Investigation of Seed transmission in Peronospora belbahrii the Causal Agent of Basil Downy Mildew
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pathogen
2.2. Plants
2.3. Inoculation
2.4. Disease Assessment
2.5. Microscopy
2.6. DNA Extraction
2.7. PCR
- Bas-F: CCGTACAACCCAATAATTTGGGGGTTAAT
- Bas-R: TTCAATTAGCTACTTGTTCAGACAAAG)
2.8. Direct Inoculation of Seeds and Roots
2.9. Seed Infection and Transmission
3. Results
3.1. Systemic Infection
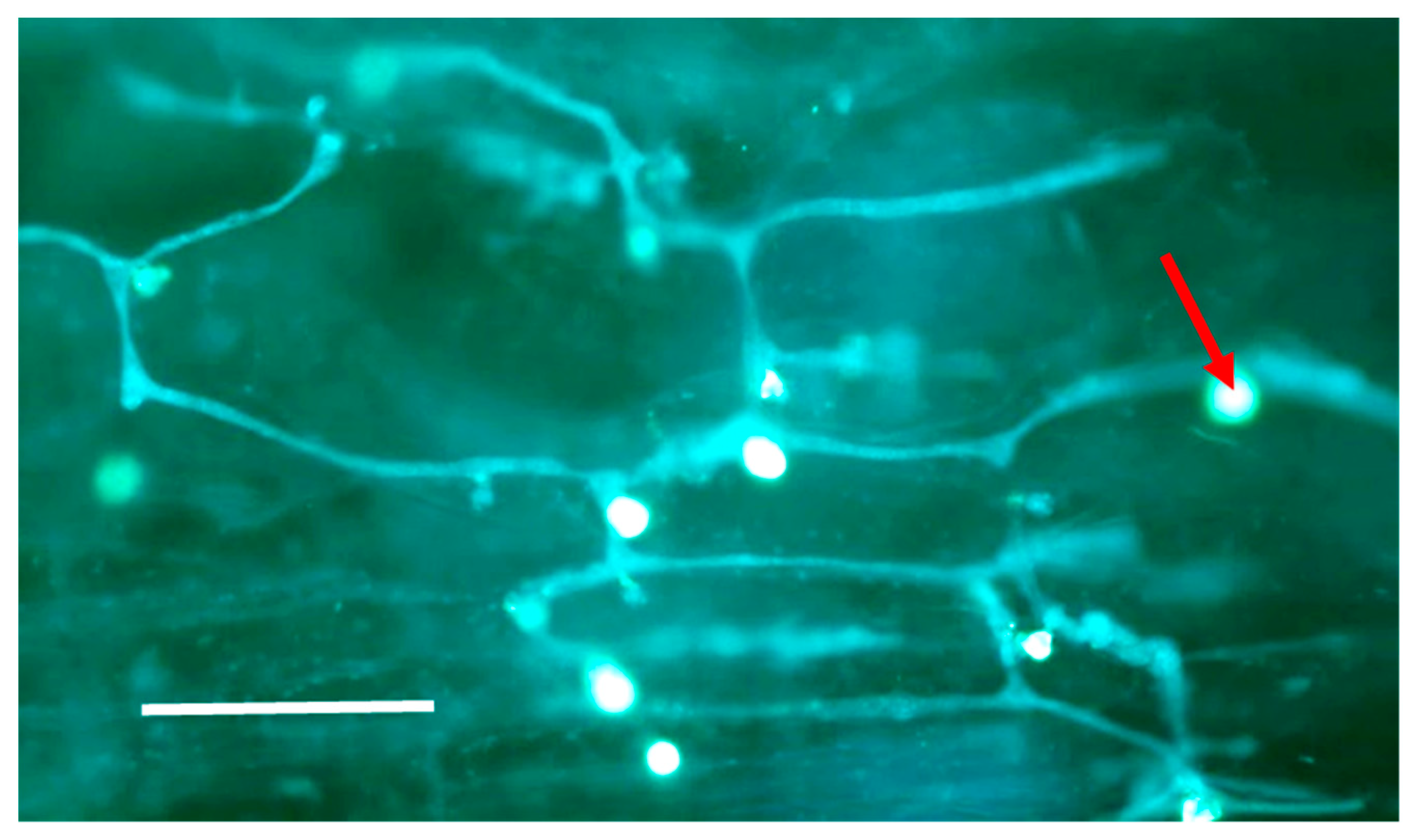
3.2. Microscopy
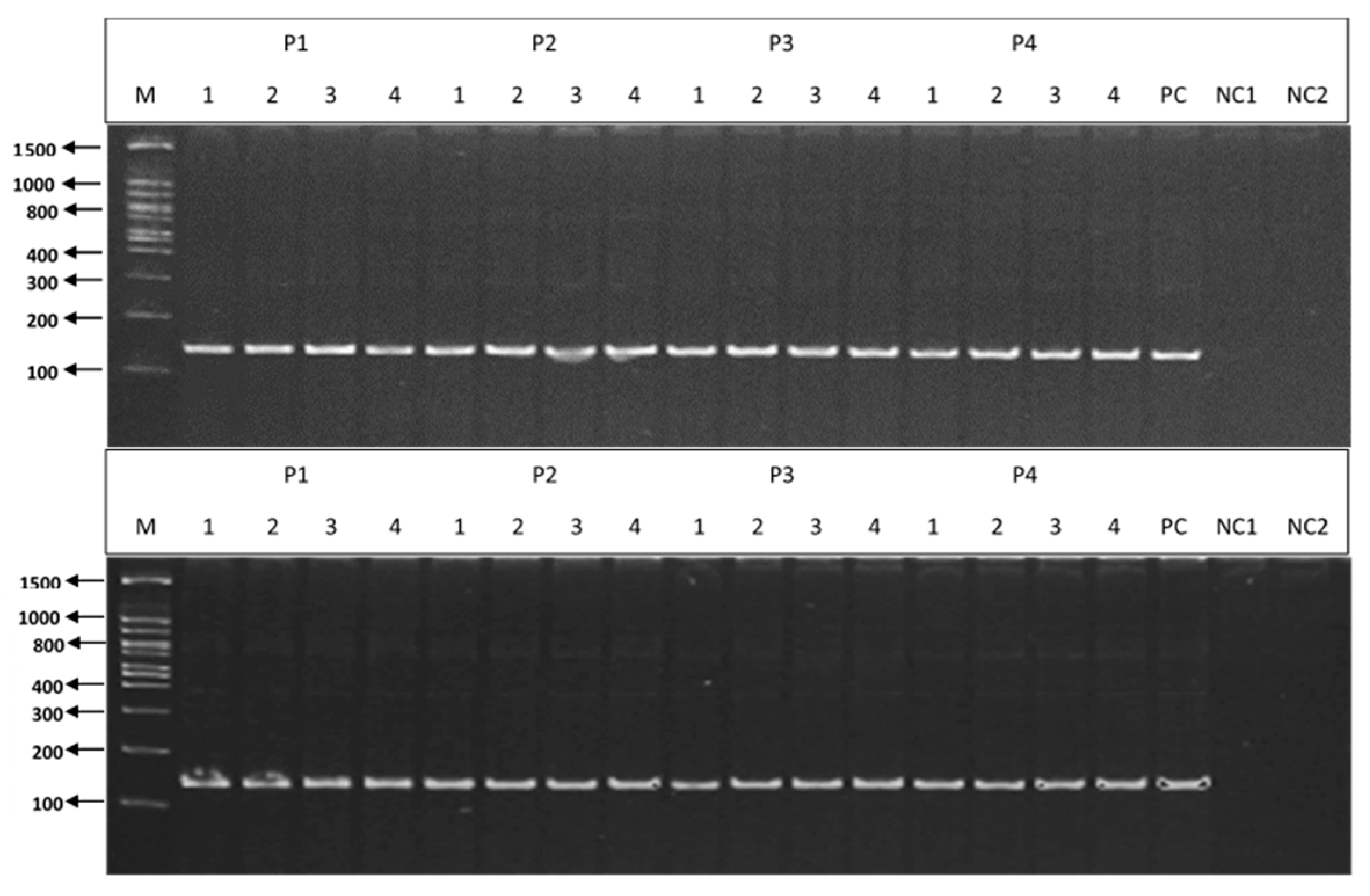
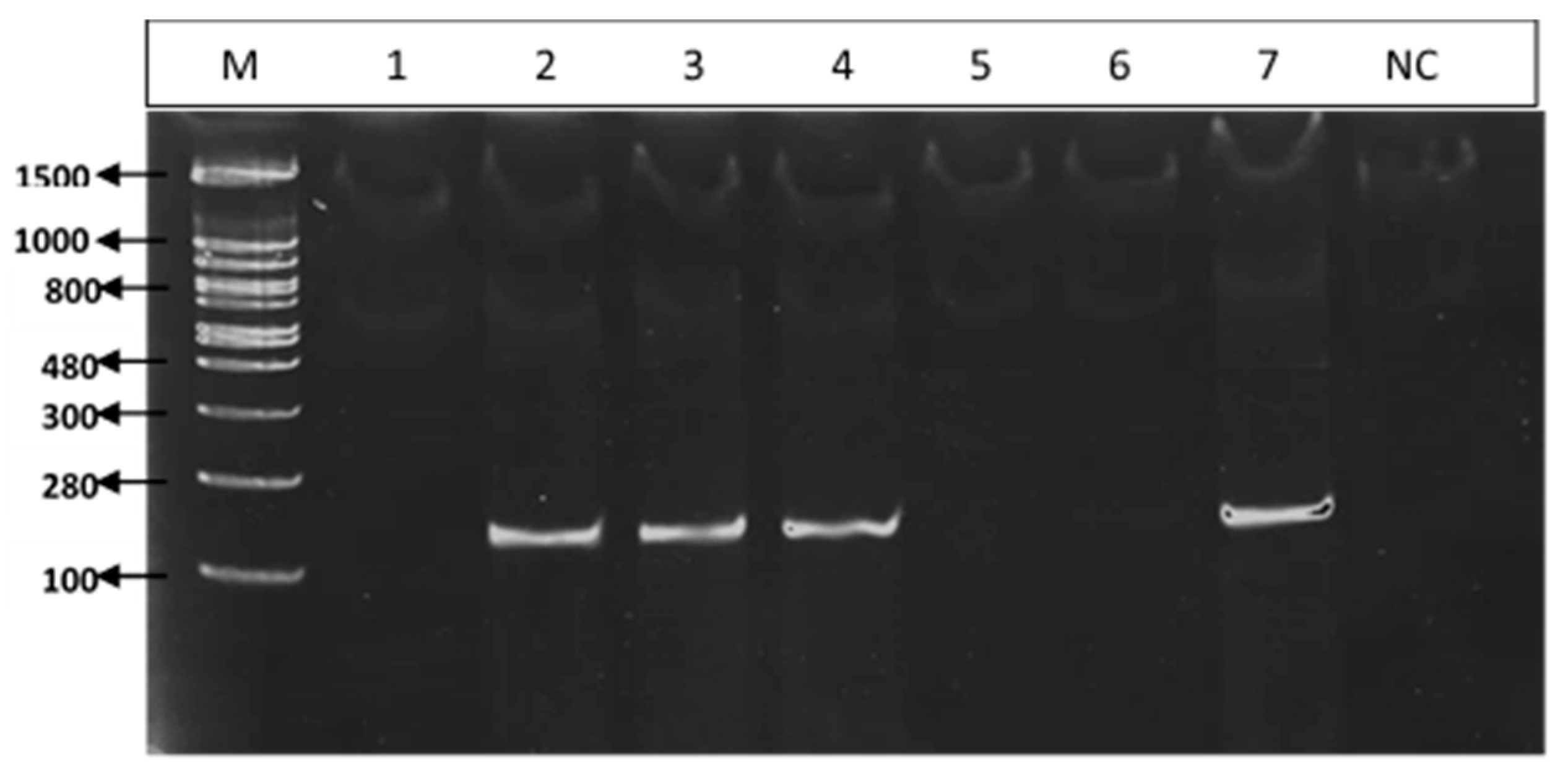
3.3. PCR
3.4. Axillary Bud Infection
3.5. Root Infection
3.6. Seed Infection and Seed Transmission
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Cohen, Y.; Ben Naim, Y.; Falach, L.; Rubin, A.E. Epidemiology of basil downy mildew. Phytopathology 2017, 107, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Hansford, C.G. Annual report of the mycologist. Rev. Appl. Mycol. 1933, 12, 421–422. [Google Scholar]
- Belbahri, L.; Calmin, G.; Pawlowski, J.; Lefort, F. Phylogenetic analysis and real time PCR detection of a presumbably undescribed Peronospora species on sweet basil and sage. Mycol. Res. 2005, 109, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.D.; Raid, R.N.; Harmon, P.F.; Jordan, S.A.; Palmateer, A.J. First report of downy mildew caused by a Peronospora sp on basil in Florida and the United States. Plant Dis. 2009, 93, 199. