A Candidate RNAi Screen Reveals Diverse RNA-Binding Protein Phenotypes in Drosophila Flight Muscle
Abstract
:1. Introduction
1.1. RNA-Binding Proteins with Diverse Structures and Functions Regulate RNA Dynamics
1.2. Regulation of RBPs Modulates RNA Processing in Muscle
1.3. Drosophila as a Model to Identify and Study Muscle-Specific RBP Function
1.4. Fiber-Types and Development of the Drosophila Adult Musculature
2. Materials and Methods
2.1. Fly Stocks and Crosses
2.2. Selection of Candidate Genes for Screen
2.3. Lethality and Behavior Assays
2.4. Immunofluorescence and Microscopy
2.5. mRNA-Seq
2.6. Bioinformatics
3. Results
3.1. Muscle RBPs Are Expressed in Distinct Spatial and Temporal Patterns
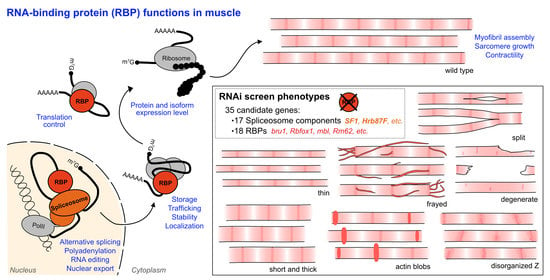
3.2. RNAi-Mediated Knockdown of Hundreds of RBPs Produces Muscle Phenotypes
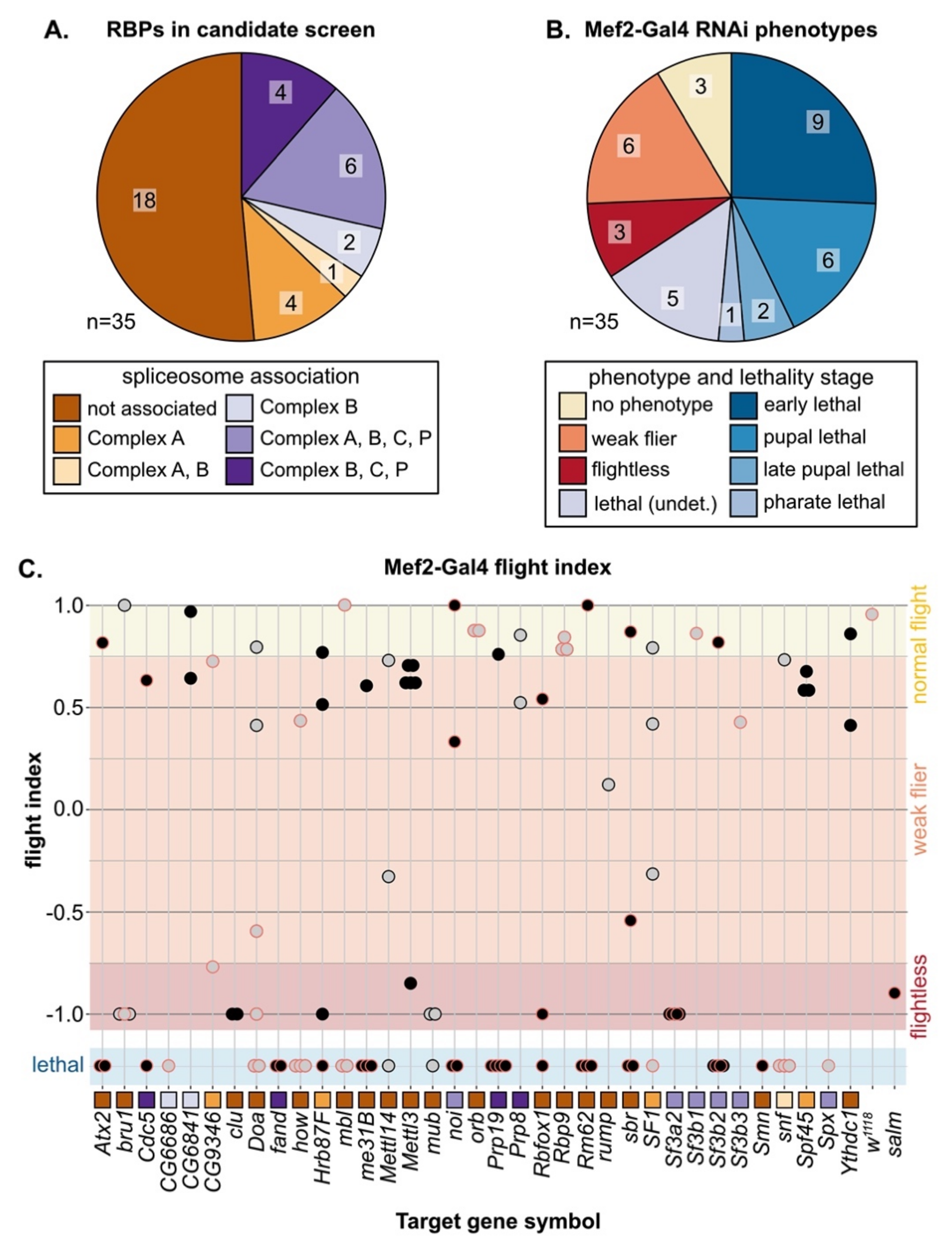
3.3. A Candidate RNAi-Screen for RBP and Spliceosome Component Phenotypes in Muscle
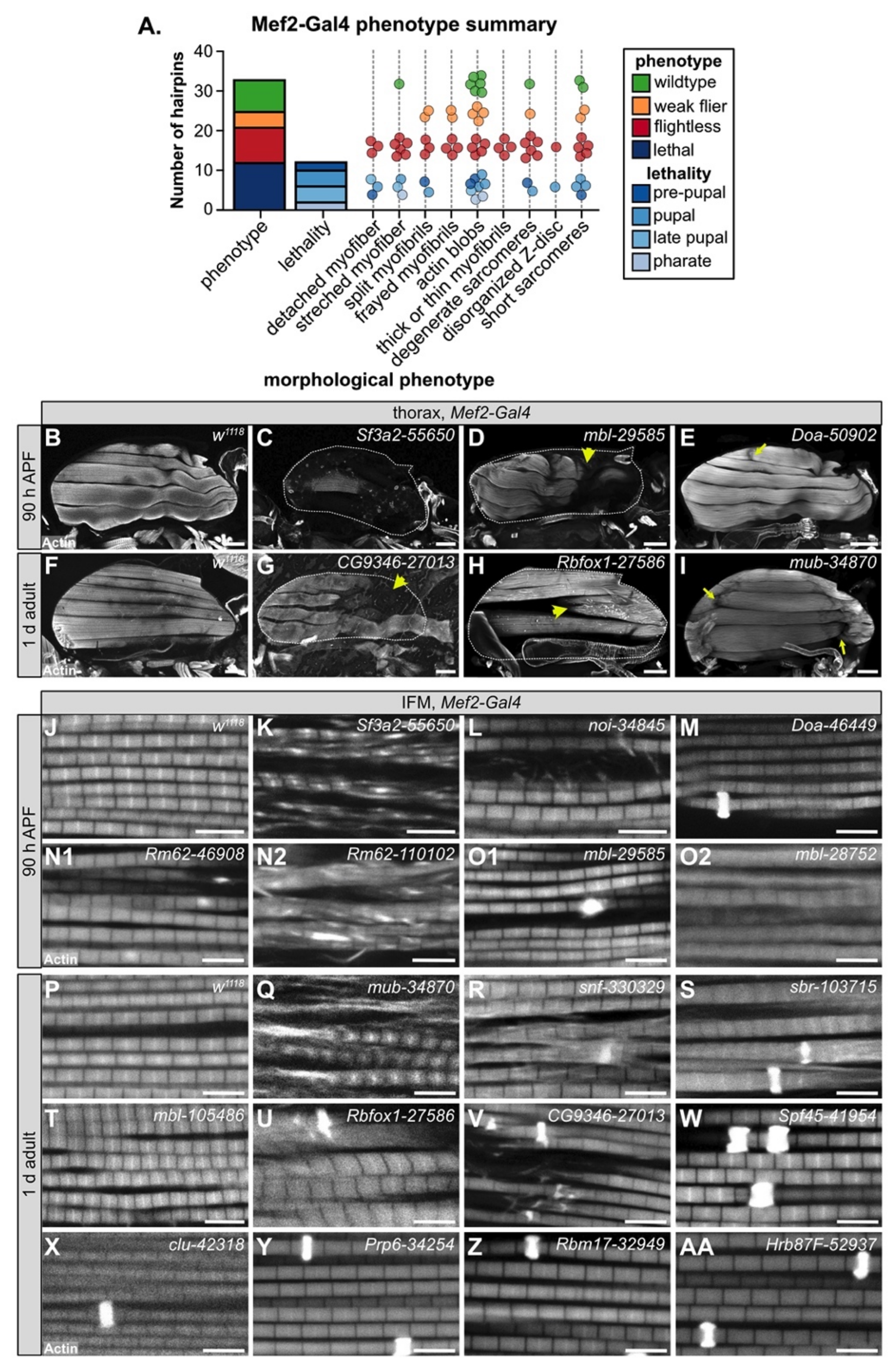
3.4. Muscle-Specific RBP Knockdown Leads to Diverse Myofibril and Sarcomere Defects
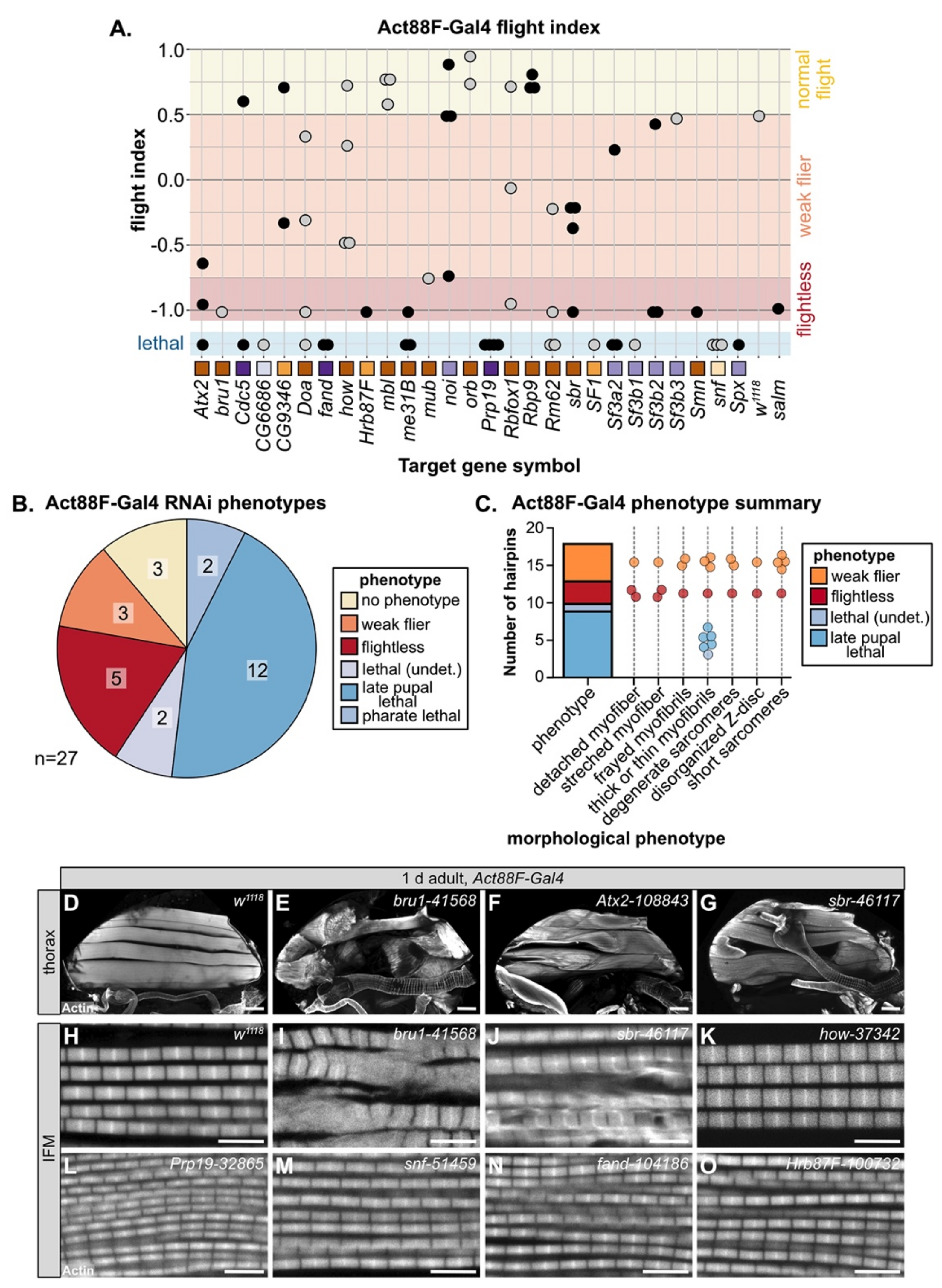
3.5. Temporally Restricted Knockdown of RBPs in IFM Using Act88F-Gal4
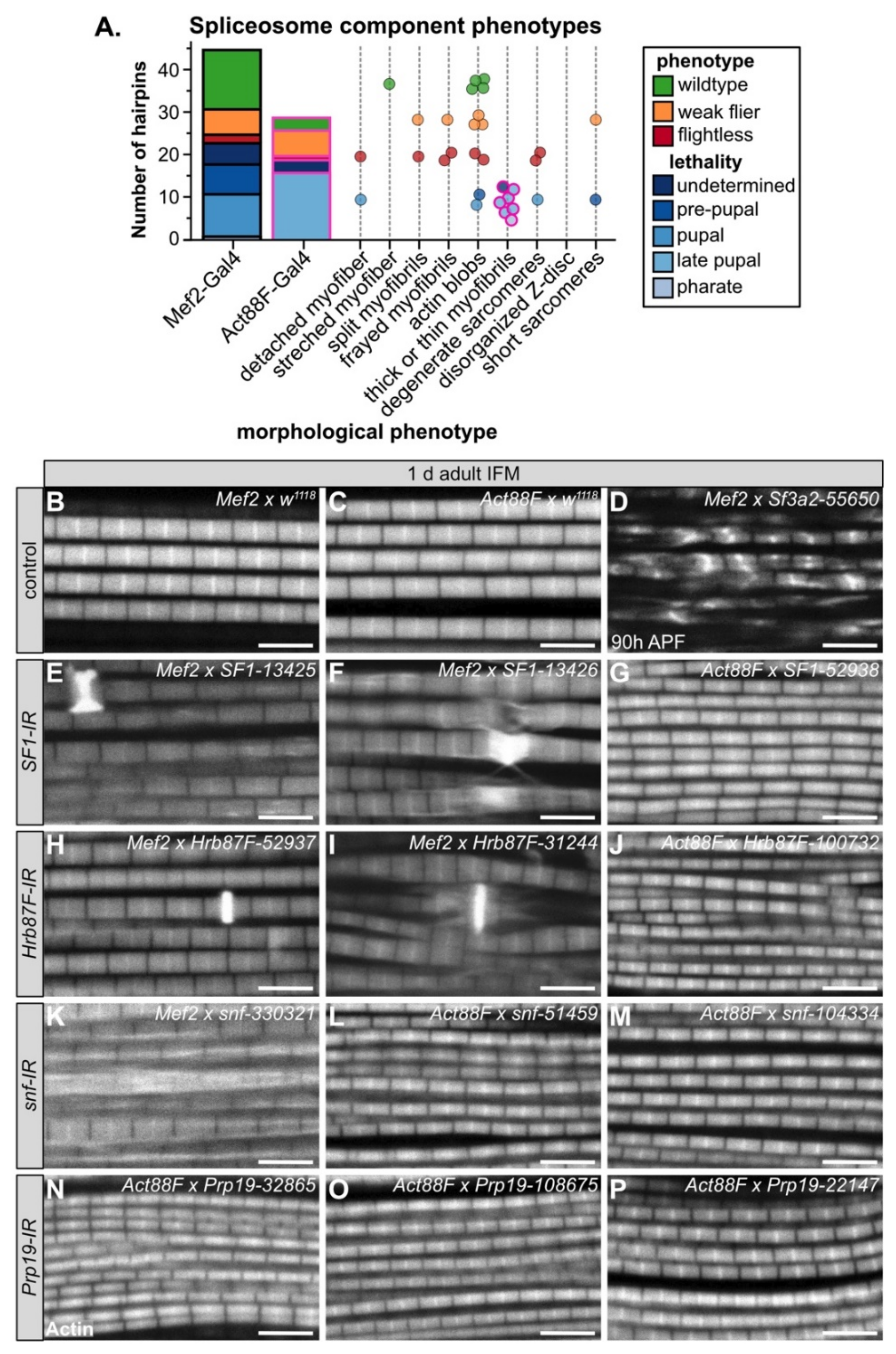
3.6. Diverse Phenotypes Associated with Altered Levels of Spliceosome Components
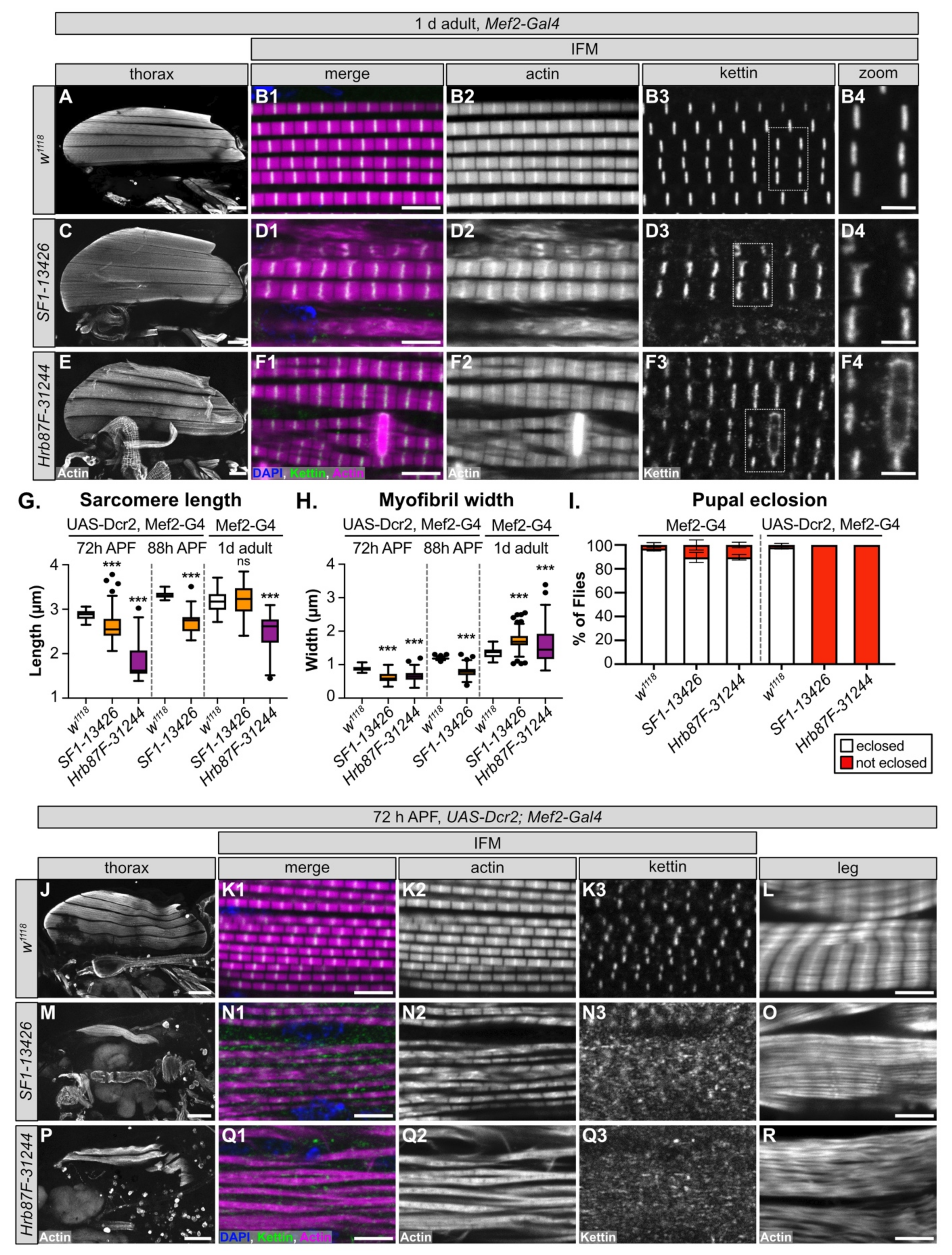
3.7. SF1 and Hrb87F Are Necessary for Myofibril Development and Z-disc Integrity
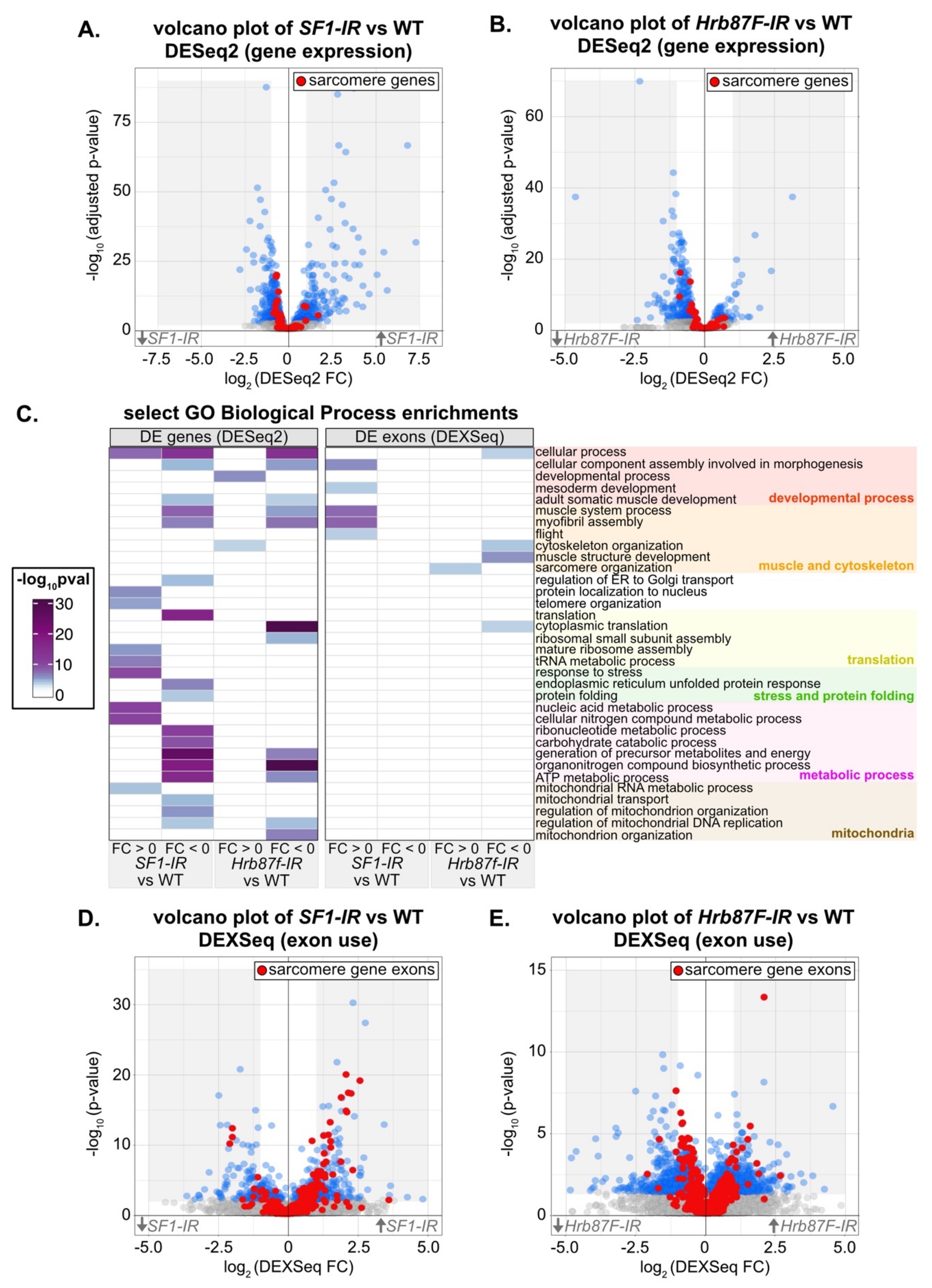
3.8. Transcriptomic Analysis Reveals Molecular Defects in SF1 and Hrb87F Knockdown IFM
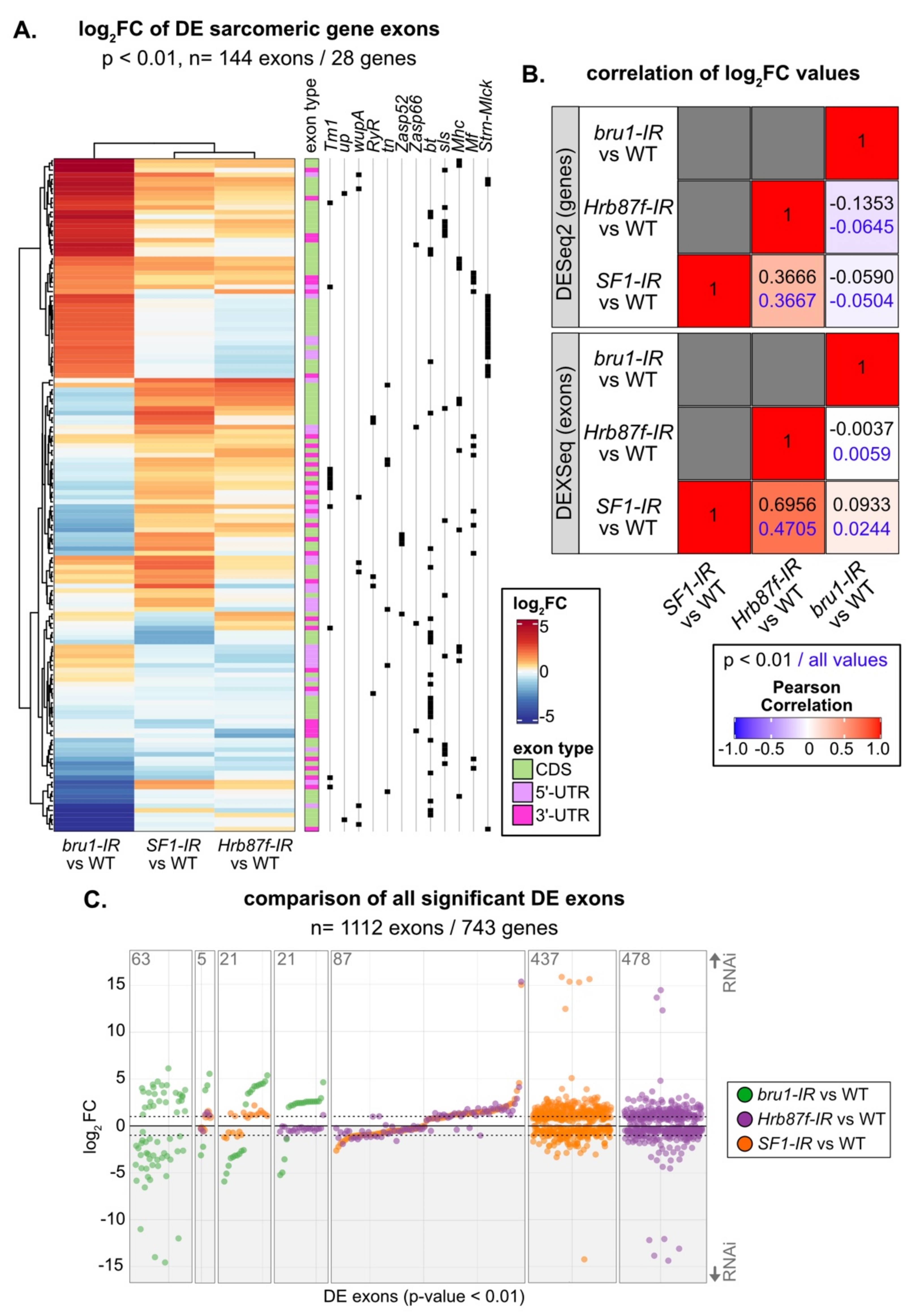
3.9. SF1, Hrb87F and Bru1 Regulate Distinct but Overlapping Splice Events
4. Discussion
4.1. Muscle Fiber-Type Specific Expression and Function of RBPs
4.2. Developmental Regulation of RBP Expression and Function during Myogenesis
4.3. Diversity in Muscle Phenotypes Likely Reflects RBP Target Specificity and Expression Level
4.4. Knockdown of Spliceosome Components Results in Alternative Splicing Defects in Muscle
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, J.; Olsen, E. Muscle, 1st ed.; Academic Press: Cambridge, MA, USA, 2012; ISBN 978-0-12-381511-8. [Google Scholar]
- Lemke, S.B.; Schnorrer, F. Mechanical Forces during Muscle Development. Mech. Dev. 2017, 144, 92–101. [Google Scholar] [CrossRef]
- Sanger, J.W.; Wang, J.; Fan, Y.; White, J.; Lei, M.-M.; Dube, D.K.; Sanger, J.M.; Pruyne, D. Assembly and Maintenance of Myofibrils in Striated Muscle. Actin Cytoskelet. 2017, 235, 39–75. [Google Scholar] [CrossRef]
- Nakka, K.; Ghigna, C.; Gabellini, D.; Dilworth, F.J. Diversification of the Muscle Proteome through Alternative Splicing. Skelet. Muscle 2018, 8, 1–18. [Google Scholar] [CrossRef]
- Narici, M.; Franchi, M.; Maganaris, C. Muscle Structural Assembly and Functional Consequences. J. Exp. Biol. 2016, 219, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Nikonova, E.; Kao, S.-Y.; Spletter, M.L. Contributions of Alternative Splicing to Muscle Type Development and Function. Semin. Cell Dev. Biol. 2020, 104, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautel, M.; Goulding, D. A Molecular Map of Titin/Connectin Elasticity Reveals Two Different Mechanisms Acting in Series. FEBS Lett. 1996, 385, 11–14. [Google Scholar] [CrossRef] [Green Version]
- Granzier, H.; Helmes, M.; Cazorla, O.; McNabb, M.; Labeit, D.; Wu, Y.; Yamasaki, R.; Redkar, A.; Kellermayer, M.; Labeit, S.; et al. Mechanical Properties of Titin Isoforms. Adv. Exp. Med. Biol. 2000, 481, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Cieply, B.; Carstens, R.P. Functional Roles of Alternative Splicing Factors in Human Disease. Wiley Interdiscip. Rev. RNA 2015, 6, 311–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikonova, E.; Kao, S.-Y.; Ravichandran, K.; Wittner, A.; Spletter, M.L. Conserved Functions of RNA-Binding Proteins in Muscle. Int. J. Biochem. Cell Biol. 2019, 110, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, V.; La Rosa, P.; Sette, C. Faulty RNA Splicing: Consequences and Therapeutic Opportunities in Brain and Muscle Disorders. Hum. Genet. 2017, 136, 1215–1235. [Google Scholar] [CrossRef]
