Genome-Wide Distribution of Novel Ta-3A1 Mini-Satellite Repeats and Its Use for Chromosome Identification in Wheat and Related Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of Tandem Repeats (TRs)
2.3. Fluorescence In Situ Hybridization (FISH)
2.4. Sequences Variation and Phylogenetic Analysis
3. Results
3.1. Ta-3A1 Was One of the Top Accumulated TRs
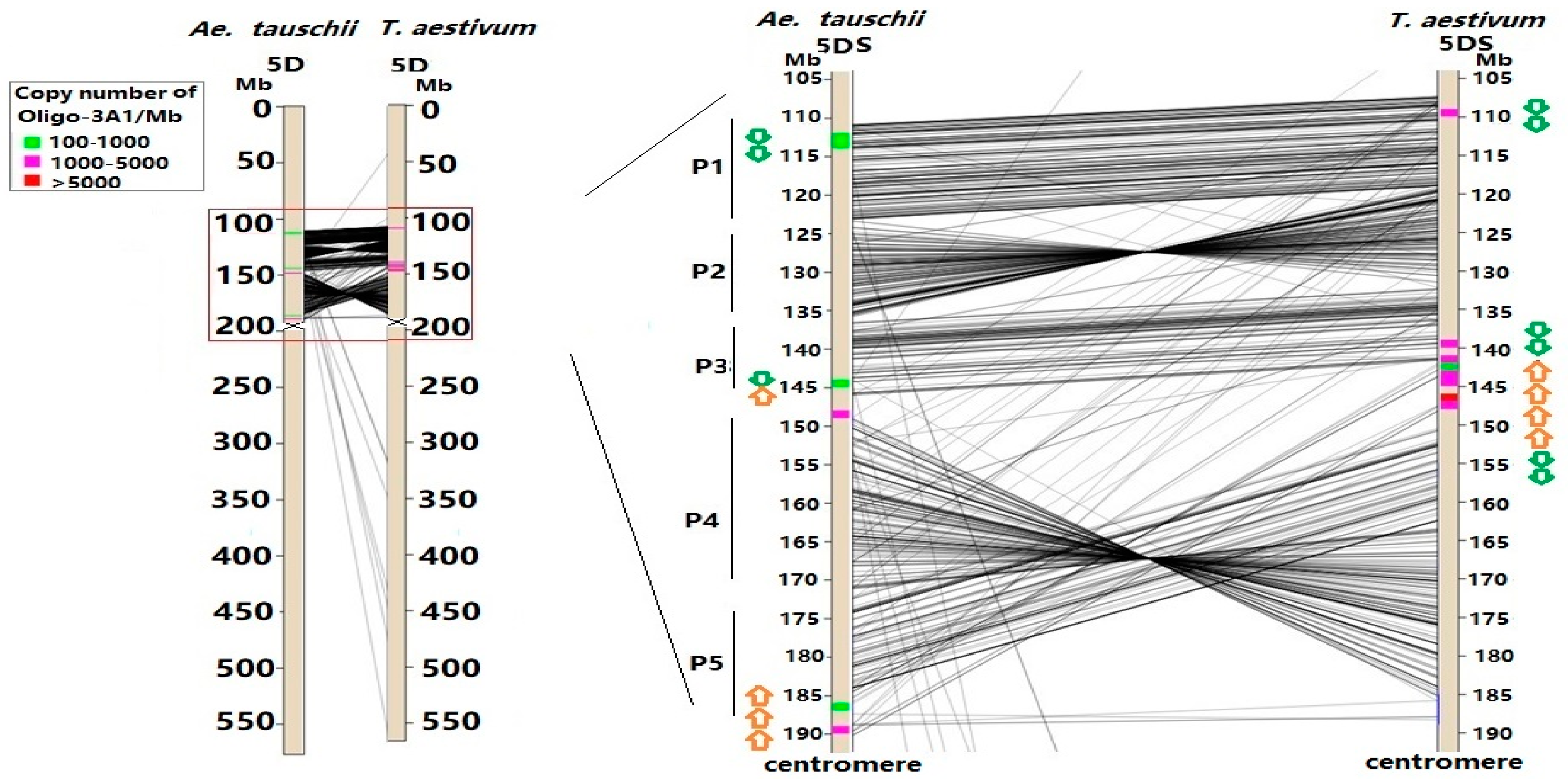
3.2. Sequence Variability of Ta-3A1 on Different Chromosomal Regions
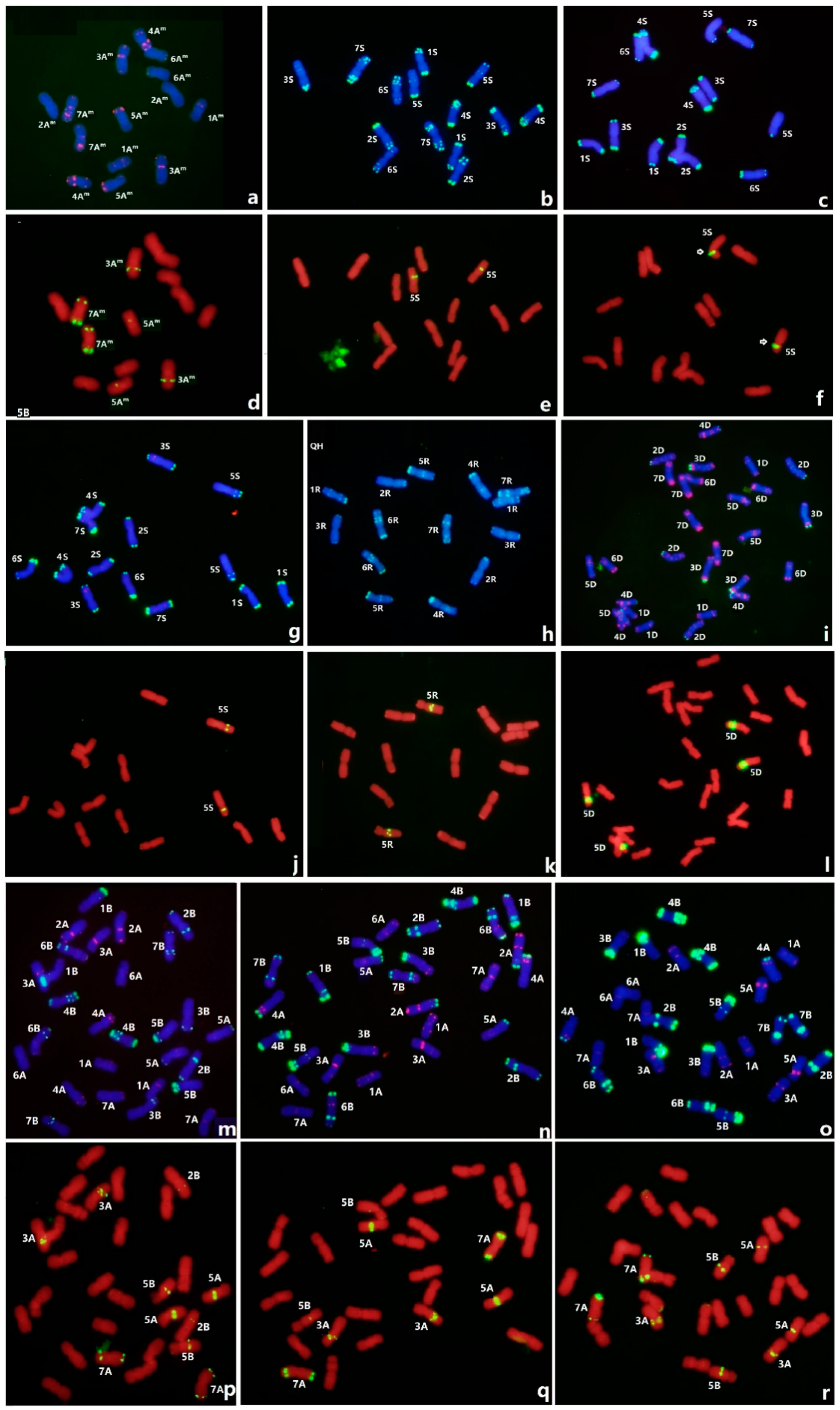
3.3. FISH of Oligo-3A1 in Wheat
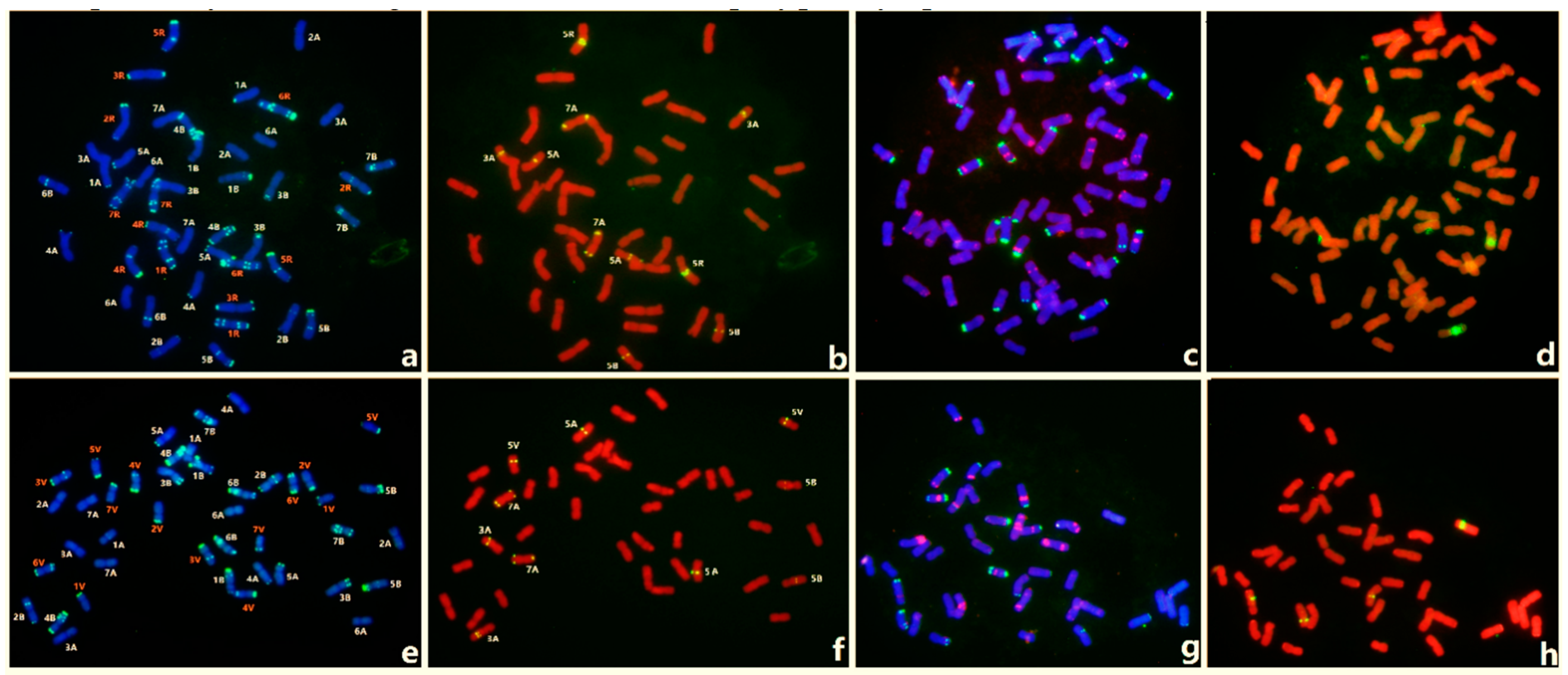
3.4. FISH of Oligo-3A1 in Representative Triticeae Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schmidt, T.; Heslop-Harrison, J.S. Genomes, genes and junk: The largescale organization of plant chromosomes. Trends Plant Sci. 1998, 3, 195–199. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Contento, A.; Heslop-Harrison, J.S.; Schwarzacher, T. Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet. Genome Res. 2005, 109, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Jeffreys, A.J.; Wilson, V.; Thein, S.L. Hypervariable “minisatellite” regions in human DNA. Nature 1985, 314, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Vergnaud, G.; Denoeud, F. Minisatellites: Mutability and genome architecture. Genome Res. 2000, 10, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Wong, Z.; Wilson, V.; Patel, I.; Povey, S.; Jeffreys, A.J. Characterization of a panel of highly variable minisatellites cloned from human DNA. Ann. Hum. Genet. 1987, 51, 269–288. [Google Scholar] [CrossRef]
