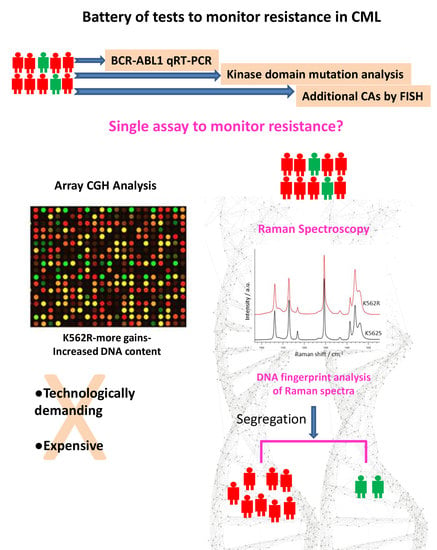
DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance
Abstract
:1. Introduction
2. Material and Methods
2.1. Development of Imatinib-Resistant Cells
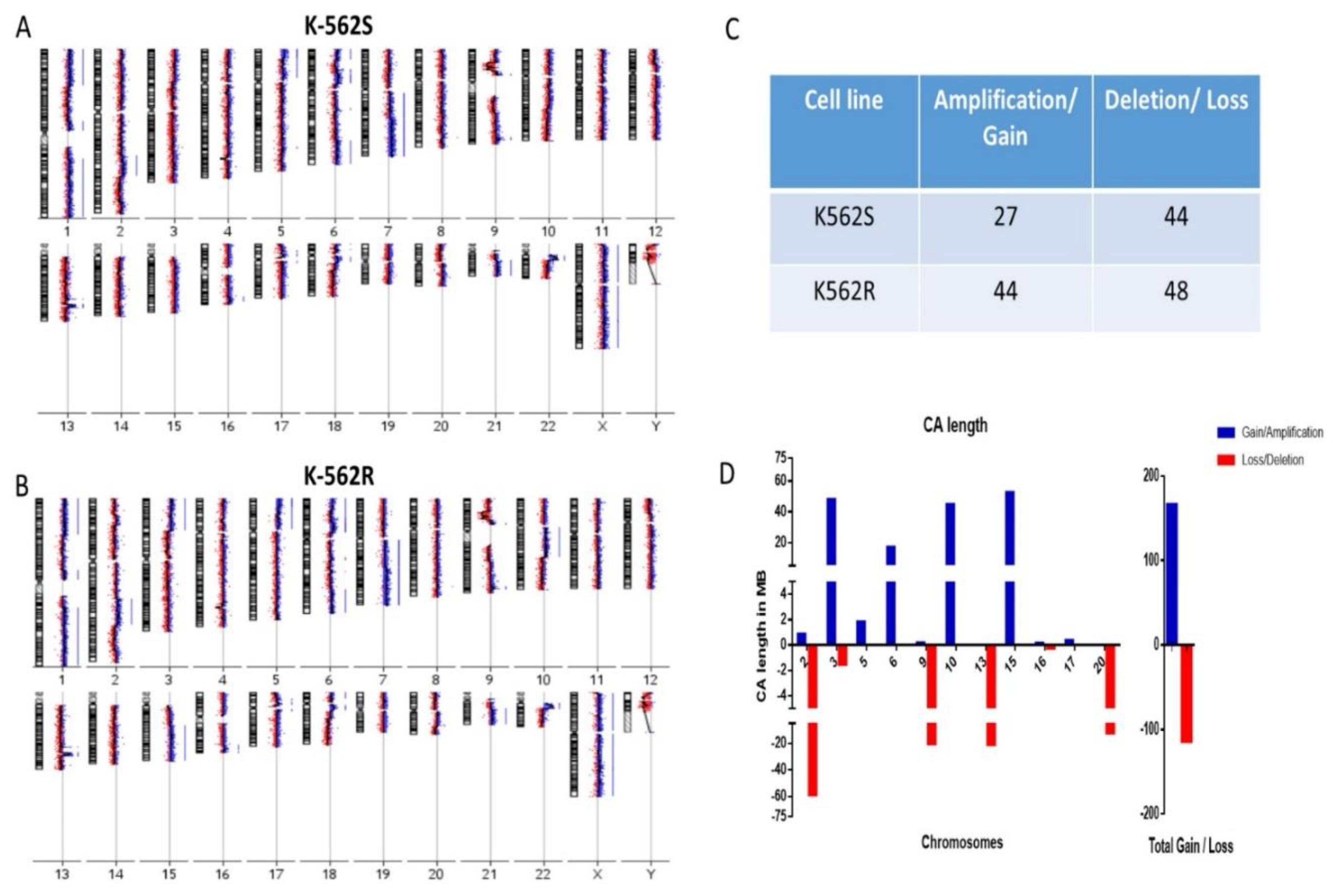
2.2. Array Comparative Genomic Hybridization Analysis
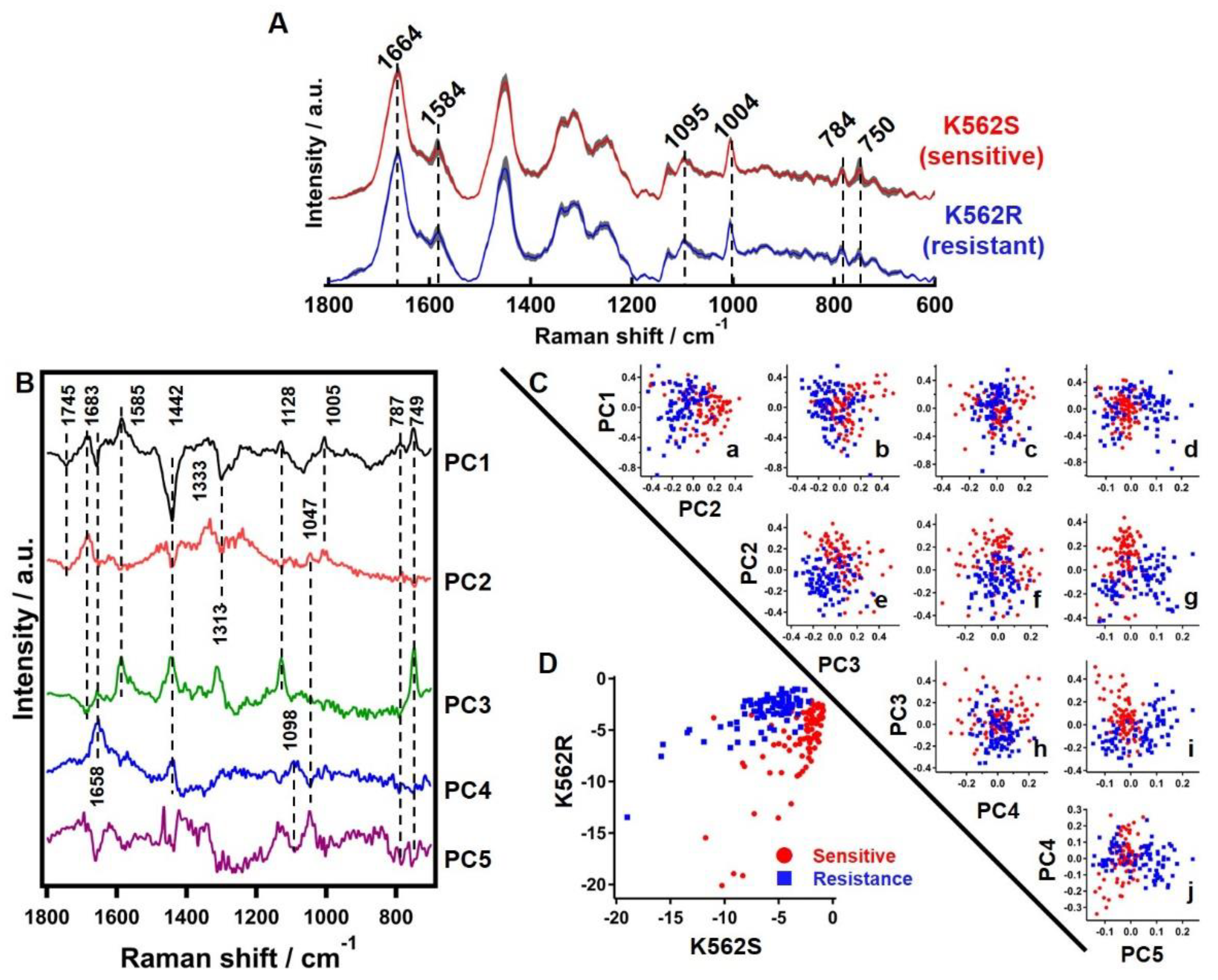
2.3. Raman Spectroscopy and Data Analysis
2.4. Multivariate Curve Resolution Analysis
3. Results
3.1. IC50 Was Ten-Fold Higher in Imatinib-Resistant Cells
3.2. Resistant Cells Showed Increased Chromosomal Gains in Genomic Analysis
3.3. Discrimination of Resistant Cells by Finger-Print Analysis of Raman Spectra
3.3.1. Average Raman Spectrum of Sensitive and Resistant Cells
3.3.2. Principle Component Analysis of Raman Hyperspectral Data
3.3.3. Linear Discriminant Analysis of Raman Hyperspectral Data
3.3.4. Multivariate Curve Resolution Analysis of Raman Spectra
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Talati, C.; Pinilla-Ibarz, J. Resistance in chronic myeloid leukemia, Definitions and novel therapeutic agents. Curr. Opin. Hematology 2018, 25, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am. J. Hematology 2018, 93, 442–459. [Google Scholar] [CrossRef] [Green Version]
- Quintas-Cardama, A.; Kantarjian, H.M.; Cortes, J.E. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control 2009, 16, 122–131. [Google Scholar] [CrossRef] [Green Version]
- An, X.; Tiwari, A.K.; Sun, Y.; Ding, P.R.; Ashby, C.R., Jr.; Chen, Z.S. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: A review. Leuk. Res. 2010, 34, 1255–1268. [Google Scholar] [CrossRef]