- Scotti, M.M.; Swanson, M.S. RNA Mis-Splicing in Disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef]
- Abreu, P.; Mendes, S.V.D.; Ceccatto, V.M.; Hirabara, S.M. Satellite Cell Activation Induced by Aerobic Muscle Adaptation in Response to Endurance Exercise in Humans and Rodents. Life Sci. 2017, 170, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Plantié, E.; Migocka-Patrzałek, M.; Daczewska, M.; Jagla, K. Model Organisms in the Fight against Muscular Dystrophy: Lessons from Drosophila and Zebrafish. Molecules 2015, 20, 6237–6253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Pelt, D.W.; Hettinger, Z.R.; Vanderklish, P.W. RNA-Binding Proteins: The next Step in Translating Skeletal Muscle Adaptations? J. Appl. Physiol. 2019, 127, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.J.; Todorova, T.T.; Chang, P. RNA Regulation by Poly(ADP-Ribose) Polymerases. Mol. Cell 2015, 58, 959–969. [Google Scholar] [CrossRef] [Green Version]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.-Y.; Cody, N.A.L.; Dominguez, D.; et al. A Large-Scale Binding and Functional Map of Human RNA-Binding Proteins. Nature 2020, 583, 711–719. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.-M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [Green Version]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A Brave New World of RNA-Binding Proteins. Nat. Rev. Mol. Cell Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Damianov, A.; Ying, Y.; Lin, C.-H.; Lee, J.-A.; Tran, D.; Vashisht, A.A.; Bahrami-Samani, E.; Xing, Y.; Martin, K.C.; Wohlschlegel, J.A.; et al. Rbfox Proteins Regulate Splicing as Part of a Large Multiprotein Complex LASR. Cell 2016, 165, 606–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Green, M.R. RS Domains Contact Splicing Signalsand Promote Splicing by a Commonmechanism in Yeast through Humans. Genes Dev. 2006, 20, 1755–1765. [Google Scholar] [CrossRef] [Green Version]
- Picchiarelli, G.; Dupuis, L. Role of RNA Binding Proteins with Prion-like Domains in Muscle and Neuromuscular Diseases. Cell Stress 2020, 4, 76–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, A.N.; Duff, M.O.; May, G.; Yang, L.; Bolisetty, M.; Landolin, J.; Wan, K.; Sandler, J.; Booth, B.W.; Celniker, S.E.; et al. Regulation of Alternative Splicing in Drosophila by 56 RNA Binding Proteins. Genome Res. 2015, 25, 1771–1780. [Google Scholar] [CrossRef] [Green Version]
- Gazzara, M.R.; Mallory, M.J.; Roytenberg, R.; Lindberg, J.P.; Jha, A.; Lynch, K.W.; Barash, Y. Ancient Antagonism between CELF and RBFOX Families Tunes MRNA Splicing Outcomes. Genome Res. 2017, 27, 1360–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.T.; Ward, A.J.; Cherone, J.M.; Giudice, J.; Wang, T.T.; Treacy, D.J.; Lambert, N.J.; Freese, P.; Saxena, T.; Cooper, T.A.; et al. Antagonistic Regulation of MRNA Expression and Splicing by CELF and MBNL Proteins. Genome Res. 2015, 25, 858–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogler, T.O.; Wheeler, J.R.; Nguyen, E.D.; Hughes, M.P.; Britson, K.A.; Lester, E.; Rao, B.; Betta, N.D.; Whitney, O.N.; Ewachiw, T.E.; et al. TDP-43 and RNA Form Amyloid-like Myo-Granules in Regenerating Muscle. Nature 2018, 563, 508–513. [Google Scholar] [CrossRef]
- Wood, A.; Gurfinkel, Y.; Polain, N.; Lamont, W.; Lyn Rea, S. Molecular Mechanisms Underlying TDP-43 Pathology in Cellular and Animal Models of ALS and FTLD. Int. J. Mol. Sci. 2021, 22, 4705. [Google Scholar] [CrossRef]
- Wang, E.T.; Cody, N.A.L.; Jog, S.; Biancolella, M.; Wang, T.T.; Treacy, D.J.; Luo, S.; Schroth, G.P.; Housman, D.E.; Reddy, S.; et al. Transcriptome-Wide Regulation of Pre-MRNA Splicing and MRNA Localization by Muscleblind Proteins. Cell 2012, 150, 710–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webster, P.J.; Liang, L.; Berg, C.A.; Lasko, P.; Macdonald, P.M. Translational Repressor Bruno Plays Multiple Roles in Development and Is Widely Conserved. Genes Dev. 1997, 11, 2510–2521. [Google Scholar] [CrossRef] [Green Version]