- Rogstad, S.H.; Patton, J.C.; Schaal, B.A. M13 repeat probe detects DNA minisatellite-like sequences in gymnosperms and angiosperms. Proc. Natl. Acad. Sci. USA 1988, 85, 9176–9178. [Google Scholar] [CrossRef]
- Broun, P.; Tanksley, S.D. Characterization of tomato DNA clones with sequence similarity to human minisatellites 33.6 and 33.15. Plant Mol. Biol. 1993, 23, 231–242. [Google Scholar] [CrossRef]
- Tourmente, S.; Deragon, J.M.; Lafleuriel, L.; Tutois, S.; Pélissier, T.; Cuvillier, C.; Espagnol, M.C.; Picard, G. Characterization of minisatellites in Arabidopsis thaliana with sequence similarity to the human minisatellite core sequence. Nucleic Acids Res. 1994, 22, 3317–3321. [Google Scholar] [CrossRef]
- Zhou, Z.; Bebeli, P.J.; Somers, D.J.; Gustafson, J.P. Direct amplification of minisatellite-region DNA with VNTR core sequences in the genus Oryza. Theory Appl. Genet. 1997, 95, 942–949. [Google Scholar] [CrossRef]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2011, 13, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Bally, P.; Grandaubert, J.; Rouxel, T.; Balesdent, M.H. FONZIE: An optimized pipeline for minisatellite marker discovery and primer design from large sequence data sets. BMC Res. Notes 2010, 3, 322. [Google Scholar] [CrossRef] [PubMed]
