-like cell differentiation can induce monocytic differentiation of human myeloid and monocytoid leukemic cell lines HL-60 and U937. Proc. Natl. Acad. Sci. USA 1986, 83, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Kiguchi, K.; Henning-Chubb, C.; Huberman, E. Glycosphingolipid patterns in human promyelocytic HL-60 leukemia cells susceptible or resistant to differentiation induction by phorbol 12-myristate 13-acetate. Biochim. Biophys. Acta 1993, 1176, 27–36. [Google Scholar] [CrossRef]
- Gracheva, E.V.; Samovilova, N.N.; Golovanova, N.K.; Kashirina, S.V.; Shevelev, A.; Rybalkin, I.; Gurskaya, T.; Vlasik, T.N.; Andreeva, E.R.; Prokazova, N.V. Enhancing of GM3 synthase expression during differentiation of human blood monocytes into macrophages as in vitro model of GM3 accumulation in atherosclerotic lesion. Mol. Cell Biochem. 2009, 330, 121–129. [Google Scholar] [CrossRef]
- Akagawa, K.S.; Momoi, T.; Nagai, Y.; Tokunaga, T. Appearance of asialo GM1 glycosphingolipid on the cell surface during lymphokine-induced differentiation of M1 cells. FEBS Lett. 1981, 130, 80–84. [Google Scholar] [CrossRef]
- Moreno-Altamirano, M.M.; Aguilar-Carmona, I.; Sanchez-Garcia, F.J. Expression of GM1, a marker of lipid rafts, defines two subsets of human monocytes with differential endocytic capacity and lipopolysaccharide responsiveness. Immunology 2007, 120, 536–543. [Google Scholar] [CrossRef]
- Aida, J.; Higuchi, S.; Hasegawa, Y.; Nagano-Ito, M.; Hirabayashi, Y.; Banba, A.; Shimizu, T.; Kikuchi, A.; Saga, M.; Ichikawa, S. Up-regulation of ceramide glucosyltransferase during the differentiation of U937 cells. J. Biochem. 2011, 150, 303–310. [Google Scholar] [CrossRef]
- Kan, C.C.; Kolesnick, R.N. A synthetic ceramide analog, D-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol, selectively inhibits adherence during macrophage differentiation of human leukemia cells. J. Biol. Chem. 1992, 267, 9663–9667. [Google Scholar] [CrossRef]
- Dressler, K.A.; Kan, C.C.; Kolesnick, R.N. Sphingomyelin synthesis is involved in adherence during macrophage differentiation of HL-60 cells. J. Biol. Chem. 1991, 266, 11522–11527. [Google Scholar] [CrossRef]
- Yamamoto, H.; Naito, Y.; Okano, M.; Kanazawa, T.; Takematsu, H.; Kozutsumi, Y. Sphingosylphosphorylcholine and lysosulfatide have inverse regulatory functions in monocytic cell differentiation into macrophages. Arch. Biochem. Biophys. 2011, 506, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Rovina, P.; Graf, C.; Bornancin, F. Modulation of ceramide metabolism in mouse primary macrophages. Biochem. Biophys. Res. Commun. 2010, 399, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Bielawska, A.; Bell, R.M.; Hannun, Y.A. Role of ceramide as a lipid mediator of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J. Biol. Chem. 1990, 265, 15823–15831. [Google Scholar] [CrossRef]
- Wallner, S.; Grandl, M.; Liebisch, G.; Peer, M.; Orso, E.; Sigruner, A.; Sobota, A.; Schmitz, G. oxLDL and eLDL Induced Membrane Microdomains in Human Macrophages. PLoS ONE 2016, 11, e0166798. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Liao, J.J.; Graeler, M.; Huang, M.C.; Goetzl, E.J. Lysophospholipid regulation of mononuclear phagocytes. Biochim. Biophys. Acta 2002, 1582, 175–177. [Google Scholar] [CrossRef]
- Hohenhaus, D.M.; Schaale, K.; Le Cao, K.A.; Seow, V.; Iyer, A.; Fairlie, D.P.; Sweet, M.J. An mRNA atlas of G protein-coupled receptor expression during primary human monocyte/macrophage differentiation and lipopolysaccharide-mediated activation identifies targetable candidate regulators of inflammation. Immunobiology 2013, 218, 1345–1353. [Google Scholar] [CrossRef]
- Muller, J.; von Bernstorff, W.; Heidecke, C.D.; Schulze, T. Differential S1P Receptor Profiles on M1- and M2-Polarized Macrophages Affect Macrophage Cytokine Production and Migration. Biomed. Res. Int. 2017, 2017, 7584621. [Google Scholar] [CrossRef] [PubMed]