- Hehlmann, R. How I treat CML blast crisis. Blood 2012, 120, 737–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, D.; Ghosh, R.; Gupta, P.; Roychoudhury, S.; Nath, S. Incidence of imatinib resistance in chronic myeloid leukemia (CML) patients: Experience from resource poor center of eastern India. Ann. Oncol. 2019, 30, 1098. [Google Scholar] [CrossRef]
- Ganesan, P.; Kumar, L. Chronic Myeloid Leukemia in India. J. Glob. Oncol. 2017, 3, 64–71. [Google Scholar] [CrossRef]
- Loscocco, F.; Visani, G.; Galimberti, S.; Curti, A.; Isidori, A. BCR-ABL Independent Mechanisms of Resistance in Chronic Myeloid Leukemia. Front. Oncol. 2019, 9, 939. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, K.; Araki, A.; Krishna, C.M.; Maruyama, R.; Yamamoto, T.; Noothalapati, H. Identification of Molecular Basis for Objective Discrimination of Breast Cancer Cells (MCF-7) from Normal Human Mammary Epithelial Cells by Raman Microspectroscopy and Multivariate Curve Resolution Analysis. Int. J. Mol. Sci. 2021, 22, 800. [Google Scholar] [CrossRef]
- Cordero, E.; Latka, I.; Matthaus, C.; Schie, I.; Popp, J. In-vivo Raman spectroscopy: From basics to applications. J. Biomed. Opt. 2018, 23, 1–23. [Google Scholar] [CrossRef]
- Sahu, A.; Krishna, C.M. Optical diagnostics in oral cancer: An update on Raman spectroscopic applications. J. Cancer Res. Ther. 2017, 13, 908–915. [Google Scholar]
- Ramos, I.R.; Malkin, A.; Lyng, F.M. Current Advances in the Application of Raman Spectroscopy for Molecular Diagnosis of Cervical Cancer. BioMed Res. Int. 2015, 2015, 561242. [Google Scholar] [CrossRef]
- Noothalapati, H.; Iwasaki, K.; Yoshimoto, C.; Yoshikiyo, K.; Nishikawa, T.; Ando, M.; Hamaguchi, H.-O.; Yamamoto, T. Imaging phospholipid conformational disorder and packing in giant multilamellar liposome by confocal Raman microspectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 187, 186–190. [Google Scholar] [CrossRef]