- Abouelhoda, M.; El-Kalioby, M.; Giegerich, R. WAMI: A web server for the analysis of minisatellite maps. BMC Evol. Biol. 2010, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Mogil, L.S.; Slowikowski, K.; Laten, H.M. Computational and experimental analyses of retrotransposon-associated minisatellite DNAs in the soybean genome. BMC Bioinform. 2012, 13, S13. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, F.; Wenke, T.; Holtgräwe, D.; Weisshaar, B.; Schmidt, T. Analysis of a c0t-1 library enables the targeted identification of minisatellite and satellite families in Beta vulgaris. BMC Plant Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Honma, Y.; Yoshida, Y.; Terachi, T.; Toriyama, K.; Mikami, T.; Kubo, T. Polymorphic minisatellites in the mitochondrial DNAs of Oryza and Brassica. Curr. Genet. 2011, 57, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.A.F.; Dantas, J.L.L.; Oliveira, E.J. Development and validation of minisatellite markers for Carica papaya. Biol. Plant. 2015, 59, 686–694. [Google Scholar] [CrossRef]
- Orzechowska, M.; Majka, M.; Weiss-Schneeweiss, H.; Kovařík, A.; Borowska-Zuchowska, N.; Kolano, B. Organization and evolution of two repetitive sequences, 18-24J and 12-13P, in the genome of Chenopodium (Amaranthaceae). Genome 2018, 61, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.A.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Puiu, D.; Hall, R.; Kingan, S.; Clavijo, B.J.; Salzberg, S.L. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef]