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Vaknin, M.; Ben-Naim, Y.; Rubin, A.E.; Galperin, M.; Silverman, D.; Bitton, S.; Adler, U. First report of the occurrence and resistance to mefenoxam of Peronospora belbahrii, causal agent of downy mildew of basil (Ocimum basilicum) in Israel. Plant Dis. 2013, 97, 692. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, A.; Minuto, G.; Bertetti, D.; Gullino, M.L. Seed transmission of Peronospora sp. of basil. J. Plant Dis. Prot. 2004, 111, 465–469. [Google Scholar]
- Farahani-Kofoet, R.D.; Roemer, P.; Grosch, R. Systemic spread of downy mildew in basil plants and detection of the pathogen in seed and plant samples. Mycol. Progress 2012, 11, 961–966. [Google Scholar] [CrossRef]
- Cohen, Y.; Sackston, W.E. Factors affecting infection of sunflowers by Plasmopara halstedii. Can. J. Bot. 1973, 51, 15–22. [Google Scholar] [CrossRef]
- Subbarao, C.S.; Anchieta, A.; Ochoa, L.; Dhar, N.; Kunjeti, S.G.; Subbarao, K.; Klosterman, S.J. Detection of latent infections of Peronospora effusa in spinach. Plant Dis. 2018, 102, 1766–1771. [Google Scholar] [CrossRef] [PubMed]