- Noothalapati, H.; Ikarashi, R.; Iwasaki, K.; Nishida, T.; Kaino, T.; Yoshikiyo, K.; Terao, K.; Nakata, D.; Ikuta, N.; Ando, M.; et al. Studying anti-oxidative properties of inclusion complexes of alpha-lipoic acid with gamma-cyclodextrin in single living fission yeast by confocal Raman microspectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 197, 237–243. [Google Scholar] [CrossRef]
- Noothalapati, H.; Shigeto, S. Stable isotope-labeled Raman imaging reveals dynamic proteome localization to lipid droplets in single fission yeast cells. Chem. Biol. 2012, 19, 1373–1380. [Google Scholar] [CrossRef] [Green Version]
- Noothalapati, H.; Iwasaki, K.; Yamamoto, T. Non-invasive diagnosis of colorectal cancer by Raman spectroscopy: Recent developments in liquid biopsy and endoscopy approaches. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 258, 119818. [Google Scholar] [CrossRef]
- Laor, D.; Sade, D.; Shaham-Niv, S.; Zaguri, D.; Gartner, M.; Basavalingappa, V.; Raveh, A.; Pichinuk, E.; Engel, H.; Iwasaki, K.; et al. Fibril formation and therapeutic targeting of amyloid-like structures in a yeast model of adenine accumulation. Nat. Commun. 2019, 10, 62. [Google Scholar] [CrossRef] [Green Version]
- Zuniga, W.C.; Jones, V.; Anderson, S.M.; Echevarria, A.; Miller, N.L.; Stashko, C.; Schmolze, D.; Cha, P.D.; Kothari, R.; Fong, Y.; et al. Raman Spectroscopy for Rapid Evaluation of Surgical Margins during Breast Cancer Lumpectomy. Sci. Rep. 2019, 9, 14639. [Google Scholar] [CrossRef] [Green Version]
