Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia
Abstract
1. Introduction
2. Materials and Methods
3. Results
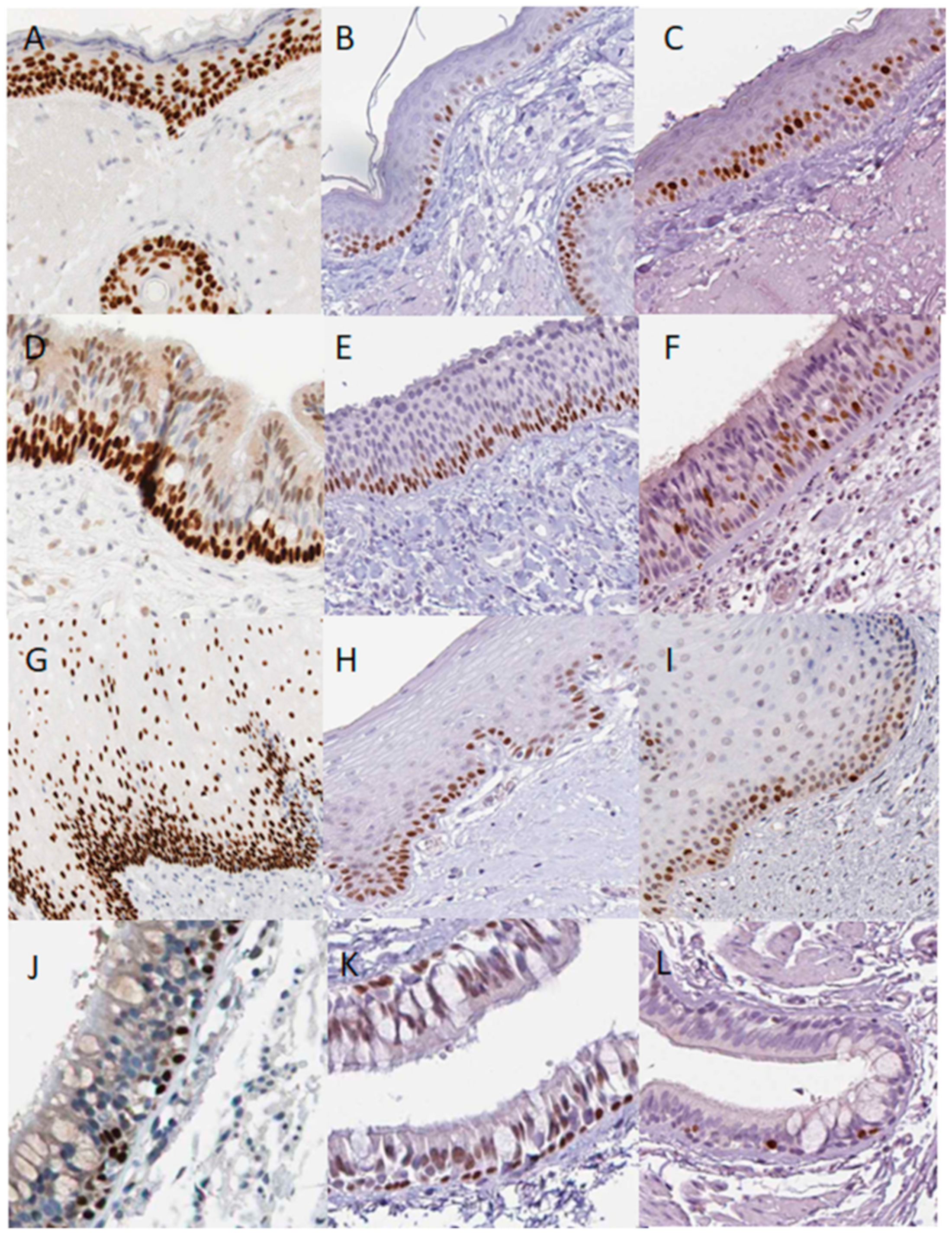
3.1. p63 and p73 Distribution in the Normal Epithelial Tissues
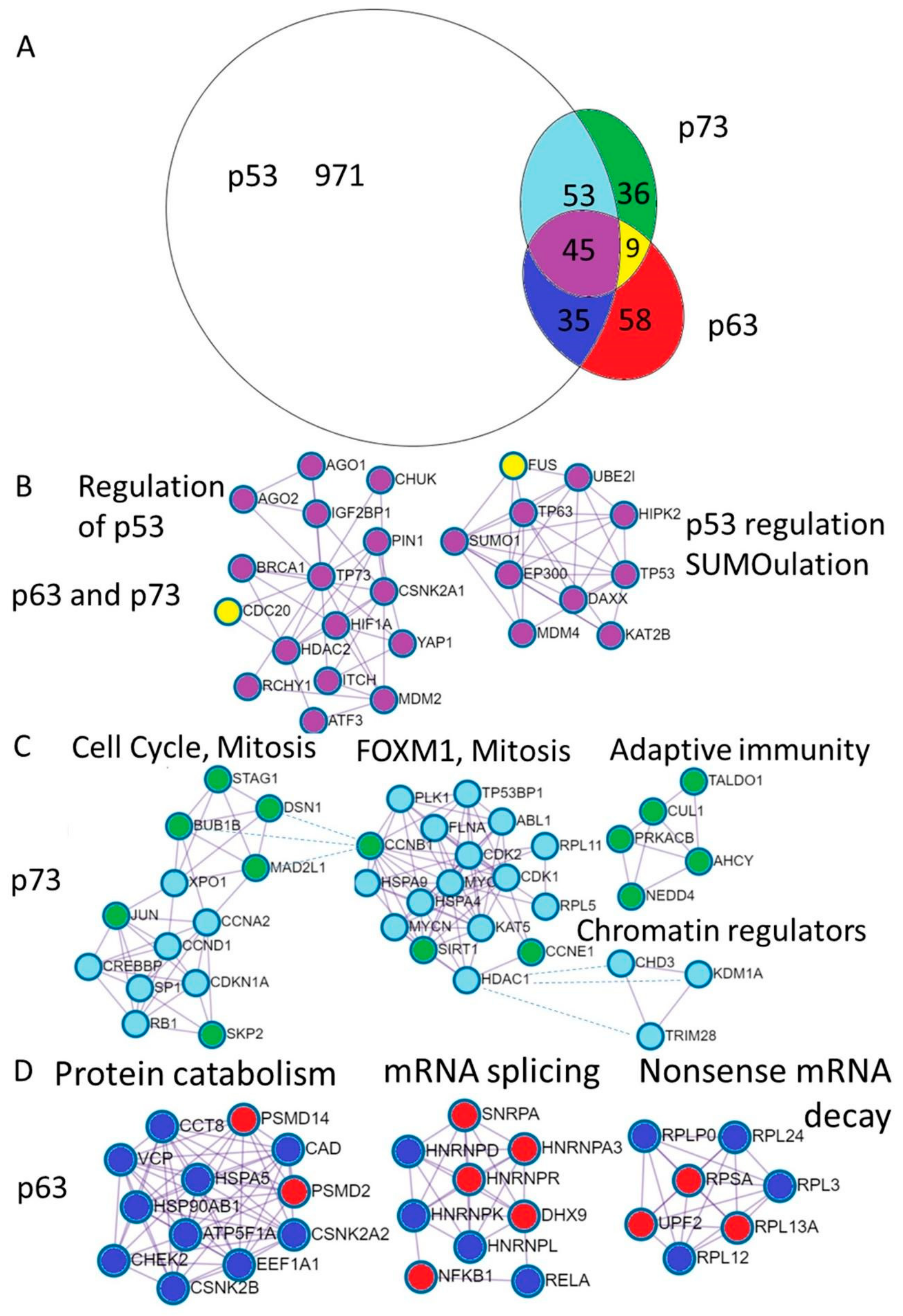
3.2. Common and Distinct Multiprotein Complexes of p53 Family Members and Their Distribution in the Epithelial Organs
3.2.1. Interaction between p63 and p73 in Gene Regulation
3.2.2. Common p63 and p73 Interactors
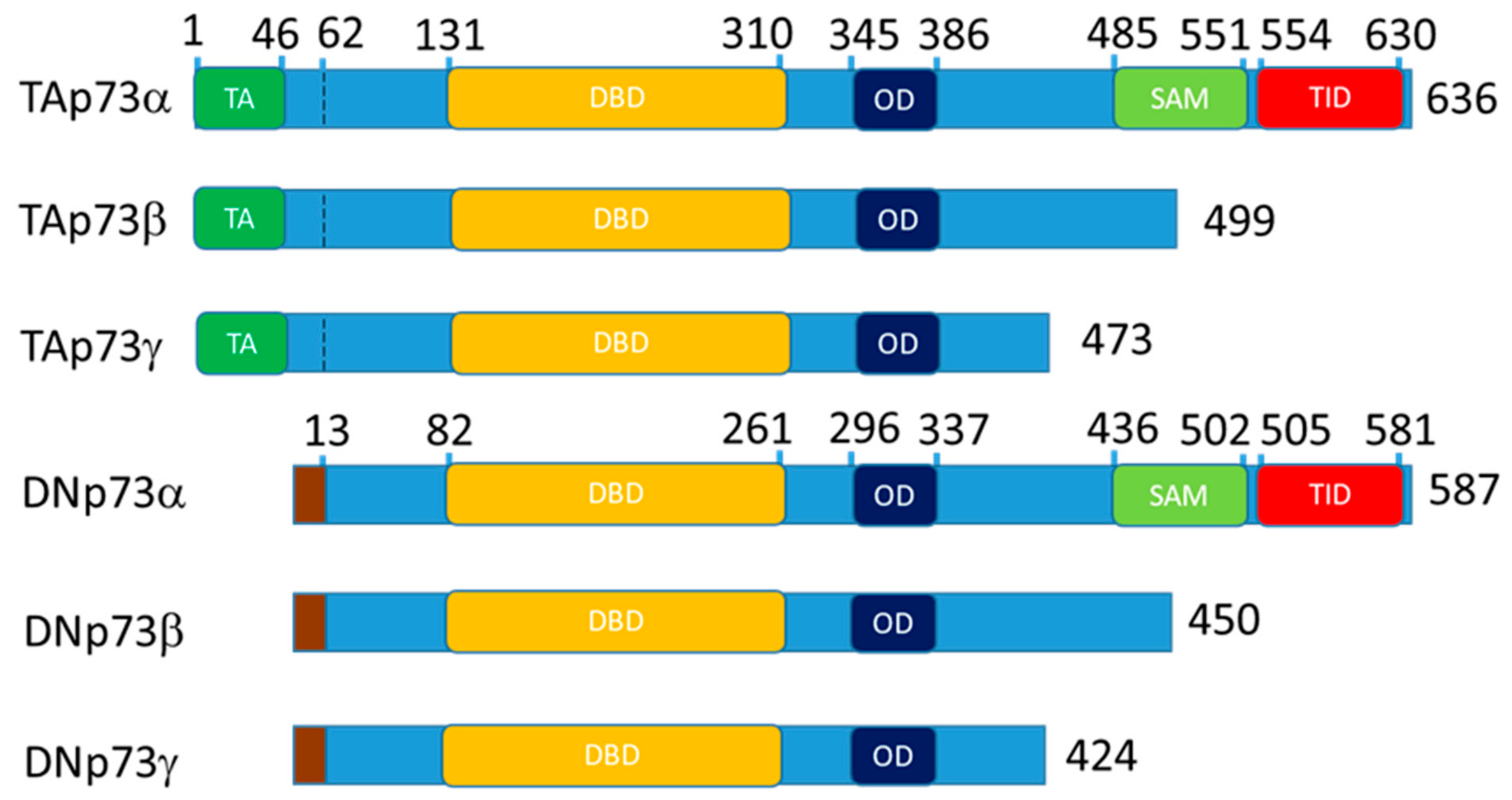
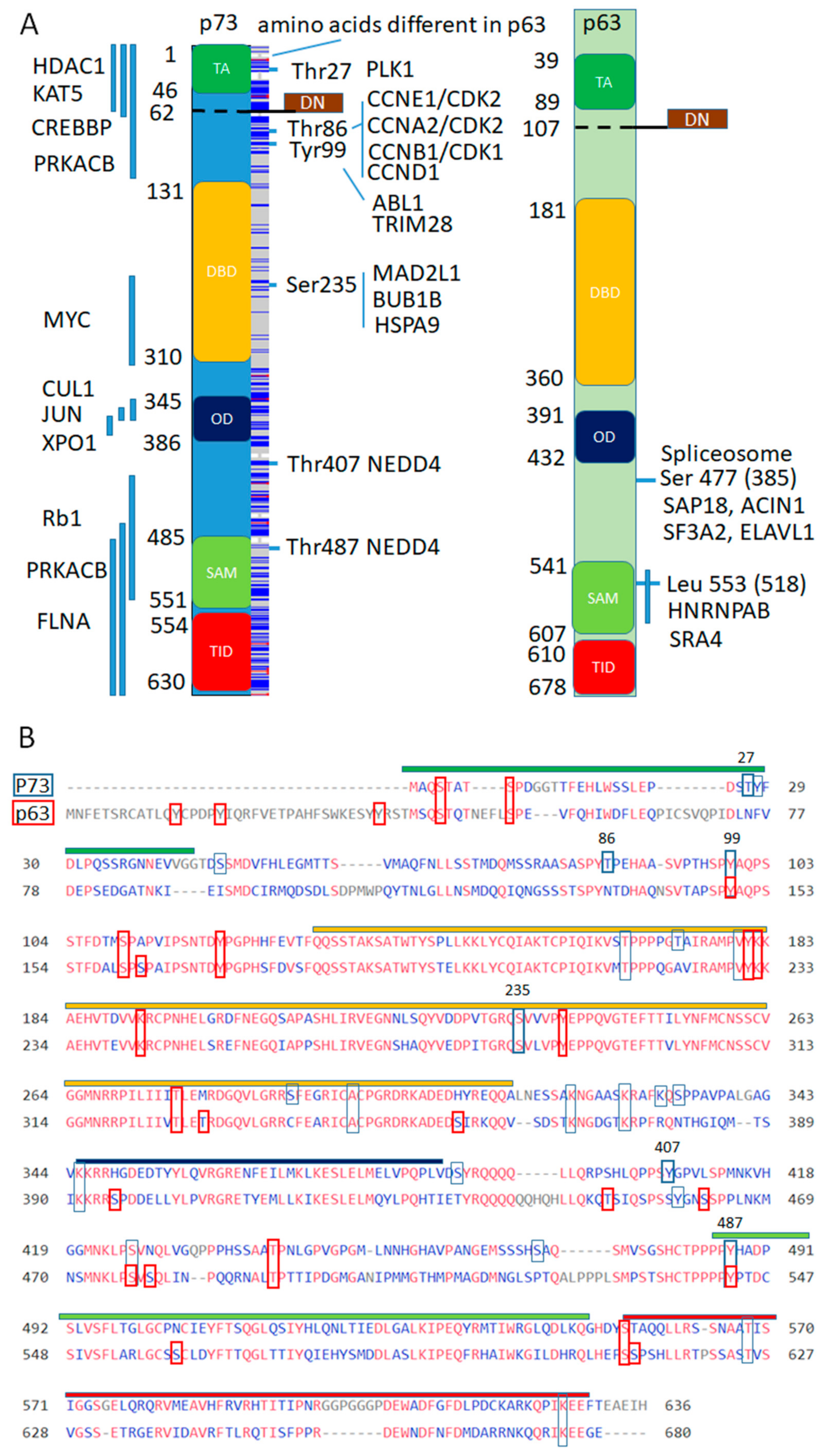
3.2.3. p73 Interactions
3.2.4. p63 Interactors
3.2.5. p53 Interactions
3.3. Distinct Roles of p63 and p73 in Epithelial Tissue
3.3.1. The Role of p73 in Mitosis and Regulation of Polyploidy
3.3.2. The Role of p63 in Transcription-Coupled Splicing
4. Discussion
5. Perspectives and Future Directions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dötsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. P63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 1998, 2, 305–316. [Google Scholar] [CrossRef]
- Kaghad, M.; Bonnet, H.; Yang, A.; Creancier, L.; Biscan, J.C.; Valent, A.; Minty, A.; Chalon, P.; Lelias, J.M.; Dumont, X.; et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90, 809–819. [Google Scholar] [CrossRef]
- Scoumanne, A.; Harms, K.L.; Chen, X. Structural basis for gene activation by p53 family members. Cancer Biol. Ther. 2005, 4, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Grob, T.J.; Novak, U.; Maisse, C.; Barcaroli, D.; Lüthi, A.U.; Pirnia, F.; Hügli, B.; Graber, H.U.; De Laurenzi, V.; Fey, M.F.; et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001, 8, 1213–1223. [Google Scholar] [CrossRef]
- Waltermann, A.; Kartasheva, N.N.; Dobbelstein, M. Differential regulation of p63 and p73 expression. Oncogene 2003, 22, 5686–5693. [Google Scholar] [CrossRef][Green Version]
- Nakagawa, T.; Takahashi, M.; Ozaki, T.; Watanabe, K.; Hayashi, S.; Hosoda, M.; Todo, S.; Nakagawara, A. Negative autoregulation of p73 and p53 by DeltaNp73 in regulating differentiation and survival of human neuroblastoma cells. Cancer Lett. 2003, 197, 105–109. [Google Scholar] [CrossRef]
- Nakagawa, T.; Takahashi, M.; Ozaki, T.; Watanabe Ki, K.; Todo, S.; Mizuguchi, H.; Hayakawa, T.; Nakagawara, A. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol. Cell. Biol. 2002, 22, 2575–2585. [Google Scholar] [CrossRef] [PubMed]
- Lanza, M.; Marinari, B.; Papoutsaki, M.; Giustizieri, M.L.; D’Alessandra, Y.; Chimenti, S.; Guerrini, L.; Costanzo, A. Cross-talks in the p53 family: DeltaNp63 is an anti-apoptotic target for deltaNp73alpha and p53 gain-of-function mutants. Cell Cycle 2006, 5, 1996–2004. [Google Scholar] [CrossRef]
- Tophkhane, C.; Yang, S.-H.; Jiang, Y.; Ma, Z.; Subramaniam, D.; Anant, S.; Yogosawa, S.; Sakai, T.; Liu, W.-G.; Edgerton, S.; et al. P53 inactivation upregulates p73 expression through E2F-1 mediated transcription. PLoS ONE 2012, 7, e43564. [Google Scholar] [CrossRef]
- Vikhreva, P.; Melino, G.; Amelio, I. P73 Alternative Splicing: Exploring a Biological Role for the C-Terminal Isoforms. J. Mol. Biol. 2018, 430, 1829–1838. [Google Scholar] [CrossRef]
- Nozell, S.; Wu, Y.; McNaughton, K.; Liu, G.; Willis, A.; Paik, J.C.; Chen, X. Characterization of p73 functional domains necessary for transactivation and growth suppression. Oncogene 2003, 22, 4333–4347. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chen, X. The C-terminal sterile alpha motif and the extreme C terminus regulate the transcriptional activity of the alpha isoform of p73. J. Biol. Chem. 2005, 280, 20111–20119. [Google Scholar] [CrossRef] [PubMed]
- Toh, W.H.; Logette, E.; Corcos, L.; Sabapathy, K. TAp73beta and DNp73beta activate the expression of the pro-survival caspase-2S. Nucleic Acids Res. 2008, 36, 4498–4509. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, M.; van Heeringen, S.J.; Kramer, D.; Smeenk, L.; Janssen-Megens, E.; Hartmann, M.; Stunnenberg, H.G.; Lohrum, M. Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Res. 2011, 39, 6069–6085. [Google Scholar] [CrossRef] [PubMed]
- Vikhanskaya, F.; Toh, W.H.; Dulloo, I.; Wu, Q.; Boominathan, L.; Ng, H.H.; Vousden, K.H.; Sabapathy, K. P73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nat. Cell Biol. 2007, 9, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Tichý, V.; Navrátilová, L.; Adámik, M.; Fojta, M.; Brázdová, M. Redox state of p63 and p73 core domains regulates sequence-specific DNA binding. Biochem. Biophys. Res. Commun. 2013, 433, 445–449. [Google Scholar] [CrossRef]
- Patel, S.; Bui, T.T.T.; Drake, A.F.; Fraternali, F.; Nikolova, P.V. The p73 DNA binding domain displays enhanced stability relative to its homologue, the tumor suppressor p53, and exhibits cooperative DNA binding. Biochemistry 2008, 47, 3235–3244. [Google Scholar] [CrossRef]