- Wyenandt, C.A.; Simon, J.E.; Pyne, R.M.; Homa, K.; McGrath, M.T.; Zhang, S.A.; Raid, R.N.; Ma, L.J.; Wick, R.; Guo, L.; et al. Basil downy mildew (Peronospora belbahrii): Discoveries and challenges relative to its control. Phytopathology 2015, 105, 885–894. [Google Scholar] [CrossRef]
- Gilardi, G.; Pintore, I.; Demarchi, S.; Gullino, M.L.; Garibaldi, A. Seed dressing to control downy mildew of basil. Phytoparasitica 2015, 43, 531–539. [Google Scholar] [CrossRef]
- Pintore, I.; Gilardi, G.; Gullino, M.L.; Garibaldi, A. Detection of mefenoxam-resistant strains of Peronospora belbahrii, the causal agent of basil downy mildew, transmitted through infected seeds. Phytoparasitica 2016, 44, 563–569. [Google Scholar] [CrossRef]
- Wyenandt, C.A.; Maimone, L.R.; Homa, K.; Madeiras, A.M.; Wick, R.L.; Simon, J.E. Detection of the downy mildew pathogen on seed of basil following field infection in Southern New Jersey. Horttechnology 2018, 28, 637–641. [Google Scholar] [CrossRef]
- Spenser, D.M.E. The Downy Mildews; Academic Press: Cambridge, MA, USA, 1981. [Google Scholar]
- Cohen, Y.; Sackston, W.E. Seed infection and latent infection of sunflowers by Plasmopara halstedii. Can. J. Bot. 1974, 52, 231–238. [Google Scholar] [CrossRef]
- Cohen, Y.; Rubin, A.E.; Galperin, M.; Ploch, S.; Runge, F.; Thines, M. Seed transmission of Pseudoperonospora cubensis. PLoS ONE 2014, 9, e109766. [Google Scholar] [CrossRef] [PubMed]