- Noothalapati, H.; Iwasaki, K.; Yamamoto, T. Biological and Medical Applications of Multivariate Curve Resolution Assisted Raman Spectroscopy. Anal. Sci. 2017, 33, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Noothalapati, H.; Sasaki, T.; Kaino, T.; Kawamukai, M.; Ando, M.; Hamaguchi, H.O.; Yamamoto, T. Label-free Chemical Imaging of Fungal Spore Walls by Raman Microscopy and Multivariate Curve Resolution Analysis. Sci. Rep. 2016, 6, 27789. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, K.; Kaneko, A.; Tanaka, Y.; Ishikawa, T.; Noothalapati, H.; Yamamoto, T. Visualizing wax ester fermentation in single Euglena gracilis cells by Raman microspectroscopy and multivariate curve resolution analysis. Biotechnol. Biofuels 2019, 12, 128. [Google Scholar] [CrossRef]
- Yasuda, M.; Takeshita, N.; Shigeto, S. Inhomogeneous Molecular Distributions and Cytochrome Types and Redox States in Fungal Cells Revealed by Raman Hyperspectral Imaging Using Multivariate Curve Resolution-Alternating Least Squares. Anal. Chem. 2019, 91, 12501–12508. [Google Scholar] [CrossRef]
- Deng, H.; Bloomfield, V.A.; Benevides, J.M.; Thomas, G.J., Jr. Dependence of the Raman signature of genomic B-DNA on nucleotide base sequence. Biopolymers 1999, 50, 656–666. [Google Scholar] [CrossRef]
- Huang, Y.S.; Karashima, T.; Yamamoto, M.; Hamaguchi, H. Molecular-level investigation of the structure, transformation, and bioactivity of single living fission yeast cells by time- and space-resolved Raman spectroscopy. Biochemistry 2005, 44, 10009–10019. [Google Scholar] [CrossRef]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Nambiar, M.; Kari, V.; Raghavan, S.C. Chromosomal translocations in cancer. Biochim. Biophys. Acta. 2008, 1786, 139–152. [Google Scholar] [CrossRef]