- Weigert, A.; Olesch, C.; Brune, B. Sphingosine-1-Phosphate and Macrophage Biology-How the Sphinx Tames the Big Eater. Front. Immunol. 2019, 10, 1706. [Google Scholar] [CrossRef]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast Cell: A Multi-Functional Master Cell. Front. Immunol. 2015, 6, 620. [Google Scholar] [CrossRef] [PubMed]
- Collington, S.J.; Williams, T.J.; Weller, C.L. Mechanisms underlying the localisation of mast cells in tissues. Trends Immunol. 2011, 32, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Boyce, J.A.; Mellor, E.A.; Perkins, B.; Lim, Y.C.; Luscinskas, F.W. Human mast cell progenitors use alpha4-integrin, VCAM-1, and PSGL-1 E-selectin for adhesive interactions with human vascular endothelium under flow conditions. Blood 2002, 99, 2890–2896. [Google Scholar] [CrossRef] [PubMed]
- Abonia, J.P.; Austen, K.F.; Rollins, B.J.; Joshi, S.K.; Flavell, R.A.; Kuziel, W.A.; Koni, P.A.; Gurish, M.F. Constitutive homing of mast cell progenitors to the intestine depends on autologous expression of the chemokine receptor CXCR2. Blood 2005, 105, 4308–4313. [Google Scholar] [CrossRef]
- Olivera, A.; Rivera, J. Sphingolipids and the balancing of immune cell function: Lessons from the mast cell. J. Immunol. 2005, 174, 1153–1158. [Google Scholar] [CrossRef]
- Irani, A.A.; Schechter, N.M.; Craig, S.S.; DeBlois, G.; Schwartz, L.B. Two types of human mast cells that have distinct neutral protease compositions. Proc. Natl. Acad. Sci. USA 1986, 83, 4464–4468. [Google Scholar] [CrossRef]
- Price, M.M.; Kapitonov, D.; Allegood, J.; Milstien, S.; Oskeritzian, C.A.; Spiegel, S. Sphingosine-1-phosphate induces development of functionally mature chymase-expressing human mast cells from hematopoietic progenitors. FASEB J. 2009, 23, 3506–3515. [Google Scholar] [CrossRef]
- Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 2010, 238, 37–46. [Google Scholar] [CrossRef]
- Kondo, M.; Scherer, D.C.; Miyamoto, T.; King, A.G.; Akashi, K.; Sugamura, K.; Weissman, I.L. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 2000, 407, 383–386. [Google Scholar] [CrossRef]
- Kondo, M.; Weissman, I.L.; Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997, 91, 661–672. [Google Scholar] [CrossRef]
- Igarashi, H.; Gregory, S.C.; Yokota, T.; Sakaguchi, N.; Kincade, P.W. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 2002, 17, 117–130. [Google Scholar] [CrossRef]
- Wallis, V.J.; Leuchars, E.; Chwalinski, S.; Davies, A.J. On the sparse seeding of bone marrow and thymus in radiation chimaeras. Transplantation 1975, 19, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Foss, D.L.; Donskoy, E.; Goldschneider, I. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J. Exp. Med. 2001, 193, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Donskoy, E.; Foss, D.; Goldschneider, I. Gated importation of prothymocytes by adult mouse thymus is coordinated with their periodic mobilization from bone marrow. J. Immunol. 2003, 171, 3568–3575. [Google Scholar] [CrossRef]
- Rocha, B. Characterization of V beta-bearing cells in athymic (nu/nu) mice suggests an extrathymic pathway for T cell differentiation. Eur. J. Immunol. 1990, 20, 919–925. [Google Scholar] [CrossRef]
- Rocha, B. The extrathymic T-cell differentiation in the murine gut. Immunol. Rev. 2007, 215, 166–177. [Google Scholar] [CrossRef]
- Masuda, K.; Itoi, M.; Amagai, T.; Minato, N.; Katsura, Y.; Kawamoto, H. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J. Immunol. 2005, 174, 2525–2532. [Google Scholar] [CrossRef]
- Vogel, P.; Donoviel, M.S.; Read, R.; Hansen, G.M.; Hazlewood, J.; Anderson, S.J.; Sun, W.; Swaffield, J.; Oravecz, T. Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS ONE 2009, 4, e4112. [Google Scholar] [CrossRef]