- Ben-Naim, Y.; Falach, L.; Cohen, Y. Transfer of downy mildew resistance from wild basil (Ocimum americanum) to sweet basil (O. basilicum). Phytopathology 2018, 108, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Ben-Naim, Y.; Falach, L.; Cohen, Y. Resistance against basil downy mildew in Ocimum species. Phytopathology 2015, 105, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Eyal, H.; Hanania, J. Ultrastructure, autofluorescence, callose deposition and lignification in susceptible and resistant muskmelon leaves infected with the powdery mildew fungus Sphaerotheca fuliginea. Physiol. Mol. Plant Pathol. 1990, 36, 191–204. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Cohen, Y.; Rubin, A.E. Daytime solar heating controls downy mildew Peronospora belbahrii in sweet basil. PLoS ONE 2015, 10, e0126103. [Google Scholar] [CrossRef]
- Cohen, Y.; Sackston, W.E. Disappearance of IAA in presence of tissues of sunflowers infected by Plasmopara halstedii. Can. J. Bot. 1974, 52, 861–866. [Google Scholar] [CrossRef]
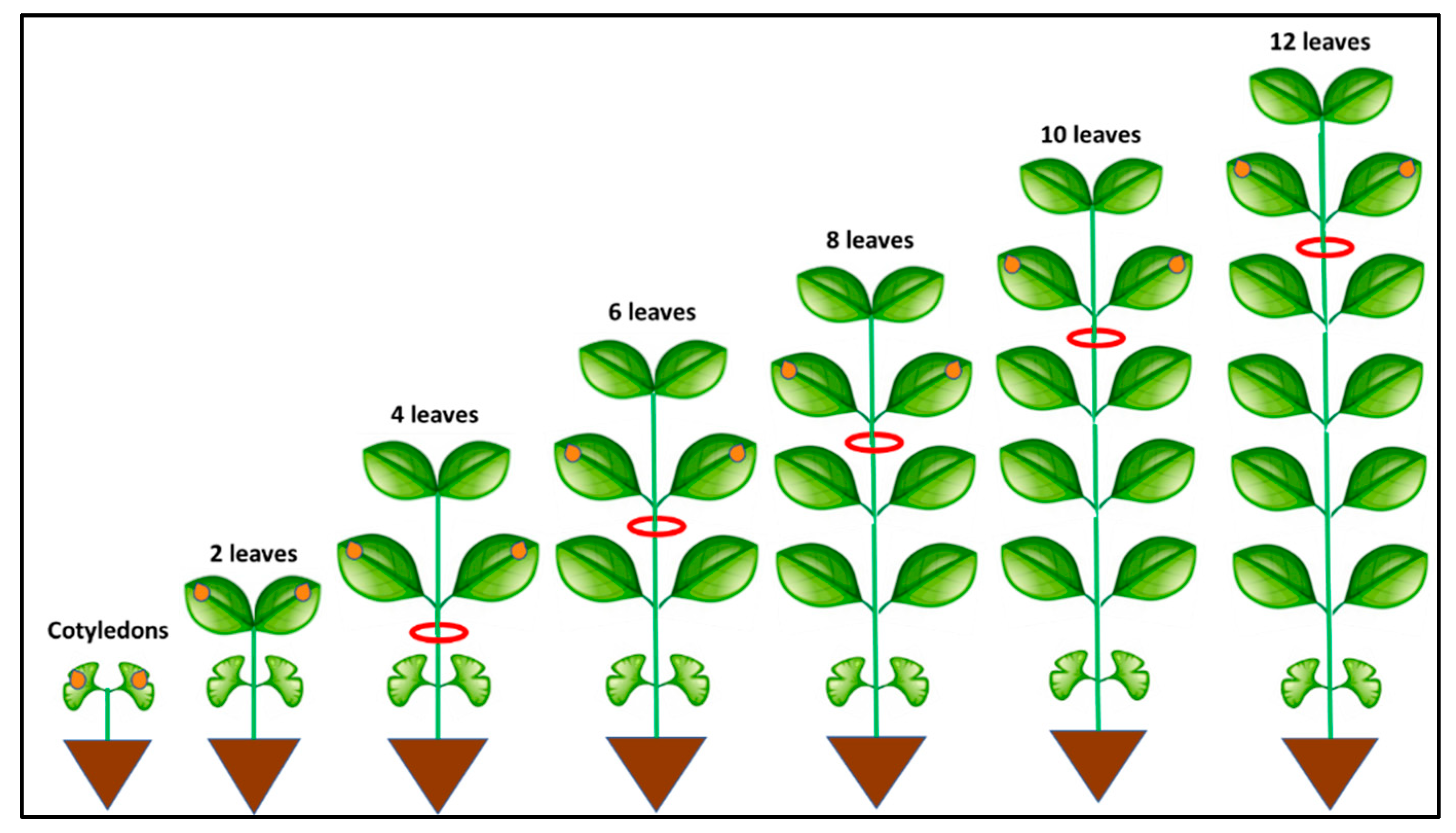



| Plant Age at Time of Inoculation | Number of Inoculated Plants | Assessment Time | Number of Systemically Infected Plants | % Systemically Infected Plants |
|---|---|---|---|---|
| Cotyledons | 147 | 7 dpi | 143 | 97.28 |
| 2 leaves | 156 | 7 dpi | 149 | 95.51 |