- Johansson, B.; Fioretos, T.; Mitelman, F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002, 107, 76–94. [Google Scholar] [CrossRef]
- Chandran, K.R.; Geetha, N.; Sakthivel, K.M.; Kumar, S.R.; Krishna, J.K.M.N.; Sreedharan, H. Impact of Additional Chromosomal Aberrations on the Disease Progression of Chronic Myelogenous Leukemia. Front. Oncol. 2019, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.T.; Ji, Y.; Kim, H.J.; Ki, C.S.; Jung, C.W.; Kim, J.W.; Kim, S.H. Sequential array comparative genomic hybridization analysis identifies copy number changes during blastic transformation of chronic myeloid leukemia. Leuk. Res. 2012, 36, 418–421. [Google Scholar] [CrossRef]
- Lu, X.; Wang, X.; Kim, Y.; Zhang, R.; Li, S.; Lee, J.Y. Acquired genomic copy number changes in CML patients with the Philadelphia chromosome (Ph+). Cancer Genet. 2012, 205, 513–518. [Google Scholar] [CrossRef]
- Marktel, S.; Marin, D.; Foot, N.; Szydlo, R.; Bua, M.; Karadimitris, A.; Melo, V.A.S.D.; Kotzampaltiris, P.; Dazzi, F.; Rahemtulla, A.; et al. Chronic myeloid leukemia in chronic phase responding to imatinib: The occurrence of additional cytogenetic abnormalities predicts disease progression. Haematologica 2003, 88, 260–267. [Google Scholar]
- Le Scouarnec, S.; Gribble, S.M. Characterising chromosome rearrangements: Recent technical advances in molecular cytogenetics. Heredity 2012, 108, 75–85. [Google Scholar] [CrossRef]
- Hemanth, N.; Suguru, U.; Naoki, O.; Yoshikazu, K.; Masahiro, A.; Hiro, H.; Yamamoto, T. Towards the development of a non-bioptic diagnostic technique for eosinophilic esophagitis using Raman spectroscopy. Vib. Spectrosc. 2016, 85, 7–10. [Google Scholar]