- Kalsotra, A.; Xiao, X.; Ward, A.J.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. A Postnatal Switch of CELF and MBNL Proteins Reprograms Alternative Splicing in the Developing Heart. Proc. Natl. Acad. Sci. USA 2008, 105, 20333–20338. [Google Scholar] [CrossRef] [Green Version]
- Klinck, R.; Fourrier, A.; Thibault, P.; Toutant, J.; Durand, M.; Lapointe, E.; Caillet-Boudin, M.-L.; Sergeant, N.; Gourdon, G.; Meola, G.; et al. RBFOX1 Cooperates with MBNL1 to Control Splicing in Muscle, Including Events Altered in Myotonic Dystrophy Type 1. PLoS ONE 2014, 9, e107324. [Google Scholar] [CrossRef] [Green Version]
- Kalsotra, A.; Cooper, T.A. Functional Consequences of Developmentally Regulated Alternative Splicing. Nat. Rev. Genet. 2011, 12, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kong, X.; Lee, Y.M.; Zhang, M.K.; Guo, L.Y.; Lin, Y.; Lim, T.K.; Lin, Q.; Xu, X.Q. Stk38 Modulates Rbm24 Protein Stability to Regulate Sarcomere Assembly in Cardiomyocytes. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Hu, Y.; Comjean, A.; Perkins, L.A.; Perrimon, N.; Mohr, S.E. GLAD: An Online Database of Gene List Annotation for Drosophila. J. Genom. 2015, 3, 75–81. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology Consortium; Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- André, L.M.; Ausems, C.R.M.; Wansink, D.G.; Wieringa, B. Abnormalities in Skeletal Muscle Myogenesis, Growth, and Regeneration in Myotonic Dystrophy. Front. Neurol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Yeo, C.J.J.; Darras, B.T. Overturning the Paradigm of Spinal Muscular Atrophy as Just a Motor Neuron Disease. Pediatr. Neurol. 2020, 109, 12–19. [Google Scholar] [CrossRef]
- Chen, S.N.; Mestroni, L.; Taylor, M.R.G. Genetics of Dilated Cardiomyopathy. Curr. Opin. Cardiol. 2021, 36, 288–294. [Google Scholar] [CrossRef]
- Blech-Hermoni, Y.; Ladd, A.N. RNA Binding Proteins in the Regulation of Heart Development. Int. J. Biochem. Cell Biol. 2013, 45, 2467–2478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.V. Comparison of Muscle Development in Drosophila and Vertebrates. In Muscle Development in Drosophila; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Jagla, K.; Kalman, B.; Boudou, T.; Hénon, S.; Batonnet-Pichon, S. Beyond Mice: Emerging and Transdisciplinary Models for the Study of Early-Onset Myopathies. Semin. Cell Dev. Biol. 2017, 64, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Jawkar, S.; Nongthomba, U. Indirect Flight Muscles in Drosophila Melanogaster as a Tractable Model to Study Muscle Development and Disease. Int. J. Dev. Biol. 2020, 64, 167–173. [Google Scholar] [CrossRef]
- Lloyd, T.E.; Taylor, J.P. Flightless Flies: Drosophila Models of Neuromuscular Disease. Ann. N. Y. Acad. Sci. 2010, 1184, e1–e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya-Çopur, A.; Schnorrer, F. RNA Interference Screening for Genes Regulating Drosophila Muscle Morphogenesis. Methods Mol. Biol. 2019, 1889, 331–348. [Google Scholar] [CrossRef] [Green Version]
- Orfanos, Z. Transgenic Tools for Drosophila Muscle Research. J. Muscle Res. Cell Motil. 2008, 29, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Weitkunat, M.; Schnorrer, F. A Guide to Study Drosophila Muscle Biology. Methods 2014, 68, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Schulman, V.K.; Dobi, K.C.; Baylies, M.K. Morphogenesis of the Somatic Musculature in Drosophila Melanogaster. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 313–334. [Google Scholar] [CrossRef] [Green Version]
- Gunage, R.D.; Dhanyasi, N.; Reichert, H.; VijayRaghavan, K. Drosophila Adult Muscle Development and Regeneration. Semin. Cell Dev. Biol. 2017, 72, 56–66. [Google Scholar] [CrossRef]