- Anamthawat-Jónsson, K.; Heslop-Harrison, J.S. Isolation and characterization of genome-specific DNA sequences in Triticeae species. Mol. Gen. Genet. 1993, 240, 151–158. [Google Scholar] [CrossRef]
- Devos, K.M.; Bryan, G.J.; Collins, A.J.; Stephenson, P.; Gale, M.D. Application of two microsatellite sequences in wheat storage proteins as molecular markers. Theory Appl. Genet. 1995, 90, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.J.; Zhou, Z.; Bebeli, P.J.; Gustafson, J.P. Repetitive, genome-specific probes in wheat (Triticum aestivum L. em Thell) amplified with minisatellite core sequences. Theory Appl. Genet. 1996, 93, 982–989. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef]
- Lang, T.; Li, G.R.; Wang, H.J.; Yu, Z.H.; Chen, Q.H.; Yang, E.N.; Fu, S.L.; Tang, Z.X.; Yang, Z.J. Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 2018. [Google Scholar] [CrossRef]
- Evtushenko, E.V.; Levitsky, V.G.; Elisafenko, E.A.; Gunbin, K.V.; Belousov, A.I.; Šafář, J.; Doležel, J.; Vershinin, A.V. The expansion of heterochromatin blocks in rye reflects the co-amplification of tandem repeats and adjacent transposable elements. BMC Genom. 2016, 17, 337. [Google Scholar] [CrossRef]
- Zeng, Z.X.; Hu, L.J.; Li, G.R.; Liu, C.; Yang, Z.J. Phenotypic and epigenetic changes occurred during the autopolyploidization of Aegilops tauschii. Cereal Res. Commun. 2012, 49, 476–485. [Google Scholar] [CrossRef]