- Iwasaki, K.; Noothalapati, H.; Yamamoto, T. Chapter 15—Recent advances in Raman spectroscopy of proteins for disease diagnosis. In Vibrational Spectroscopy in Protein Research; Ozaki, Y., Baranska, M., Lednev, I.K., Wood, B.R., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 435–459. [Google Scholar]
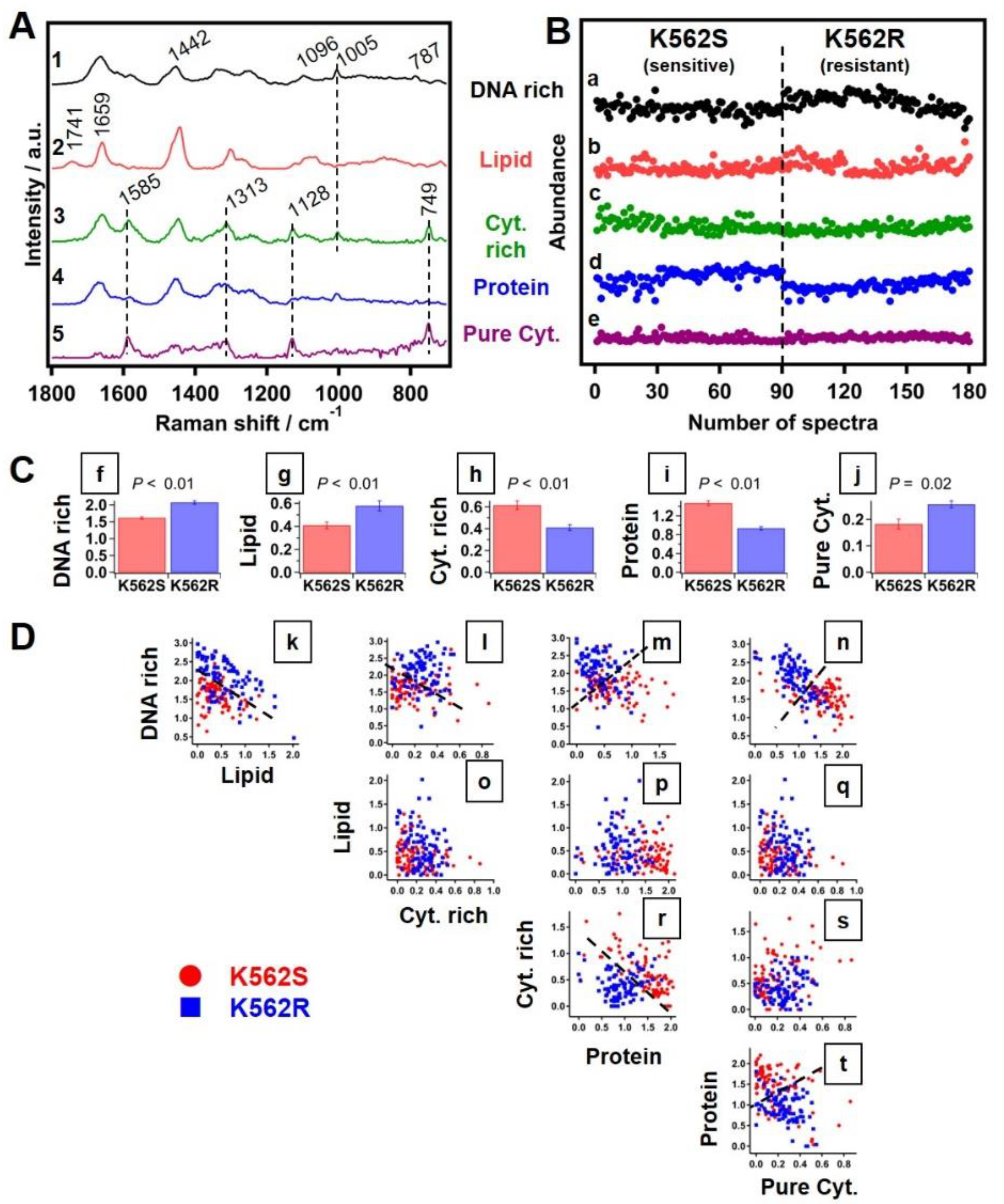
| Biomolecules | Wavenumbers |
|---|---|
| Proteins | 1005 and 1683 cm−1 |
| Lipids | 1442, 1658 and 1745 cm−1 |
| Cytochromes | 749, 1128, 1313 and 1585 cm−1 |
| DNA | 784 cm−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mojidra, R.; Hole, A.; Iwasaki, K.; Noothalapati, H.; Yamamoto, T.; C, M.K.; Govekar, R. DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance. Cells 2021, 10, 2506. https://doi.org/10.3390/cells10102506
Mojidra R, Hole A, Iwasaki K, Noothalapati H, Yamamoto T, C MK, Govekar R. DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance. Cells. 2021; 10(10):2506. https://doi.org/10.3390/cells10102506
Chicago/Turabian StyleMojidra, Rahul, Arti Hole, Keita Iwasaki, Hemanth Noothalapati, Tatsuyuki Yamamoto, Murali Krishna C, and Rukmini Govekar. 2021. "DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance" Cells 10, no. 10: 2506. https://doi.org/10.3390/cells10102506
APA StyleMojidra, R., Hole, A., Iwasaki, K., Noothalapati, H., Yamamoto, T., C, M. K., & Govekar, R. (2021). DNA Fingerprint Analysis of Raman Spectra Captures Global Genomic Alterations in Imatinib-Resistant Chronic Myeloid Leukemia: A Potential Single Assay for Screening Imatinib Resistance. Cells, 10(10), 2506. https://doi.org/10.3390/cells10102506