- Gunage, R.D.; Reichert, H.; Vijay Raghavan, K. Identification of a New Stem Cell Population That Generates Drosophila Flight Muscles. Elife 2014, 3, e03126. [Google Scholar] [CrossRef]
- Kreipke, R.E.; Kwon, Y.V.; Shcherbata, H.R.; Ruohola-Baker, H. Drosophila Melanogaster as a Model of Muscle Degeneration Disorders. Curr. Top. Dev. Biol. 2017, 121, 83–109. [Google Scholar] [CrossRef]
- Deng, S.; Azevedo, M.; Baylies, M. Acting on Identity: Myoblast Fusion and the Formation of the Syncytial Muscle Fiber. Semin. Cell Dev. Biol. 2017, 72, 45–55. [Google Scholar] [CrossRef]
- Kim, J.H.; Jin, P.; Duan, R.; Chen, E.H. Mechanisms of Myoblast Fusion during Muscle Development. Curr. Opin. Genet. Dev. 2015, 32, 162–170. [Google Scholar] [CrossRef] [Green Version]
- Dobi, K.C.; Schulman, V.K.; Baylies, M.K. Specification of the Somatic Musculature in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 357–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weitkunat, M.; Kaya-Çopur, A.; Grill, S.W.; Schnorrer, F. Tension and Force-Resistant Attachment Are Essential for Myofibrillogenesis in Drosophila Flight Muscle. Curr. Biol. 2014, 24, 705–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasbiswas, K.; Hu, S.; Schnorrer, F.; Safran, S.A.; Bershadsky, A.D. Ordering of Myosin II Filaments Driven by Mechanical Forces: Experiments and Theory. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spletter, M.L.; Barz, C.; Yeroslaviz, A.; Zhang, X.; Lemke, S.B.; Bonnard, A.; Brunner, E.; Cardone, G.; Basler, K.; Habermann, B.H.; et al. A Transcriptomics Resource Reveals a Transcriptional Transition during Ordered Sarcomere Morphogenesis in Flight Muscle. Elife 2018, 7, e34058. [Google Scholar] [CrossRef] [PubMed]
- Loison, O.; Weitkunat, M.; Kaya-Çopur, A.; Nascimento Alves, C.; Matzat, T.; Spletter, M.L.; Luschnig, S.; Brasselet, S.; Lenne, P.-F.; Schnorrer, F. Polarization-Resolved Microscopy Reveals a Muscle Myosin Motor-Independent Mechanism of Molecular Actin Ordering during Sarcomere Maturation. PLoS Biol. 2018, 16, e2004718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weitkunat, M.; Brasse, M.; Bausch, A.R.; Schnorrer, F. Mechanical Tension and Spontaneous Muscle Twitching Precede the Formation of Cross-Striated Muscle in Vivo. Development 2017, 144, 1261–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya-Çopur, A.; Marchiano, F.; Hein, M.Y.; Alpern, D.; Russeil, J.; Luis, N.M.; Mann, M.; Deplancke, B.; Habermann, B.H.; Schnorrer, F. The Hippo Pathway Controls Myofibril Assembly and Muscle Fiber Growth by Regulating Sarcomeric Gene Expression. Elife 2021, 10, e63726. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C.; Murgia, M. Fiber Type Diversity in Skeletal Muscle Explored by Mass Spectrometry-Based Single Fiber Proteomics. Histol. Histopathol. 2020, 35, 239–246. [Google Scholar] [CrossRef]
- Marden, J.H. Functional and Ecological Effects of Isoform Variation in Insect Flight Muscle. In Nature’s Versatile Engine: Insect Flight Muscle Inside and Out; Vigoreaux, J.O., Ed.; Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2006; pp. 214–229. ISBN 978-0-387-31213-2. [Google Scholar]
- Spletter, M.L.; Schnorrer, F. Transcriptional Regulation and Alternative Splicing Cooperate in Muscle Fiber-Type Specification in Flies and Mammals. Exp. Cell Res. 2014, 321, 90–98. [Google Scholar] [CrossRef] [Green Version]
- Vigoreaux, J.O. Genetics of the Drosophila Flight Muscle Myofibril: A Window into the Biology of Complex Systems. Bioessays 2001, 23, 1047–1063. [Google Scholar] [CrossRef] [PubMed]
- Josephson, R.K. Comparative Physiology of Insect Flight Muscle. In Nature’s Versatile Engine: Insect Flight Muscle Inside and Out; Vigoreaux, J.O., Ed.; Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2006; pp. 34–43. ISBN 978-0-387-31213-2. [Google Scholar]
- Josephson, R.K.; Malamud, J.G.; Stokes, D.R. Asynchronous Muscle: A Primer. J. Exp. Biol. 2000, 203, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Maughan, D.; Swank, D. Insect Flight Muscle Chemomechanics. In Nature’s Versatile Engine: Insect Flight Muscle Inside and Out; Vigoreaux, J.O., Ed.; Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2006; pp. 251–269. ISBN 978-0-387-31213-2. [Google Scholar]