- Yang, Z.J.; Li, G.R.; Feng, J.; Jiang, H.R.; Ren, Z.L. Molecular cytogenetic characterization and disease resistance observation of wheat—Dasypyrum breviaristatum partial amphiploid and its derivatives. Hereditas 2005, 142, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Li, W.Z.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 2014, 55, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.L.; Chen, L.; Wang, Y.Y.; Li, M.; Yang, Z.J.; Qiu, L.; Yan, B.J.; Ren, Z.L.; Tang, Z.X. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, G.R.; Gao, D.; Zhang, H.J.; Li, J.B.; Wang, H.J.; La, S.X.; Ma, J.W.; Yang, Z.J. Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol. Cytogenet. 2016, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Lang, T.; Li, B.; Yu, Z.H.; Wang, H.J.; Li, G.R.; Yang, E.N.; Yang, Z.J. Introduction of Thinopyrum intermedium ssp. trichophorum chromosomes to wheat by trigeneric hybridization involving Triticum, Secale and Thinopyrum genera. Planta 2017, 245, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; De Oliveira, R.; International Wheat Genome Sequencing Consortium (IWGSC); Mayer, K.F.X.; Paux, E.; et al. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Martienssen, R.A.; Baulcombe, D.C. An unusual wheat insertion sequence (WIS1) lies upstream of an α-amylase gene in hexaploid wheat, and carries a “minisatellite” array. Mol. Gen. Genet. 1989, 217, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Y.; Tang, Z.X.; Qiu, L.; Yang, Z.J.; Li, G.R.; Lang, T.; Zhu, W.Q.; Zhang, J.H.; Fu, S.L. Developing new Oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 2018, 9, 1104. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Peluso, P.; Shi, J.; Liang, T.; Stitzer, M.C.; Wang, B.; Campbell, M.S.; Stein, J.C.; Wei, X.; Chin, C.S.; et al. Improved maize reference genome with single-molecule technologies. Nature 2017, 546, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S.; Heslop-Harrison, J.S. Characterization and genomic organization of PERI, a repetitive DNA in the Drosophila buzzatii cluster related to DINE-1 transposable elements and highly abundant in the sex chromosomes. Cytogenet. Genome Res. 2011, 132, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.H.C.; Grenier, J.K.; Barbash, D.A.; Clark, A.G. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 18793–18798. [Google Scholar] [CrossRef]
- Macas, J.; Navrátilová, A.; Koblížková, A. Sequence homogenization and chromosomal localization of VicTR-B satellites differ between closely related Vicia species. Chromosoma 2006, 115, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S.; Sene, F.M.; Moreira-Filho, O.; Schwarzacher, T.; Heslop-Harrison, J.S. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res. 2008, 16, 307–324. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, L.; Venanzetti, F.; Johnsen, A.; Sbordoni, V.; Bachmann, L. Molecular evolution of the pDo500 satellite DNA family in Dolichopoda cave crickets (Rhaphidophoridae). BMC Evol. Biol. 