- Cai, B.-H.; Chao, C.-F.; Lu, M.-H.; Lin, H.-C.; Chen, J.-Y. A half-site of the p53-binding site on the keratin 14 promoter is specifically activated by p63. J. Biochem. 2012, 152, 99–110. [Google Scholar] [CrossRef]
- Klein, C.; Georges, G.; Künkele, K.P.; Huber, R.; Engh, R.A.; Hansen, S. High thermostability and lack of cooperative DNA binding distinguish the p63 core domain from the homologous tumor suppressor p53. J. Biol. Chem. 2001, 276, 37390–37401. [Google Scholar] [CrossRef]
- Di Como, C.J.; Gaiddon, C.; Prives, C. P73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 1999, 19, 1438–1449. [Google Scholar] [CrossRef]
- Si, H.; Lu, H.; Yang, X.; Mattox, A.; Jang, M.; Bian, Y.; Sano, E.; Viadiu, H.; Yan, B.; Yau, C.; et al. TNF-α modulates genome-wide redistribution of ΔNp63α/TAp73 and NF-κB cREL interactive binding on TP53 and AP-1 motifs to promote an oncogenic gene program in squamous cancer. Oncogene 2016, 35, 5781–5794. [Google Scholar] [CrossRef] [PubMed]
- Zdzalik, M.; Pustelny, K.; Kedracka-Krok, S.; Huben, K.; Pecak, A.; Wladyka, B.; Jankowski, S.; Dubin, A.; Potempa, J.; Dubin, G. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle 2010, 9, 4584–4591. [Google Scholar] [CrossRef] [PubMed]
- Talos, F.; Nemajerova, A.; Flores, E.R.; Petrenko, O.; Moll, U.M. P73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol. Cell 2007, 27, 647–659. [Google Scholar] [CrossRef]
- Tomasini, R.; Tsuchihara, K.; Wilhelm, M.; Fujitani, M.; Rufini, A.; Cheung, C.C.; Khan, F.; Itie-Youten, A.; Wakeham, A.; Tsao, M.-S.; et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008, 22, 2677–2691. [Google Scholar] [CrossRef]
- Nicolai, S.; Rossi, A.; Di Daniele, N.; Melino, G.; Annicchiarico-Petruzzelli, M.; Raschellà, G. DNA repair and aging: The impact of the p53 family. Aging 2015, 7, 1050–1065. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, M.; Sato, R.; Yamashita, T. Loss of p73 in ependymal cells during the perinatal period leads to aqueductal stenosis. Sci. Rep. 2017, 7, 12007. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Sengupta, S.; Gurdziel, K.; Bell, G.W.; Jacks, T.; Flores, E.R. P63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet. 2009, 5, e1000680. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lin, M.; Zhang, C.; Wang, D.; Feng, Z.; Hu, W. TAp63γ enhances nucleotide excision repair through transcriptional regulation of DNA repair genes. DNA Repair 2012, 11, 167–176. [Google Scholar] [CrossRef]
- Zaika, E.; Wei, J.; Yin, D.; Andl, C.; Moll, U.; El-Rifai, W.; Zaika, A.I. P73 protein regulates DNA damage repair. FASEB J. 2011, 25, 4406–4414. [Google Scholar] [CrossRef]
- Jackson, P.K.; Attardi, L.D. P73 and foxj1: Programming multiciliated epithelia. Trends Cell Biol. 2016, 26, 239–240. [Google Scholar] [CrossRef]
- Marshall, C.B.; Mays, D.J.; Beeler, J.S.; Rosenbluth, J.M.; Boyd, K.L.; Santos Guasch, G.L.; Shaver, T.M.; Tang, L.J.; Liu, Q.; Shyr, Y.; et al. P73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep. 2016, 14, 2289–2300. [Google Scholar] [CrossRef] [PubMed]
- Fuertes-Alvarez, S.; Maeso-Alonso, L.; Villoch-Fernandez, J.; Wildung, M.; Martin-Lopez, M.; Marshall, C.; Villena-Cortes, A.J.; Diez-Prieto, I.; Pietenpol, J.A.; Tissir, F.; et al. P73 regulates ependymal planar cell polarity by modulating actin and microtubule cytoskeleton. Cell Death Dis. 2018, 9, 1183. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Schweitzer, R.; Sun, D.; Kaghad, M.; Walker, N.; Bronson, R.T.; Tabin, C.; Sharpe, A.; Caput, D.; Crum, C.; et al. P63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999, 398, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Mills, A.A.; Zheng, B.; Wang, X.J.; Vogel, H.; Roop, D.R.; Bradley, A. P63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999, 398, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Lechler, T.; Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 2005, 437, 275–280. [Google Scholar] [CrossRef]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef]
- LeBoeuf, M.; Terrell, A.; Trivedi, S.; Sinha, S.; Epstein, J.A.; Olson, E.N.; Morrisey, E.E.; Millar, S.E. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell 2010, 19, 807–818. [Google Scholar] [CrossRef]
- Candi, E.; Rufini, A.; Terrinoni, A.; Dinsdale, D.; Ranalli, M.; Paradisi, A.; De Laurenzi, V.; Spagnoli, L.G.; Catani, M.V.; Ramadan, S.; et al. Differential roles of p63 isoforms in epidermal development: Selective genetic complementation in p63 null mice. Cell Death Differ. 2006, 13, 1037–1047. [Google Scholar] [CrossRef]
- Shalom-Feuerstein, R.; Lena, A.M.; Zhou, H.; De La Forest Divonne, S.; Van Bokhoven, H.; Candi, E.; Melino, G.; Aberdam, D. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011, 18, 887–896. [Google Scholar] [CrossRef]
- Poulson, N.D.; Lechler, T. Robust control of mitotic spindle orientation in the developing epidermis. J. Cell Biol. 2010, 191, 915–922. [Google Scholar] [CrossRef]