- Sarov, M.; Barz, C.; Jambor, H.; Hein, M.Y.; Schmied, C.; Suchold, D.; Stender, B.; Janosch, S.; KJ, V.V.; Krishnan, R.T.; et al. A Genome-Wide Resource for the Analysis of Protein Localisation in Drosophila. Elife 2016, 5, e12068. [Google Scholar] [CrossRef] [PubMed]
- Chechenova, M.B.; Maes, S.; Oas, S.T.; Nelson, C.; Kiani, K.G.; Bryantsev, A.L.; Cripps, R.M. Functional Redundancy and Nonredundancy between Two Troponin C Isoforms in Drosophila Adult Muscles. MBoC 2017, 28, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Eldred, C.C.; Simeonov, D.R.; Koppes, R.A.; Yang, C.; Corr, D.T.; Swank, D.M. The Mechanical Properties of Drosophila Jump Muscle Expressing Wild-Type and Embryonic Myosin Isoforms. Biophys. J. 2010, 98, 1218–1226. [Google Scholar] [CrossRef] [Green Version]
- Avellaneda, J.; Rodier, C.; Daian, F.; Brouilly, N.; Rival, T.; Luis, N.M.; Schnorrer, F. Myofibril and Mitochondria Morphogenesis Are Coordinated by a Mechanical Feedback Mechanism in Muscle. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Katti, P.; Rai, M.; Srivastava, S.; D’Silva, P.; Nongthomba, U. Marf-Mediated Mitochondrial Fusion Is Imperative for the Development and Functioning of Indirect Flight Muscles (IFMs) in Drosophila. Exp. Cell Res. 2021, 399, 112486. [Google Scholar] [CrossRef]
- Schönbauer, C.; Distler, J.; Jährling, N.; Radolf, M.; Dodt, H.-U.; Frasch, M.; Schnorrer, F. Spalt Mediates an Evolutionarily Conserved Switch to Fibrillar Muscle Fate in Insects. Nature 2011, 479, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Reedy, M.C.; Beall, C. Ultrastructure of Developing Flight Muscle in Drosophila. I. Assembly of Myofibrils. Dev. Biol. 1993, 160, 443–465. [Google Scholar] [CrossRef]
- Spletter, M.L.; Barz, C.; Yeroslaviz, A.; Schönbauer, C.; Ferreira, I.R.S.; Sarov, M.; Gerlach, D.; Stark, A.; Habermann, B.H.; Schnorrer, F. The RNA-Binding Protein Arrest (Bruno) Regulates Alternative Splicing to Enable Myofibril Maturation in Drosophila Flight Muscle. EMBO Rep. 2015, 16, 178–191. [Google Scholar] [CrossRef]
- De Joussineau, C.; Bataillé, L.; Jagla, T.; Jagla, K. Diversification of Muscle Types in Drosophila: Upstream and Downstream of Identity Genes. Curr. Top. Dev. Biol. 2012, 98, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, T.; Soler, C.; Jagla, T.; Daczewska, M.; Lodha, N.; Palliyil, S.; VijayRaghavan, K.; Jagla, K. Shaping Leg Muscles in Drosophila: Role of Ladybird, a Conserved Regulator of Appendicular Myogenesis. PLoS ONE 2006, 1, e122. [Google Scholar] [CrossRef] [PubMed]
- Sudarsan, V.; Anant, S.; Guptan, P.; VijayRaghavan, K.; Skaer, H. Myoblast Diversification and Ectodermal Signaling in Drosophila. Dev. Cell 2001, 1, 829–839. [Google Scholar] [CrossRef]
- Bernard, F.; Lalouette, A.; Gullaud, M.; Jeantet, A.Y.; Cossard, R.; Zider, A.; Ferveur, J.F.; Silber, J. Control of Apterous by Vestigial Drives Indirect Flight Muscle Development in Drosophila. Dev. Biol. 2003, 260, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Bryantsev, A.L.; Duong, S.; Brunetti, T.M.; Chechenova, M.B.; Lovato, T.L.; Nelson, C.; Shaw, E.; Uhl, J.D.; Gebelein, B.; Cripps, R.M. Extradenticle and Homothorax Control Adult Muscle Fiber Identity in Drosophila. Dev. Cell 2012, 23, 664–673. [Google Scholar] [CrossRef] [Green Version]
- Oas, S.T.; Bryantsev, A.L.; Cripps, R.M. Arrest Is a Regulator of Fiber-Specific Alternative Splicing in the Indirect Flight Muscles of Drosophila. J. Cell Biol. 2014, 206, 895–908. [Google Scholar] [CrossRef] [Green Version]
- Venables, J.P.; Tazi, J.; Juge, F. Regulated Functional Alternative Splicing in Drosophila. Nucleic Acids Res. 2012, 40, 1–10. [Google Scholar] [CrossRef]
- Ranganayakulu, G.; Schulz, R.A.; Olson, E.N. Wingless Signaling Induces Nautilus Expression in the Ventral Mesoderm of the Drosophila Embryo. Dev. Biol. 1996, 176, 143–148. [Google Scholar] [CrossRef] [Green Version]