- Weber, C.; Krueger, A.; Munk, A.; Bode, C.; Van Veldhoven, P.P.; Graler, M.H. Discontinued postnatal thymocyte development in sphingosine 1-phosphate-lyase-deficient mice. J. Immunol. 2009, 183, 4292–4301. [Google Scholar] [CrossRef]
- Thangada, S.; Khanna, K.M.; Blaho, V.A.; Oo, M.L.; Im, D.S.; Guo, C.; Lefrancois, L.; Hla, T. Cell-surface residence of sphingosine 1-phosphate receptor 1 on lymphocytes determines lymphocyte egress kinetics. J. Exp. Med. 2010, 207, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Seki, N.; Sato, N.; Sugahara, K.; Chiba, K. Sphingosine 1-phosphate receptor type 1 regulates egress of mature T cells from mouse bone marrow. Int. Immunol. 2010, 22, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Pyne, N.J.; Pyne, S. Sphingosine 1-Phosphate Receptor 1 Signaling in Mammalian Cells. Molecules 2017, 22, 344. [Google Scholar] [CrossRef]
- Blaho, V.A.; Galvani, S.; Engelbrecht, E.; Liu, C.; Swendeman, S.L.; Kono, M.; Proia, R.L.; Steinman, L.; Han, M.H.; Hla, T. HDL-bound sphingosine-1-phosphate restrains lymphopoiesis and neuroinflammation. Nature 2015, 523, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Wiese, T.; Dennstadt, F.; Hollmann, C.; Stonawski, S.; Wurst, C.; Fink, J.; Gorte, E.; Mandasari, P.; Domschke, K.; Hommers, L.; et al. Inhibition of acid sphingomyelinase increases regulatory T cells in humans. Brain Commun. 2021, 3, fcab020. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Salker, M.S.; Walker, B.; Munzer, P.; Borst, O.; Gawaz, M.; Gulbins, E.; Singh, Y.; Lang, F. Acid Sphingomyelinase (ASM) is a Negative Regulator of Regulatory T Cell (Treg) Development. Cell Physiol. Biochem. 2016, 39, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, C.; Werner, S.; Avota, E.; Reuter, D.; Japtok, L.; Kleuser, B.; Gulbins, E.; Becker, K.A.; Schneider-Schaulies, J.; Beyersdorf, N. Inhibition of Acid Sphingomyelinase Allows for Selective Targeting of CD4+ Conventional versus Foxp3+ Regulatory T Cells. J. Immunol. 2016, 197, 3130–3141. [Google Scholar] [CrossRef]
- Goldfinger, M.; Laviad, E.L.; Hadar, R.; Shmuel, M.; Dagan, A.; Park, H.; Merrill, A.H., Jr.; Ringel, I.; Futerman, A.H.; Tirosh, B. De novo ceramide synthesis is required for N-linked glycosylation in plasma cells. J. Immunol. 2009, 182, 7038–7047. [Google Scholar] [CrossRef]
- Chen, Q.; Mosovsky, K.L.; Ross, A.C. Retinoic acid and alpha-galactosylceramide regulate the expression of costimulatory receptors and transcription factors responsible for B cell activation and differentiation. Immunobiology 2013, 218, 1477–1487. [Google Scholar] [CrossRef]
- Kleinwort, A.; Luhrs, F.; Heidecke, C.D.; Lipp, M.; Schulze, T. S1P Signalling Differentially Affects Migration of Peritoneal B Cell Populations In Vitro and Influences the Production of Intestinal IgA In Vivo. Int. J. Mol. Sci. 2018, 19, 391. [Google Scholar] [CrossRef]
- Saroha, A.; Pewzner-Jung, Y.; Ferreira, N.S.; Sharma, P.; Jouan, Y.; Kelly, S.L.; Feldmesser, E.; Merrill, A.H., Jr.; Trottein, F.; Paget, C.; et al. Critical Role for Very-Long Chain Sphingolipids in Invariant Natural Killer T Cell Development and Homeostasis. Front. Immunol. 2017, 8, 1386. [Google Scholar] [CrossRef]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, Y.; Salman, H.; Luberto, C. Sphingolipids in Hematopoiesis: Exploring Their Role in Lineage Commitment. Cells 2021, 10, 2507. https://doi.org/10.3390/cells10102507
Raza Y, Salman H, Luberto C. Sphingolipids in Hematopoiesis: Exploring Their Role in Lineage Commitment. Cells. 2021; 10(10):2507. https://doi.org/10.3390/cells10102507
Chicago/Turabian StyleRaza, Yasharah, Huda Salman, and Chiara Luberto. 2021. "Sphingolipids in Hematopoiesis: Exploring Their Role in Lineage Commitment" Cells 10, no. 10: 2507. https://doi.org/10.3390/cells10102507
APA StyleRaza, Y., Salman, H., & Luberto, C. (2021). Sphingolipids in Hematopoiesis: Exploring Their Role in Lineage Commitment. Cells, 10(10), 2507. https://doi.org/10.3390/cells10102507