- Yang, A.; Walker, N.; Bronson, R.; Kaghad, M.; Oosterwegel, M.; Bonnin, J.; Vagner, C.; Bonnet, H.; Dikkes, P.; Sharpe, A.; et al. P73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000, 404, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Beeler, J.S.; Marshall, C.B.; Gonzalez-Ericsson, P.I.; Shaver, T.M.; Santos Guasch, G.L.; Lea, S.T.; Johnson, K.N.; Jin, H.; Venters, B.J.; Sanders, M.E.; et al. P73 regulates epidermal wound healing and induced keratinocyte programming. PLoS ONE 2019, 14, e0218458. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.T.; Rufini, A.; Wetzel, M.K.; Tsuchihara, K.; Inoue, S.; Tomasini, R.; Itie-Youten, A.; Wakeham, A.; Arsenian-Henriksson, M.; Melino, G.; et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010, 24, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Lecue, P.; de Pedro, I.; Coulon, V.; Molinuevo, R.; Lorz, C.; Segrelles, C.; Ceballos, L.; López-Aventín, D.; García-Valtuille, A.; Bernal, J.M.; et al. Inefficient differentiation response to cell cycle stress leads to genomic instability and malignant progression of squamous carcinoma cells. Cell Death Dis. 2017, 8, e2901. [Google Scholar] [CrossRef] [PubMed]
- Freije, A.; Ceballos, L.; Coisy, M.; Barnes, L.; Rosa, M.; De Diego, E.; Blanchard, J.M.; Gandarillas, A. Cyclin E drives human keratinocyte growth into differentiation. Oncogene 2012, 31, 5180–5192. [Google Scholar] [CrossRef]
- Gandarillas, A.; Sanz-Gómez, N.; Freije, A. Polyploidy and the mitosis path to epidermal cell fate. Cell Cycle 2019, 18, 359–362. [Google Scholar] [CrossRef]
- Zanet, J.; Freije, A.; Ruiz, M.; Coulon, V.; Sanz, J.R.; Chiesa, J.; Gandarillas, A. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS ONE 2010, 5, e15701. [Google Scholar] [CrossRef]
- Gebel, J.; Luh, L.M.; Coutandin, D.; Osterburg, C.; Löhr, F.; Schäfer, B.; Frombach, A.-S.; Sumyk, M.; Buchner, L.; Krojer, T.; et al. Mechanism of TAp73 inhibition by ΔNp63 and structural basis of p63/p73 hetero-tetramerization. Cell Death Differ. 2016, 23, 1930–1940. [Google Scholar] [CrossRef]
- Aylon, Y.; Oren, M. P53: Guardian of ploidy. Mol. Oncol. 2011, 5, 315–323. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Micallef, L.; Rodgers, P. EulerAPE: Drawing area-proportional 3-Venn diagrams using ellipses. PLoS ONE 2014, 9, e101717. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Cheng, J.B.; Sedgewick, A.J.; Finnegan, A.I.; Harirchian, P.; Lee, J.; Kwon, S.; Fassett, M.S.; Golovato, J.; Gray, M.; Ghadially, R.; et al. Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. 2018, 25, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-Y.; Lee, T.-Y.; Kao, H.-J.; Ma, C.-T.; Lee, C.-C.; Lin, T.-H.; Chang, W.-C.; Huang, H.-D. DbPTM in 2019: Exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019, 47, D298–D308. [Google Scholar] [CrossRef]
- Finnegan, A.; Cho, R.J.; Luu, A.; Harirchian, P.; Lee, J.; Cheng, J.B.; Song, J.S. Single-cell transcriptomics reveals spatial and temporal turnover of keratinocyte differentiation regulators. Front. Genet. 2019, 10, 775. [Google Scholar] [CrossRef]
- Trost, A.; Bauer, J.W.; Lanschützer, C.; Laimer, M.; Emberger, M.; Hintner, H.; Onder, K. Rapid, high-quality and epidermal-specific isolation of RNA from human skin. Exp. Dermatol. 2007, 16, 185–190. [Google Scholar] [CrossRef]
- Reimann, E.; Abram, K.; Kõks, S.; Kingo, K.; Fazeli, A. Identification of an optimal method for extracting RNA from human skin biopsy, using domestic pig as a model system. Sci. Rep. 2019, 9, 20111. [Google Scholar] [CrossRef]
- Santos Guasch, G.L.; Beeler, J.S.; Marshall, C.B.; Shaver, T.M.; Sheng, Q.; Johnson, K.N.; Boyd, K.L.; Venters, B.J.; Cook, R.S.; Pietenpol, J.A. P73 is required for ovarian follicle development and regulates a gene network involved in cell-to-cell adhesion. iScience 2018, 8, 236–249. [Google Scholar] [CrossRef]
- Rozenberg, J.M.; Bhattacharya, P.; Chatterjee, R.; Glass, K.; Vinson, C. Combinatorial recruitment of CREB, C/EBPβ and c-Jun determines activation of promoters upon keratinocyte differentiation. PLoS ONE 2013, 8, e78179. [Google Scholar] [CrossRef]
- Suda, N.; Itoh, T.; Nakato, R.; Shirakawa, D.; Bando, M.; Katou, Y.; Kataoka, K.; Shirahige, K.; Tickle, C.; Tanaka, M. Dimeric combinations of MafB, cFos and cJun control the apoptosis-survival balance in limb morphogenesis. Development 2014, 141, 2885–2894. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, D.; Bunjobpol, W.; Sabapathy, K. Interplay between TAp73 protein and selected activator protein-1 (AP-1) Family Members Promotes AP-1 target gene activation and cellular growth. J. Biol. Chem. 2015, 290, 18636–18649. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, J.M.; Taylor, J.M.; Mack, C.P. RBPJ binds to consensus and methylated cis elements within phased nucleosomes and controls gene expression in human aortic smooth muscle cells in cooperation with SRF. Nucleic Acids Res. 2018, 46, 8232–8244. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Guo, M.; Yu, J.; Yan, C. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics 2016, 17, 335. [Google Scholar] [CrossRef]