| 4 leaves | 159 | 7 dpi | 140 | 88.05 |
| 6 leaves | 113 | 10 dpi | 79 | 69.91 |
| 8 leaves | 117 | 10 dpi | 72 | 61.54 |
| 10 leaves | 229 | 14 dpi | 73 | 31.88 |
| 12 leaves | 117 | 14 dpi | 31 | 26.50 |
| Experiment | Year | Cultivar | Supplier | Production | Harvest | Isolate | Calyx |
|---|---|---|---|---|---|---|---|
| 1 | 2012 | Peri | Volcani center | Petah Tikva, IL | healthy | ||
| 2 | 2012 | Peri | Volcani center | Petah Tikva, IL | infected | Unknown | removed |
| 3 | 2016 | Sweet Basil | Genesis | Bar Ilan University, IL | infected | Knafo 3 | removed |
| 4 | 2016 | Peri | Volcani center | Newe Yaar, IL | healthy | ||
| 5 | 2016 | Sweet Basil | Genesis | Ashalim, IL | healthy | ||
| 6 | 2017 | Sweet Basil | Genesis | Bar Ilan University, IL | infected | Knafo 3 | removed |
| 7 | 2017 | Sweet Basil | Genesis | Bar Ilan University, IL | infected | Knafo 3 | attached |
| 8 | 2017 | Peri | Volcani center | Newe Yaar, IL | healthy | ||
| 9 | 2017 | Sweet Basil | Genesis | Ashalim, IL | healthy | ||
| 10 | 2017 | Genoveser | Ball Straathof | South Africa | healthy | ||
| 11 | 2018 | Five cultivars * | Genesis | Ashalim, IL | infected | M | removed |
| Experiment | Year | Disease Transmission | Plants Assayed by PCR | ||
|---|---|---|---|---|---|
| Seeds Sown | Infected Plants | Total * | PCR Positive | ||