2009, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Petrović, V.; Luchetti, A.; Ricci, A.; Šatović, E.; Passamonti, M.; Mantovani, B. Long-term conservation vs high sequence divergence: The case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 2010, 104, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Li, G.R.; Lang, T.; Yang, E.N.; Liu, C.; Yang, Z.J. Characterization and phylogenetic analysis of α-gliadin gene sequences reveals significant genomic divergence in Triticeae species. J. Genet. 2014, 93, 725–731. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Friebe, B.; Gill, B.S. Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 1996, 39, 293–306. [Google Scholar] [CrossRef]
- Ruban, A.S.; Badaeva, E.D. Evolution of the S-Genomes in Triticum-Aegilops alliance: Evidences from chromosome analysis. Front. Plant Sci. 2018, 9, 1756. [Google Scholar] [CrossRef]
- Luchetti, A.; Cesari, M.; Carrara, G.; Cavicchi, S.; Passamonti, M.; Scali, V.; Mantovani, B. Unisexuality and molecular drive: Bag320 sequence diversity in Bacillus taxa (Insecta Phasmatodea). J. Mol. Evol. 2003, 56, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Robles, F.; de la Herrán, R.; Ludwig, A.; Ruiz Rejón, C.; Ruiz Rejón, M.; Garrido-Ramos, M.A. Evolution of ancient satellite DNAs in sturgeon genomes. Gene 2004, 338, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Meštrović, N.; Castagnone-Sereno, P.; Plohl, M. Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Mol. Biol. Evol. 2006, 23, 2316–2325. [Google Scholar] [CrossRef] [PubMed]