- Kouwenhoven, E.N.; Oti, M.; Niehues, H.; van Heeringen, S.J.; Schalkwijk, J.; Stunnenberg, H.G.; van Bokhoven, H.; Zhou, H. Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 2015, 16, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yi, G.; Zhou, H. P63 cooperates with CTCF to modulate chromatin architecture in skin keratinocytes. Epigenet. Chromatin 2019, 12, 31. [Google Scholar] [CrossRef]
- Qu, J.; Tanis, S.E.J.; Smits, J.P.H.; Kouwenhoven, E.N.; Oti, M.; van den Bogaard, E.H.; Logie, C.; Stunnenberg, H.G.; van Bokhoven, H.; Mulder, K.W.; et al. Mutant p63 affects epidermal cell identity through rewiring the enhancer landscape. Cell Rep. 2018, 25, 3490–3503. [Google Scholar] [CrossRef] [PubMed]
- Deb, D.; Lanyi, A.; Scian, M.; Keiger, J.; Brown, D.R.; Le Roith, D.; Deb, S.P.; Deb, S. Differential modulation of cellular and viral promoters by p73 and p53. Int. J. Oncol. 2001, 18, 401–409. [Google Scholar]
- Martin, A.G.; Trama, J.; Crighton, D.; Ryan, K.M.; Fearnhead, H.O. Activation of p73 and induction of Noxa by DNA damage requires NF-kappa B. Aging 2009, 1, 335–349. [Google Scholar] [CrossRef]
- Lu, H.; Yang, X.; Duggal, P.; Allen, C.T.; Yan, B.; Cohen, J.; Nottingham, L.; Romano, R.-A.; Sinha, S.; King, K.E.; et al. TNF-α promotes c-REL/ΔNp63α interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res. 2011, 71, 6867–6877. [Google Scholar] [CrossRef] [PubMed]
- De Laurenzi, V.; Rossi, A.; Terrinoni, A.; Barcaroli, D.; Levrero, M.; Costanzo, A.; Knight, R.A.; Guerrieri, P.; Melino, G. P63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem. Biophys. Res. Commun. 2000, 273, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Rishi, V.; Bhattacharya, P.; Chatterjee, R.; Rozenberg, J.; Zhao, J.; Glass, K.; Fitzgerald, P.; Vinson, C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. USA 2010, 107, 20311–20316. [Google Scholar] [CrossRef] [PubMed]
- Ratovitski, E.A. LKB1/PEA3/ΔNp63 pathway regulates PTGS-2 (COX-2) transcription in lung cancer cells upon cigarette smoke exposure. Oxid. Med. Cell. Longev. 2010, 3, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Innocente, S.A.; Lee, J.M. p73 is a p53-independent, Sp1-dependent repressor of cyclin B1 transcription. Biochem. Biophys. Res. Commun. 2005, 329, 713–718. [Google Scholar] [CrossRef]
- Ozaki, T.; Watanabe, K.; Nakagawa, T.; Miyazaki, K.; Takahashi, M.; Nakagawara, A. Function of p73, not of p53, is inhibited by the physical interaction with RACK1 and its inhibitory effect is counteracted by pRB. Oncogene 2003, 22, 3231–3242. [Google Scholar] [CrossRef][Green Version]
- Zhang, J.; Chen, X. DeltaNp73 modulates nerve growth factor-mediated neuronal differentiation through repression of TrkA. Mol. Cell. Biol. 2007, 27, 3868–3880. [Google Scholar] [CrossRef]
- Yang, A.; Zhu, Z.; Kettenbach, A.; Kapranov, P.; McKeon, F.; Gingeras, T.R.; Struhl, K. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar dna-binding profiles in vivo. PLoS ONE 2010, 5, e11572. [Google Scholar] [CrossRef]
- Wu, H.; Zeinab, R.A.; Flores, E.R.; Leng, R.P. Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol. Cancer Res. 2011, 9, 1780–1790. [Google Scholar] [CrossRef]
- Oberst, A.; Malatesta, M.; Aqeilan, R.I.; Rossi, M.; Salomoni, P.; Murillas, R.; Sharma, P.; Kuehn, M.R.; Oren, M.; Croce, C.M.; et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc. Natl. Acad. Sci. USA 2007, 104, 11280–11285. [Google Scholar] [CrossRef]
- Rossi, M.; Aqeilan, R.I.; Neale, M.; Candi, E.; Salomoni, P.; Knight, R.A.; Croce, C.M.; Melino, G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc. Natl. Acad. Sci. USA 2006, 103, 12753–12758. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-P.; Wu, G.; Guo, Z.; Osada, M.; Fomenkov, T.; Park, H.L.; Trink, B.; Sidransky, D.; Fomenkov, A.; Ratovitski, E.A. Altered sumoylation of p63alpha contributes to the split-hand/foot malformation phenotype. Cell Cycle 2004, 3, 1587–1596. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Arooz, T.; Siu, W.Y.; Chiu, C.H.; Lau, A.; Yamashita, K.; Poon, R.Y. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001, 490, 202–208. [Google Scholar] [CrossRef]
- Bálint, E.; Bates, S.; Vousden, K.H. Mdm2 binds p73 alpha without targeting degradation. Oncogene 1999, 18, 3923–3929. [Google Scholar] [CrossRef] [PubMed]
- Minty, A.; Dumont, X.; Kaghad, M.; Caput, D. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 2000, 275, 36316–36323. [Google Scholar] [CrossRef]
- Asatsuma-Okumura, T.; Ando, H.; De Simone, M.; Yamamoto, J.; Sato, T.; Shimizu, N.; Asakawa, K.; Yamaguchi, Y.; Ito, T.; Guerrini, L.; et al. P63 is a cereblon substrate involved in thalidomide teratogenicity. Nat. Chem. Biol. 2019, 15, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.-M.; Zhang, Y.; Liao, W.; Zeng, S.X.; Su, X.; Flores, E.R.; Lu, H. IκB kinase β (IKKβ) inhibits p63 isoform γ (TAp63γ) transcriptional activity. J. Biol. Chem. 2013, 288, 18184–18193. [Google Scholar] [CrossRef]