- Bryantsev, A.L.; Baker, P.W.; Lovato, T.L.; Jaramillo, M.S.; Cripps, R.M. Differential Requirements for Myocyte Enhancer Factor-2 during Adult Myogenesis in Drosophila. Dev. Biol. 2012, 361, 191–207. [Google Scholar] [CrossRef] [Green Version]
- Cvitkovic, I.; Jurica, M.S. Spliceosome Database: A Tool for Tracking Components of the Spliceosome. Nucleic Acids Res. 2013, 41, D132–D141. [Google Scholar] [CrossRef] [Green Version]
- Schnorrer, F.; Schönbauer, C.; Langer, C.C.H.; Dietzl, G.; Novatchkova, M.; Schernhuber, K.; Fellner, M.; Azaryan, A.; Radolf, M.; Stark, A.; et al. Systematic Genetic Analysis of Muscle Morphogenesis and Function in Drosophila. Nature 2010, 464, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, E.; Kamble, K.; Mukherjee, A.; Barz, C.; Nongthomba, U.; Spletter, M.L. Rbfox1 Is Required for Myofibril Development and Maintaining Fiber-Type Specific Isoform Expression in Drosophila Muscles. bioRxiv 2021. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A Web-Server to Retrieve and Display the Repeatedly Occurring Neighbourhood of a Gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Lence, T.; Soller, M.; Roignant, J.-Y. A Fly View on the Roles and Mechanisms of the M6A MRNA Modification and Its Players. RNA Biol. 2017, 14, 1232–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Wei, L.; Zhang, B.; Goering, R.; Majumdar, S.; Wen, J.; Taliaferro, J.M.; Lai, E.C. ELAV/Hu RNA Binding Proteins Determine Multiple Programs of Neural Alternative Splicing. PLoS Genet. 2021, 17, e1009439. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, S.-Y.; Nikonova, E.; Ravichandran, K.; Spletter, M.L. Dissection of Drosophila Melanogaster Flight Muscles for Omics Approaches. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [Green Version]
- Kumar, L.; Futschik, M.E. Mfuzz: A Software Package for Soft Clustering of Microarray Data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting Differential Usage of Exons from RNA-Seq Data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A Tool for Discovery and Visualization of Enriched GO Terms in Ranked Gene Lists. BMC Bioinform. 2009, 10, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montana, E.S.; Littleton, J.T. Expression Profiling of a Hypercontraction-Induced Myopathy in Drosophila Suggests a Compensatory Cytoskeletal Remodeling Response. J. Biol. Chem. 2006, 281, 8100–8109. [Google Scholar] [CrossRef] [Green Version]
- Nongthomba, U.; Cummins, M.; Clark, S.; Vigoreaux, J.O.; Sparrow, J.C. Suppression of Muscle Hypercontraction by Mutations in the Myosin Heavy Chain Gene of Drosophila Melanogaster. Genetics 2003, 164, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, A.M.; Cygan, K.J.; Brown, B.A.; Fairbrother, W.G. RNA-Binding Proteins: Splicing Factors and Disease. Biomolecules 2015, 5, 893–909. [Google Scholar] [CrossRef] [Green Version]
- Mazroui, R.; Puoti, A.; Krämer, A. Splicing Factor SF1 from Drosophila and Caenorhabditis: Presence of an N-Terminal RS Domain and Requirement for Viability. RNA 1999, 5, 1615–1631. [Google Scholar] [CrossRef] [Green Version]
- Rymond, B.C. The Branchpoint Binding Protein: In and out of the Spliceosome Cycle. Adv. Exp. Med. Biol. 2010, 693, 123–141. [Google Scholar] [PubMed]
- Howard, J.M.; Lin, H.; Wallace, A.J.; Kim, G.; Draper, J.M.; Haeussler, M.; Katzman, S.; Toloue, M.; Liu, Y.; Sanford, J.R. HNRNPA1 Promotes Recognition of Splice Site Decoys by U2AF2 In Vivo. Genome Res. 2018, 28, 689–698. [Google Scholar] [CrossRef] [Green Version]
- Tavanez, J.P.; Madl, T.; Kooshapur, H.; Sattler, M.; Valcárcel, J. HnRNP A1 Proofreads 3′ Splice Site Recognition by U2AF. Mol. Cell 2012, 45, 314–329. [Google Scholar] [CrossRef]
- Kaya-Çopur, A.; Schnorrer, F. A Guide to Genome-Wide In Vivo RNAi Applications in Drosophila. Methods Mol. Biol. 2016, 1478, 117–143. [Google Scholar] [CrossRef]