| 1 | 2012 | 1000 | 0 | 50 | 0 |
| 2 | 2012 | 900 | 0 | 50 | 0 |
| 3 | 2016 | 4700 | 0 | 150 | 0 |
| 4 | 2016 | 1000 | 0 | 50 | 0 |
| 5 | 2016 | 1000 | 0 | 50 | 0 |
| 6 | 2017 | 9400 | 0 | 220 | 0 |
| 7 | 2017 | 4300 | 0 | 150 | 0 |
| 8 | 2017 | 1000 | 0 | 50 | 0 |
| 9 | 2017 | 1000 | 0 | 50 | 0 |
| 10 | 2017 | 900 | 0 | 20 | 0 |
| 11 | 2018 | 500/cultivar | 0 | 50/cultivar | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falach-Block, L.; Ben-Naim, Y.; Cohen, Y. Investigation of Seed transmission in Peronospora belbahrii the Causal Agent of Basil Downy Mildew. Agronomy 2019, 9, 205. https://doi.org/10.3390/agronomy9040205
Falach-Block L, Ben-Naim Y, Cohen Y. Investigation of Seed transmission in Peronospora belbahrii the Causal Agent of Basil Downy Mildew. Agronomy. 2019; 9(4):205. https://doi.org/10.3390/agronomy9040205
Chicago/Turabian StyleFalach-Block, Lidan, Yariv Ben-Naim, and Yigal Cohen. 2019. "Investigation of Seed transmission in Peronospora belbahrii the Causal Agent of Basil Downy Mildew" Agronomy 9, no. 4: 205. https://doi.org/10.3390/agronomy9040205
APA StyleFalach-Block, L., Ben-Naim, Y., & Cohen, Y. (2019). Investigation of Seed transmission in Peronospora belbahrii the Causal Agent of Basil Downy Mildew. Agronomy, 9(4), 205. https://doi.org/10.3390/agronomy9040205