- MacPartlin, M.; Zeng, S.; Lee, H.; Stauffer, D.; Jin, Y.; Thayer, M.; Lu, H. P300 regulates p63 transcriptional activity. J. Biol. Chem. 2005, 280, 30604–30610. [Google Scholar] [CrossRef]
- Strano, S.; Monti, O.; Pediconi, N.; Baccarini, A.; Fontemaggi, G.; Lapi, E.; Mantovani, F.; Damalas, A.; Citro, G.; Sacchi, A.; et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol. Cell 2005, 18, 447–459. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Liu, Y.; Bertos, N.R.; Yang, X.-J.; Liao, D. PCAF is a coactivator for p73-mediated transactivation. Oncogene 2003, 22, 8316–8329. [Google Scholar] [CrossRef][Green Version]
- Buckley, N.E.; Conlon, S.J.; Jirstrom, K.; Kay, E.W.; Crawford, N.T.; O’Grady, A.; Sheehan, K.; Mc Dade, S.S.; Wang, C.-W.; McCance, D.J.; et al. The DeltaNp63 proteins are key allies of BRCA1 in the prevention of basal-like breast cancer. Cancer Res. 2011, 71, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.; He, L.; Leong, C.-O.; Xing, D.; Karlan, B.Y.; Swisher, E.M.; Rueda, B.R.; Orsulic, S.; Ellisen, L.W. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer Res. 2010, 70, 7155–7165. [Google Scholar] [CrossRef] [PubMed]
- Strano, S.; Munarriz, E.; Rossi, M.; Castagnoli, L.; Shaul, Y.; Sacchi, A.; Oren, M.; Sudol, M.; Cesareni, G.; Blandino, G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 2001, 276, 15164–15173. [Google Scholar] [CrossRef] [PubMed]
- Senoo, M.; Matsumura, Y.; Habu, S. TAp63gamma (p51A) and dNp63alpha (p73L), two major isoforms of the p63 gene, exert opposite effects on the vascular endothelial growth factor (VEGF) gene expression. Oncogene 2002, 21, 2455–2465. [Google Scholar] [CrossRef]
- Amelio, I.; Inoue, S.; Markert, E.K.; Levine, A.J.; Knight, R.A.; Mak, T.W.; Melino, G. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1α degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 226–231. [Google Scholar] [CrossRef]
- Katayama, H.; Wang, J.; Treekitkarnmongkol, W.; Kawai, H.; Sasai, K.; Zhang, H.; Wang, H.; Adams, H.P.; Jiang, S.; Chakraborty, S.N.; et al. Aurora kinase-A inactivates DNA damage-induced apoptosis and spindle assembly checkpoint response functions of p73. Cancer Cell 2012, 21, 196–211. [Google Scholar] [CrossRef]
- Rokudai, S.; Li, Y.; Otaka, Y.; Fujieda, M.; Owens, D.M.; Christiano, A.M.; Nishiyama, M.; Prives, C. STXBP4 regulates APC/C-mediated p63 turnover and drives squamous cell carcinogenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E4806–E4814. [Google Scholar] [CrossRef]
- Amoresano, A.; Di Costanzo, A.; Leo, G.; Di Cunto, F.; La Mantia, G.; Guerrini, L.; Calabrò, V. Identification of DeltaNp63alpha protein interactions by mass spectrometry. J. Proteome Res. 2010, 9, 2042–2048. [Google Scholar] [CrossRef]
- Wang, T.; Jiang, X.; Chen, G.; Xu, J. Interaction of amyotrophic lateral sclerosis/frontotemporal lobar degeneration-associated fused-in-sarcoma with proteins involved in metabolic and protein degradation pathways. Neurobiol. Aging 2015, 36, 527–535. [Google Scholar] [CrossRef]
- Su, X.; Cho, M.S.; Gi, Y.-J.; Ayanga, B.A.; Sherr, C.J.; Flores, E.R. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009, 28, 1904–1915. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Enge, M.; Bao, W.; Hedström, E.; Jackson, S.P.; Moumen, A.; Selivanova, G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell 2009, 15, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, R.; Tsuchihara, K.; Tsuda, C.; Lau, S.K.; Wilhelm, M.; Rufini, A.; Tsao, M.; Iovanna, J.L.; Jurisicova, A.; Melino, G.; et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc. Natl. Acad. Sci. USA 2009, 106, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Gebel, J.; Tuppi, M.; Krauskopf, K.; Coutandin, D.; Pitzius, S.; Kehrloesser, S.; Osterburg, C.; Dötsch, V. Control mechanisms in germ cells mediated by p53 family proteins. J. Cell Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mikulenkova, E.; Neradil, J.; Zitterbart, K.; Sterba, J.; Veselska, R. Overexpression of the ∆Np73 isoform is associated with centrosome amplification in brain tumor cell lines. Tumour Biol. 2015, 36, 7483–7491. [Google Scholar] [CrossRef]
- Gaiddon, C.; Lokshin, M.; Gross, I.; Levasseur, D.; Taya, Y.; Loeffler, J.-P.; Prives, C. Cyclin-dependent kinases phosphorylate p73 at threonine 86 in a cell cycle-dependent manner and negatively regulate p73. J. Biol. Chem. 2003, 278, 27421–27431. [Google Scholar] [CrossRef]
- Lunardi, A.; Di Minin, G.; Provero, P.; Dal Ferro, M.; Carotti, M.; Del Sal, G.; Collavin, L. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl. Acad. Sci. USA 2010, 107, 6322–6327. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The bioplex network: A systematic exploration of the human interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef]