- Katzemich, A.; Liao, K.A.; Czerniecki, S.; Schöck, F. Alp/Enigma Family Proteins Cooperate in Z-Disc Formation and Myofibril Assembly. PLoS Genet. 2013, 9, e1003342. [Google Scholar] [CrossRef] [Green Version]
- Liao, K.A.; González-Morales, N.; Schöck, F. Zasp52, a Core Z-Disc Protein in Drosophila Indirect Flight Muscles, Interacts with α-Actinin via an Extended PDZ Domain. PLoS Genet. 2016, 12, e1006400. [Google Scholar] [CrossRef] [PubMed]
- Szikora, S.; Gajdos, T.; Novák, T.; Farkas, D.; Földi, I.; Lenart, P.; Erdélyi, M.; Mihály, J. Nanoscopy Reveals the Layered Organization of the Sarcomeric H-Zone and I-Band Complexes. J. Cell Biol. 2020, 219, e201907026. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, E.R.; Wiedner, H.J.; Black, A.J.; Giudice, J. RNA Processing in Skeletal Muscle Biology and Disease. Transcription 2019, 10, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kino, Y.; Washizu, C.; Kurosawa, M.; Oma, Y.; Hattori, N.; Ishiura, S.; Nukina, N. Nuclear Localization of MBNL1: Splicing-Mediated Autoregulation and Repression of Repeat-Derived Aberrant Proteins. Hum. Mol. Genet. 2015, 24, 740–756. [Google Scholar] [CrossRef] [Green Version]
- Fardaei, M.; Larkin, K.; Brook, J.D.; Hamshere, M.G. In Vivo Co-Localisation of MBNL Protein with DMPK Expanded-Repeat Transcripts. Nucleic Acids Res. 2001, 29, 2766–2771. [Google Scholar] [CrossRef] [Green Version]
- Timchenko, N.A.; Cai, Z.-J.; Welm, A.L.; Reddy, S.; Ashizawa, T.; Timchenko, L.T. RNA CUG Repeats Sequester CUGBP1 and Alter Protein Levels and Activity of CUGBP1. J. Biol. Chem. 2001, 276, 7820–7826. [Google Scholar] [CrossRef] [Green Version]
- Ozimski, L.L.; Sabater-Arcis, M.; Bargiela, A.; Artero, R. The Hallmarks of Myotonic Dystrophy Type 1 Muscle Dysfunction. Biol. Rev. 2021, 96, 716–730. [Google Scholar] [CrossRef]
- Imbriano, C.; Molinari, S. Alternative Splicing of Transcription Factors Genes in Muscle Physiology and Pathology. Genes 2018, 9, 107. [Google Scholar] [CrossRef] [Green Version]
- Bachinski, L.L.; Sirito, M.; Böhme, M.; Baggerly, K.A.; Udd, B.; Krahe, R. Altered MEF2 Isoforms in Myotonic Dystrophy and Other Neuromuscular Disorders. Muscle Nerve 2010, 42, 856–863. [Google Scholar] [CrossRef]
- Rampalli, S.; Li, L.; Mak, E.; Ge, K.; Brand, M.; Tapscott, S.J.; Dilworth, F.J. P38 MAPK Signaling Regulates Recruitment of Ash2L-Containing Methyltransferase Complexes to Specific Genes during Differentiation. Nat. Struct. Mol. Biol. 2007, 14, 1150–1156. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, B.; Davie, J. Alternative Splicing of MEF2C Pre-MRNA Controls Its Activity in Normal Myogenesis and Promotes Tumorigenicity in Rhabdomyosarcoma Cells. J. Biol. Chem. 2015, 290, 310–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zappia, M.P.; Frolov, M.V. E2F Function in Muscle Growth Is Necessary and Sufficient for Viability in Drosophila. Nat. Commun. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orfanos, Z.; Sparrow, J.C. Myosin Isoform Switching during Assembly of the Drosophila Flight Muscle Thick Filament Lattice. J. Cell Sci. 2013, 126, 139–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Morales, N.; Xiao, Y.S.; Schilling, M.A.; Marescal, O.; Liao, K.A.; Schöck, F. Myofibril Diameter Is Set by a Finely Tuned Mechanism of Protein Oligomerization in Drosophila. Elife 2019, 8, e50496. [Google Scholar] [CrossRef]
- Artero, R.; Prokop, A.; Paricio, N.; Begemann, G.; Pueyo, I.; Mlodzik, M.; Perez-Alonso, M.; Baylies, M.K. The Muscleblind Gene Participates in the Organization of Z-Bands and Epidermal Attachments of Drosophila Muscles and Is Regulated by Dmef2. Dev. Biol. 1998, 195, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Nir, R.; Grossman, R.; Paroush, Z.; Volk, T. Phosphorylation of the Drosophila Melanogaster RNA-Binding Protein HOW by MAPK/ERK Enhances Its Dimerization and Activity. PLoS Genet. 2012, 8, e1002632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvinge, H.; Kim, E.; Abdel-Wahab, O.; Bradley, R.K. RNA Splicing Factors as Oncoproteins and Tumour Suppressors. Nat. Rev. Cancer 2016, 16, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Braunschweig, U.; Gonatopoulos-Pournatzis, T.; Weatheritt, R.J.; Hirsch, C.L.; Ha, K.C.H.; Radovani, E.; Nabeel-Shah, S.; Sterne-Weiler, T.; Wang, J.; et al. Multilayered Control of Alternative Splicing Regulatory Networks by Transcription Factors. Mol. Cell 2017, 65, 539–553. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, K.; Nimura, K. Regulation of RNA Splicing: Aberrant Splicing Regulation and Therapeutic Targets in Cancer. Cells 2021, 10, 923. [Google Scholar] [CrossRef] [PubMed]
- Dvinge, H.; Guenthoer, J.; Porter, P.L.; Bradley, R.K. RNA Components of the Spliceosome Regulate Tissue- and Cancer-Specific Alternative Splicing. Genome Res. 2019, 29, 1591–1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chechenova, M.B.; Bryantsev, A.L.; Cripps, R.M. The Drosophila Z-Disc Protein Z(210) Is an Adult Muscle Isoform of Zasp52, Which Is Required for Normal Myofibril Organization in Indirect Flight Muscles. J. Biol. Chem. 2013, 288, 3718–3726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Morales, N.; Holenka, T.K.; Schöck, F. Filamin Actin-Binding and Titin-Binding Fulfill Distinct Functions in Z-Disc Cohesion. PLoS Genet. 2017, 13, e1006880. [Google Scholar] [CrossRef]
- Bullard, B.; Leake, M.C.; Leonard, K. Some Functions of Proteins from the Drosophila sallimus (sls) Gene. In Nature’s Versatile Engine: Insect Flight Muscle Inside and Out; Vigoreaux, J.O., Ed.; Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2006; pp. 177–186. ISBN 978-0-387-31213-2. [Google Scholar]
- Liu, T.-Y.; Chen, Y.-C.; Jong, Y.-J.; Tsai, H.-J.; Lee, C.-C.; Chang, Y.-S.; Chang, J.-G.; Chang, Y.-F. Muscle Developmental Defects in Heterogeneous Nuclear Ribonucleoprotein A1 Knockout Mice. Open Biol. 2017, 7, 160303. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Shen, L.; Ouyang, Z.; Li, M.; Deng, G.; Yang, C.; Zheng, W.; Kong, L.; Wu, X.; Wu, X.; et al. Loss of HnRNP A1 in Murine Skeletal Muscle Exacerbates High-Fat Diet-Induced Onset of Insulin Resistance and Hepatic Steatosis. J. Mol. Cell Biol. 2020, 12, 277–290. [Google Scholar] [CrossRef]
- Li, M.; Zhuang, Y.; Batra, R.; Thomas, J.D.; Li, M.; Nutter, C.A.; Scotti, M.M.; Carter, H.A.; Wang, Z.J.; Huang, X.-S.; et al. HNRNPA1-Induced Spliceopathy in a Transgenic Mouse Model of Myotonic Dystrophy. Proc. Natl. Acad. Sci. USA 2020, 117, 5472–5477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corioni, M.; Antih, N.; Tanackovic, G.; Zavolan, M.; Krämer, A. Analysis of in Situ Pre-MRNA Targets of Human Splicing Factor SF1 Reveals a Function in Alternative Splicing. Nucleic Acids Res. 2011, 39, 1868–1879. [Google Scholar] [CrossRef] [Green Version]
- Crisci, A.; Raleff, F.; Bagdiul, I.; Raabe, M.; Urlaub, H.; Rain, J.-C.; Krämer, A. Mammalian Splicing Factor SF1 Interacts with SURP Domains of U2 SnRNP-Associated Proteins. Nucleic Acids Res. 2015, 43, 10456–10473. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kao, S.-Y.; Nikonova, E.; Chaabane, S.; Sabani, A.; Martitz, A.; Wittner, A.; Heemken, J.; Straub, T.; Spletter, M.L. A Candidate RNAi Screen Reveals Diverse RNA-Binding Protein Phenotypes in Drosophila Flight Muscle. Cells 2021, 10, 2505. https://doi.org/10.3390/cells10102505
Kao S-Y, Nikonova E, Chaabane S, Sabani A, Martitz A, Wittner A, Heemken J, Straub T, Spletter ML. A Candidate RNAi Screen Reveals Diverse RNA-Binding Protein Phenotypes in Drosophila Flight Muscle. Cells. 2021; 10(10):2505. https://doi.org/10.3390/cells10102505
Chicago/Turabian StyleKao, Shao-Yen, Elena Nikonova, Sabrina Chaabane, Albiona Sabani, Alexandra Martitz, Anja Wittner, Jakob Heemken, Tobias Straub, and Maria L. Spletter. 2021. "A Candidate RNAi Screen Reveals Diverse RNA-Binding Protein Phenotypes in Drosophila Flight Muscle" Cells 10, no. 10: 2505. https://doi.org/10.3390/cells10102505