- Uramoto, H.; Wetterskog, D.; Hackzell, A.; Matsumoto, Y.; Funa, K. P73 competes with co-activators and recruits histone deacetylase to NF-Y in the repression of PDGF beta-receptor. J. Cell Sci. 2004, 117, 5323–5331. [Google Scholar] [CrossRef]
- Lemasson, I.; Nyborg, J.K. Human T-cell leukemia virus type I tax repression of p73beta is mediated through competition for the C/H1 domain of CBP. J. Biol. Chem. 2001, 276, 15720–15727. [Google Scholar] [CrossRef]
- Kim, J.-W.; Song, P.I.; Jeong, M.-H.; An, J.-H.; Lee, S.-Y.; Jang, S.-M.; Song, K.-H.; Armstrong, C.A.; Choi, K.-H. TIP60 represses transcriptional activity of p73beta via an MDM2-bridged ternary complex. J. Biol. Chem. 2008, 283, 20077–20086. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-W.; Nguyen, T.T.; Shi, Y.; Barton, M.C. p53-targeted LSD1 functions in repression of chromatin structure and transcription in vivo. Mol. Cell. Biol. 2008, 28, 5139–5146. [Google Scholar] [CrossRef] [PubMed]
- Chau, B.N.; Chen, T.-T.; Wan, Y.Y.; DeGregori, J.; Wang, J.Y.J. Tumor necrosis factor alpha-induced apoptosis requires p73 and c-ABL activation downstream of RB degradation. Mol. Cell. Biol. 2004, 24, 4438–4447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koutsodontis, G.; Vasilaki, E.; Chou, W.-C.; Papakosta, P.; Kardassis, D. Physical and functional interactions between members of the tumour suppressor p53 and the Sp families of transcription factors: Importance for the regulation of genes involved in cell-cycle arrest and apoptosis. Biochem. J. 2005, 389, 443–455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uramoto, H.; Izumi, H.; Ise, T.; Tada, M.; Uchiumi, T.; Kuwano, M.; Yasumoto, K.; Funa, K.; Kohno, K. P73 Interacts with c-Myc to regulate Y-box-binding protein-1 expression. J. Biol. Chem. 2002, 277, 31694–31702. [Google Scholar] [CrossRef] [PubMed]
- Fulco, M.; Costanzo, A.; Merlo, P.; Mangiacasale, R.; Strano, S.; Blandino, G.; Balsano, C.; Lavia, P.; Levrero, M. P73 is regulated by phosphorylation at the G2/M transition. J. Biol. Chem. 2003, 278, 49196–49202. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Belkhiri, A.; Ecsedy, J.; Zaika, A.; El-Rifai, W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008, 68, 8998–9004. [Google Scholar] [CrossRef]
- Tentler, J.J.; Ionkina, A.A.; Tan, A.C.; Newton, T.P.; Pitts, T.M.; Glogowska, M.J.; Kabos, P.; Sartorius, C.A.; Sullivan, K.D.; Espinosa, J.M.; et al. P53 family members regulate phenotypic response to aurora kinase A inhibition in triple-negative breast cancer. Mol. Cancer Ther. 2015, 14, 1117–1129. [Google Scholar] [CrossRef]
- Kim, E.M.; Jung, C.-H.; Kim, J.; Hwang, S.-G.; Park, J.K.; Um, H.-D. The p53/p21 complex regulates cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017, 77, 3092–3100. [Google Scholar] [CrossRef]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Fogeron, M.-L.; Müller, H.; Schade, S.; Dreher, F.; Lehmann, V.; Kühnel, A.; Scholz, A.-K.; Kashofer, K.; Zerck, A.; Fauler, B.; et al. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat. Commun. 2013, 4, 1531. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Salokas, K.; Tamene, F.; Jiu, Y.; Weldatsadik, R.G.; Öhman, T.; Varjosalo, M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Peschiaroli, A.; Scialpi, F.; Bernassola, F.; El Sherbini, E.S.; Melino, G. The E3 ubiquitin ligase WWP1 regulates ΔNp63-dependent transcription through Lys63 linkages. Biochem. Biophys. Res. Commun. 2010, 402, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, X.; Guo, P.; Dong, X.-Y.; Sethi, P.; Cheng, X.; Zhou, J.; Ling, J.; Simons, J.W.; Lingrel, J.B.; et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J. Biol. Chem. 2005, 280, 41553–41561. [Google Scholar] [CrossRef]
- Ge, F.; Chen, W.; Qin, J.; Zhou, Z.; Liu, R.; Liu, L.; Tan, J.; Zou, T.; Li, H.; Ren, G.; et al. Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Krüppel-like factor 5 (KLF5). Oncotarget 2015, 6, 21369–21378. [Google Scholar] [CrossRef] [PubMed]
- Morén, A.; Imamura, T.; Miyazono, K.; Heldin, C.-H.; Moustakas, A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 2005, 280, 22115–22123. [Google Scholar] [CrossRef]
- Tripp, C.S.; Blomme, E.A.G.; Chinn, K.S.; Hardy, M.M.; LaCelle, P.; Pentland, A.P. Epidermal COX-2 induction following ultraviolet irradiation: Suggested mechanism for the role of COX-2 inhibition in photoprotection. J. Investig. Dermatol. 2003, 121, 853–861. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Chen, Y.; Mistry, D.S.; Sen, G.L. Highly rapid and efficient conversion of human fibroblasts to keratinocyte-like cells. J. Investig. Dermatol. 2014, 134, 335–344. [Google Scholar] [CrossRef]
- Alexandrova, E.M.; Petrenko, O.; Nemajerova, A.; Romano, R.A.; Sinha, S.; Moll, U.M. ΔNp63 regulates select routes of reprogramming via multiple mechanisms. Cell Death Differ. 2013, 20, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Matta, A.; Masui, O.; Srivastava, G.; Kaur, J.; Thakar, A.; Shukla, N.K.; RoyChoudhury, A.; Sharma, M.; Walfish, P.G.; et al. Nuclear heterogeneous nuclear ribonucleoprotein D is associated with poor prognosis and interactome analysis reveals its novel binding partners in oral cancer. J. Transl. Med. 2015, 13, 285. [Google Scholar] [CrossRef] [PubMed]
- Fomenkov, A.; Huang, Y.-P.; Topaloglu, O.; Brechman, A.; Osada, M.; Fomenkova, T.; Yuriditsky, E.; Trink, B.; Sidransky, D.; Ratovitski, E. P63 alpha mutations lead to aberrant splicing of keratinocyte growth factor receptor in the Hay-Wells syndrome. J. Biol. Chem. 2003, 278, 23906–23914. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cui, Y.; Liu, X.; Zhang, F.; Lu, L.-Y.; Yu, X. The RNF20/40 complex regulates p53-dependent gene transcription and mRNA splicing. J. Mol. Cell Biol. 2020, 12, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S.; Dey, S. Induction of p73, Δ133p53, Δ160p53, pAKT lead to neuroprotection via DNA repair by 5-LOX inhibition. Mol. Biol. Rep. 2020, 47, 269–274. [Google Scholar] [CrossRef]
- Hall, C.; Muller, P.A.J. The diverse functions of mutant 53, its family members and isoforms in cancer. Int. J. Mol. Sci. 2019, 20, 6188. [Google Scholar] [CrossRef]
- Merlo, P.; Fulco, M.; Costanzo, A.; Mangiacasale, R.; Strano, S.; Blandino, G.; Taya, Y.; Lavia, P.; Levrero, M. A role of p73 in mitotic exit. J. Biol. Chem. 2005, 280, 30354–30360. [Google Scholar] [CrossRef]
- Toh, W.H.; Nam, S.Y.; Sabapathy, K. An essential role for p73 in regulating mitotic cell death. Cell Death Differ. 2010, 17, 787–800. [Google Scholar] [CrossRef]
- Marrazzo, E.; Marchini, S.; Tavecchio, M.; Alberio, T.; Previdi, S.; Erba, E.; Rotter, V.; Broggini, M. The expression of the DeltaNp73beta isoform of p73 leads to tetraploidy. Eur. J. Cancer 2009, 45, 443–453. [Google Scholar] [CrossRef]
- Sasai, K.; Treekitkarnmongkol, W.; Kai, K.; Katayama, H.; Sen, S. Functional significance of aurora kinases-p53 protein family interactions in cancer. Front. Oncol. 2016, 6, 247. [Google Scholar] [CrossRef]
- Gregson, H.C.; Schmiesing, J.A.; Kim, J.S.; Kobayashi, T.; Zhou, S.; Yokomori, K. A potential role for human cohesin in mitotic spindle aster assembly. J. Biol. Chem. 2001, 276, 47575–47582. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Ball, A.R.; Pham, H.X.; Zeng, W.; Chen, H.-Y.; Schmiesing, J.A.; Kim, J.-S.; Berns, M.; Yokomori, K. Distinct functions of human cohesin-SA1 and cohesin-SA2 in double-strand break repair. Mol. Cell. Biol. 2014, 34, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Kiyomitsu, T.; Obuse, C.; Yanagida, M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 2007, 13, 663–676. [Google Scholar] [CrossRef]
- Dou, Z.; Prifti, D.K.; Gui, P.; Liu, X.; Elowe, S.; Yao, X. Recent progress on the localization of the spindle assembly checkpoint machinery to kinetochores. Cells 2019, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Vernole, P.; Neale, M.H.; Barcaroli, D.; Munarriz, E.; Knight, R.A.; Tomasini, R.; Mak, T.W.; Melino, G.; De Laurenzi, V. TAp73alpha binds the kinetochore proteins Bub1 and Bub3 resulting in polyploidy. Cell Cycle 2009, 8, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Bid, H.K.; Roberts, R.D.; Cam, M.; Audino, A.; Kurmasheva, R.T.; Lin, J.; Houghton, P.J.; Cam, H. ΔNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumor angiogenesis. Cancer Res. 2014, 74, 320–329. [Google Scholar] [CrossRef]
- Monti, P.; Ciribilli, Y.; Foggetti, G.; Menichini, P.; Bisio, A.; Cappato, S.; Inga, A.; Divizia, M.T.; Lerone, M.; Bocciardi, R.; et al. P63 modulates the expression of the WDFY2 gene which is implicated in cancer regulation and limb development. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Gatti, V.; Fierro, C.; Annicchiarico-Petruzzelli, M.; Melino, G.; Peschiaroli, A. ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 2019, 13, 981–1001. [Google Scholar] [CrossRef]
- Tanis, S.E.J.; Jansen, P.W.T.C.; Zhou, H.; van Heeringen, S.J.; Vermeulen, M.; Kretz, M.; Mulder, K.W. Splicing and chromatin factors jointly regulate epidermal differentiation. Cell Rep. 2018, 25, 1292–1303. [Google Scholar] [CrossRef]
- Mohibi, S.; Zhang, J.; Chen, X. PABPN1, a target of p63, modulates keratinocyte differentiation through regulation of p63α mRNA translation. J. Investig. Dermatol. 2020. [Google Scholar] [CrossRef]
- Ranieri, D.; Rosato, B.; Nanni, M.; Belleudi, F.; Torrisi, M.R. Expression of the FGFR2c mesenchymal splicing variant in human keratinocytes inhibits differentiation and promotes invasion. Mol. Carcinog. 2018, 57, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Vorovich, E.; Ratovitski, E.A. Dual regulation of TERT activity through transcription and splicing by DeltaNP63alpha. Aging 2008, 1, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Aktaş, T.; Avşar Ilık, İ.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Okholm, T.L.H.; Venø, M.T.; Kjems, J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018, 15, 280–291. [Google Scholar] [CrossRef]
- Yu, Y.; Reed, R. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Natl. Acad. Sci. USA 2015, 112, 8608–8613. [Google Scholar] [CrossRef]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef]
- Huang, Y.; Sen, T.; Nagpal, J.; Upadhyay, S.; Trink, B.; Ratovitski, E.; Sidransky, D. ATM kinase is a master switch for the Delta Np63 alpha phosphorylation/degradation in human head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 2008, 7, 2846–2855. [Google Scholar] [CrossRef]
- Huang, Y.; Jeong, J.S.; Okamura, J.; Sook-Kim, M.; Zhu, H.; Guerrero-Preston, R.; Ratovitski, E.A. Global tumor protein p53/p63 interactome: Making a case for cisplatin chemoresistance. Cell Cycle 2012, 11, 2367–2379. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Kim, Y.; Li, Z.; Fomenkov, T.; Fomenkov, A.; Ratovitski, E.A. AEC-associated p63 mutations lead to alternative splicing/protein stabilization of p63 and modulation of Notch signaling. Cell Cycle 2005, 4, 1440–1447. [Google Scholar] [CrossRef]
- Marini, A.; Rotblat, B.; Sbarrato, T.; Niklison-Chirou, M.V.; Knight, J.R.P.; Dudek, K.; Jones, C.; Bushell, M.; Knight, R.A.; Amelio, I.; et al. TAp73 contributes to the oxidative stress response by regulating protein synthesis. Proc. Natl. Acad. Sci. USA 2018, 115, 6219–6224. [Google Scholar] [CrossRef]
- Horvilleur, E.; Bauer, M.; Goldschneider, D.; Mergui, X.; de la Motte, A.; Bénard, J.; Douc-Rasy, S.; Cappellen, D. P73alpha isoforms drive opposite transcriptional and post-transcriptional regulation of MYCN expression in neuroblastoma cells. Nucleic Acids Res. 2008, 36, 4222–4232. [Google Scholar] [CrossRef] [PubMed]
- Gonfloni, S.; Di Tella, L.; Caldarola, S.; Cannata, S.M.; Klinger, F.G.; Di Bartolomeo, C.; Mattei, M.; Candi, E.; De Felici, M.; Melino, G.; et al. Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat. Med. 2009, 15, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; De Laurenzi, V.; Munarriz, E.; Green, D.R.; Liu, Y.-C.; Vousden, K.H.; Cesareni, G.; Melino, G. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005, 24, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, Z.; Chen, C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008, 15, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.C.; Marques, M.M. Novel role of p73 as a regulator of developmental angiogenesis: Implication for cancer therapy. Mol. Cell. Oncol. 2016, 3, e1019973. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sabapathy, K. P73: A positive or negative regulator of angiogenesis, or both? Mol. Cell. Biol. 2015, 36, 848–854. [Google Scholar] [CrossRef]
- Napoli, M.; Flores, E.R. The p53 family orchestrates the regulation of metabolism: Physiological regulation and implications for cancer therapy. Br. J. Cancer 2017, 116, 149–155. [Google Scholar] [CrossRef]
- Itahana, Y.; Itahana, K. Emerging roles of p53 family members in glucose metabolism. Int. J. Mol. Sci. 2018, 19, 776. [Google Scholar] [CrossRef]
- Engelmann, D.; Meier, C.; Alla, V.; Pützer, B.M. A balancing act: Orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 2015, 34, 4287–4299. [Google Scholar] [CrossRef]
- Nemajerova, A.; Amelio, I.; Gebel, J.; Dötsch, V.; Melino, G.; Moll, U.M. Non-oncogenic roles of TAp73: From multiciliogenesis to metabolism. Cell Death Differ. 2018, 25, 144–153. [Google Scholar] [CrossRef]
- Cam, M.; Charan, M.; Welker, A.M.; Dravid, P.; Studebaker, A.W.; Leonard, J.R.; Pierson, C.R.; Nakano, I.; Beattie, C.E.; Hwang, E.I.; et al. ΔNp73/ETS2 complex drives glioblastoma pathogenesis- targeting downstream mediators by rebastinib prolongs survival in preclinical models of glioblastoma. Neuro Oncol. 2020, 22, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Fürst, K.; Steder, M.; Logotheti, S.; Angerilli, A.; Spitschak, A.; Marquardt, S.; Schumacher, T.; Engelmann, D.; Herchenröder, O.; Rupp, R.A.W.; et al. DNp73-induced degradation of tyrosinase links depigmentation with EMT-driven melanoma progression. Cancer Lett. 2019, 442, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozenberg, J.M.; Rogovaya, O.S.; Melino, G.; Barlev, N.A.; Kagansky, A. Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia. Cells 2021, 10, 25. https://doi.org/10.3390/cells10010025
Rozenberg JM, Rogovaya OS, Melino G, Barlev NA, Kagansky A. Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia. Cells. 2021; 10(1):25. https://doi.org/10.3390/cells10010025
Chicago/Turabian StyleRozenberg, Julian M., Olga S. Rogovaya, Gerry Melino, Nickolai A. Barlev, and Alexander Kagansky. 2021. "Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia" Cells 10, no. 1: 25. https://doi.org/10.3390/cells10010025
APA StyleRozenberg, J. M., Rogovaya, O. S., Melino, G., Barlev, N. A., & Kagansky, A. (2021). Distinct p63 and p73 Protein Interactions Predict Specific Functions in mRNA Splicing and Polyploidy Control in Epithelia. Cells, 10(1), 25. https://doi.org/10.3390/cells10010025