- Navajas-Pérez, R.; Quesada del Bosque, M.E.; Garrido-Ramos, M.A. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Mol. Genet. Genom. 2009, 282, 2316–2325. [Google Scholar] [CrossRef] [PubMed]
- Giovannotti, M.; Cerioni, P.N.; Splendiani, A.; Ruggeri, P.; Olmo, E.; Barucchi, V.C. Slow evolving satellite DNAs: The case of a centromeric satellite in Chalcides ocellatus (Forskål, 1775) (Reptilia, Scincidae). Amphibia-Reptilia 2013, 34, 401–411. [Google Scholar] [CrossRef]
- Komuro, S.; Endo, R.; Shikata, K.; Kato, A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 2013, 56, 131–137. [Google Scholar] [CrossRef]
- Danilova, T.V.; Akhunova, A.R.; Akhunov, E.D.; Friebe, B.; Gill, B.S. Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 2017, 92, 317–330. [Google Scholar] [CrossRef]
- Lang, T.; La, S.X.; Li, B.; Yu, Z.H.; Chen, Q.H.; Li, J.B.; Yang, E.N.; Li, G.R.; Yang, Z.J. Precise identification of wheat—Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 2018, 61, 177–185. [Google Scholar] [CrossRef]
- Yu, Z.H.; Wang, H.J.; Xu, Y.F.; Li, Y.S.; Lang, T.; Yang, Z.J.; Li, G.R. Characterization of chromosomal rearrangement in new wheat—Thinopyrum intermedium addition lines carrying Thinopyrum—Specific grain hardness genes. Agronomy 2019, 9, 18. [Google Scholar] [CrossRef]
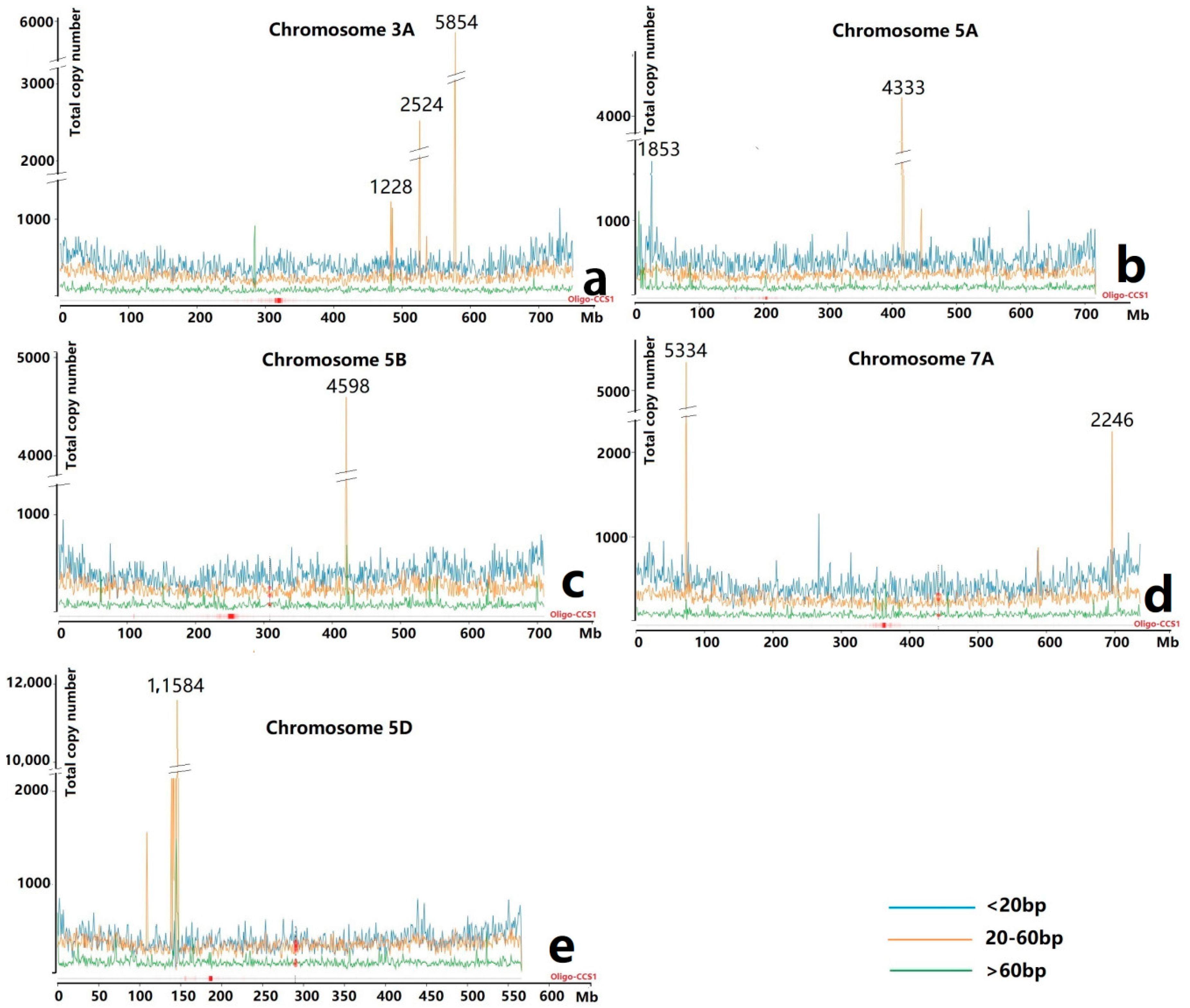
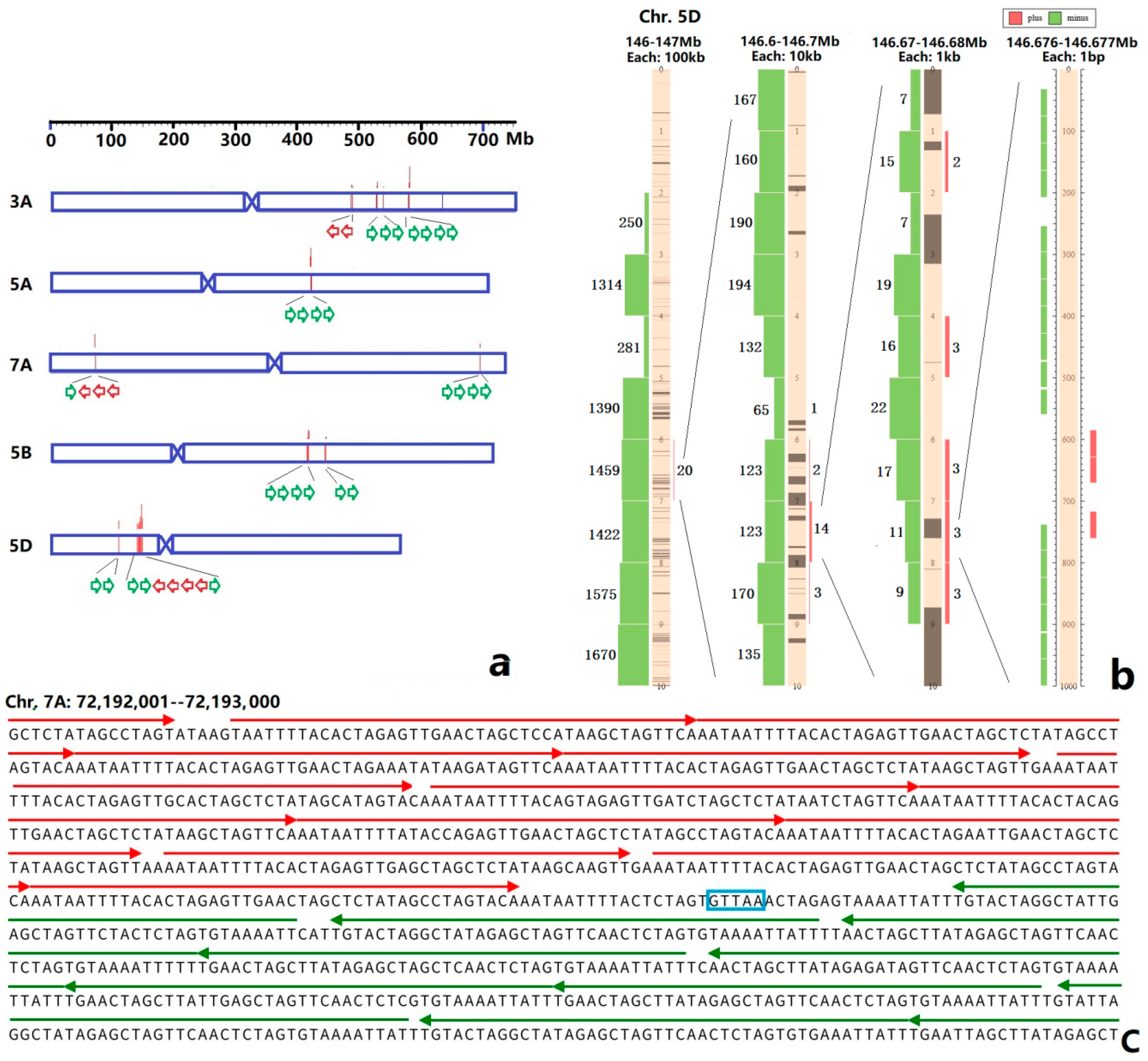
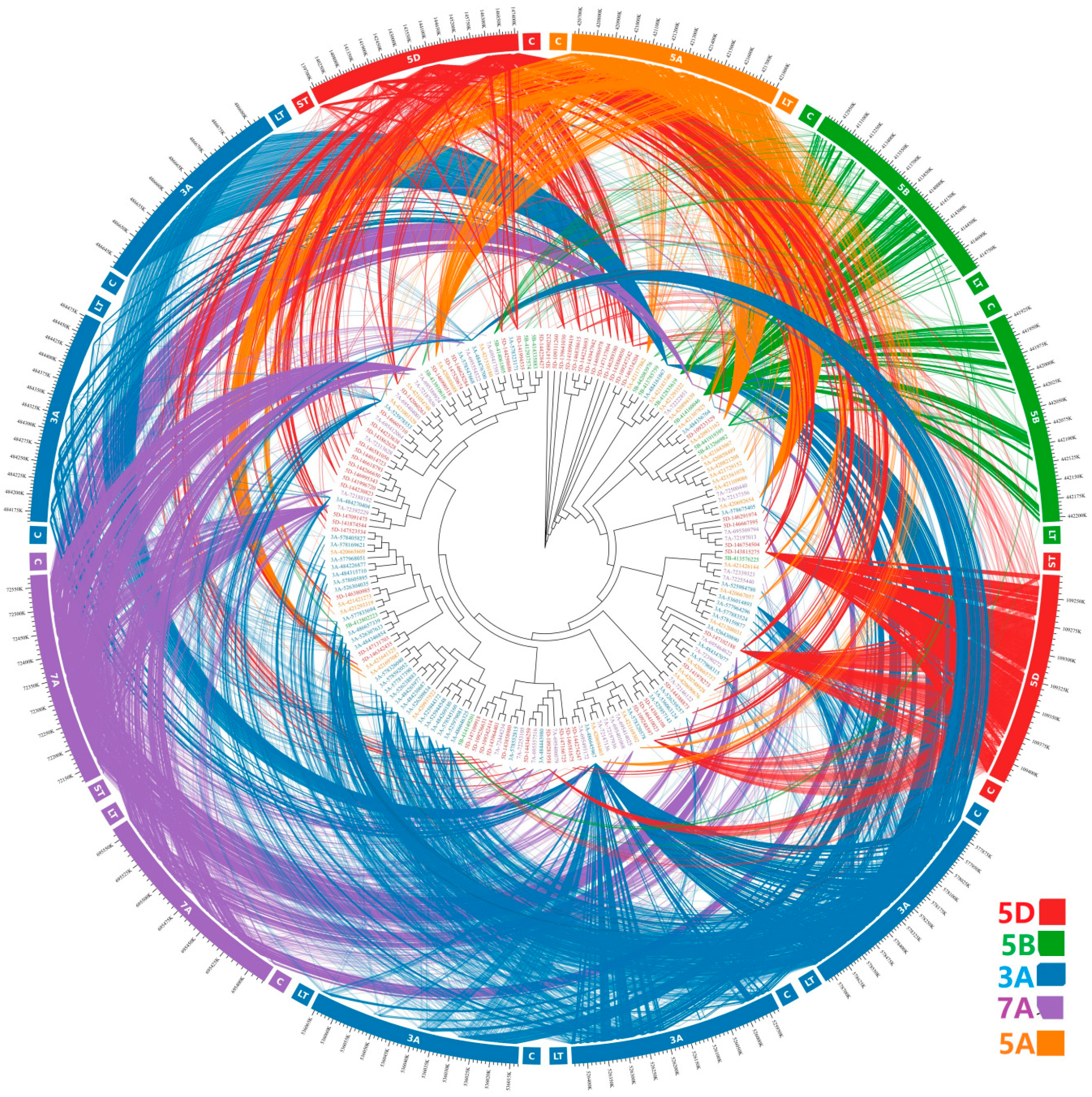
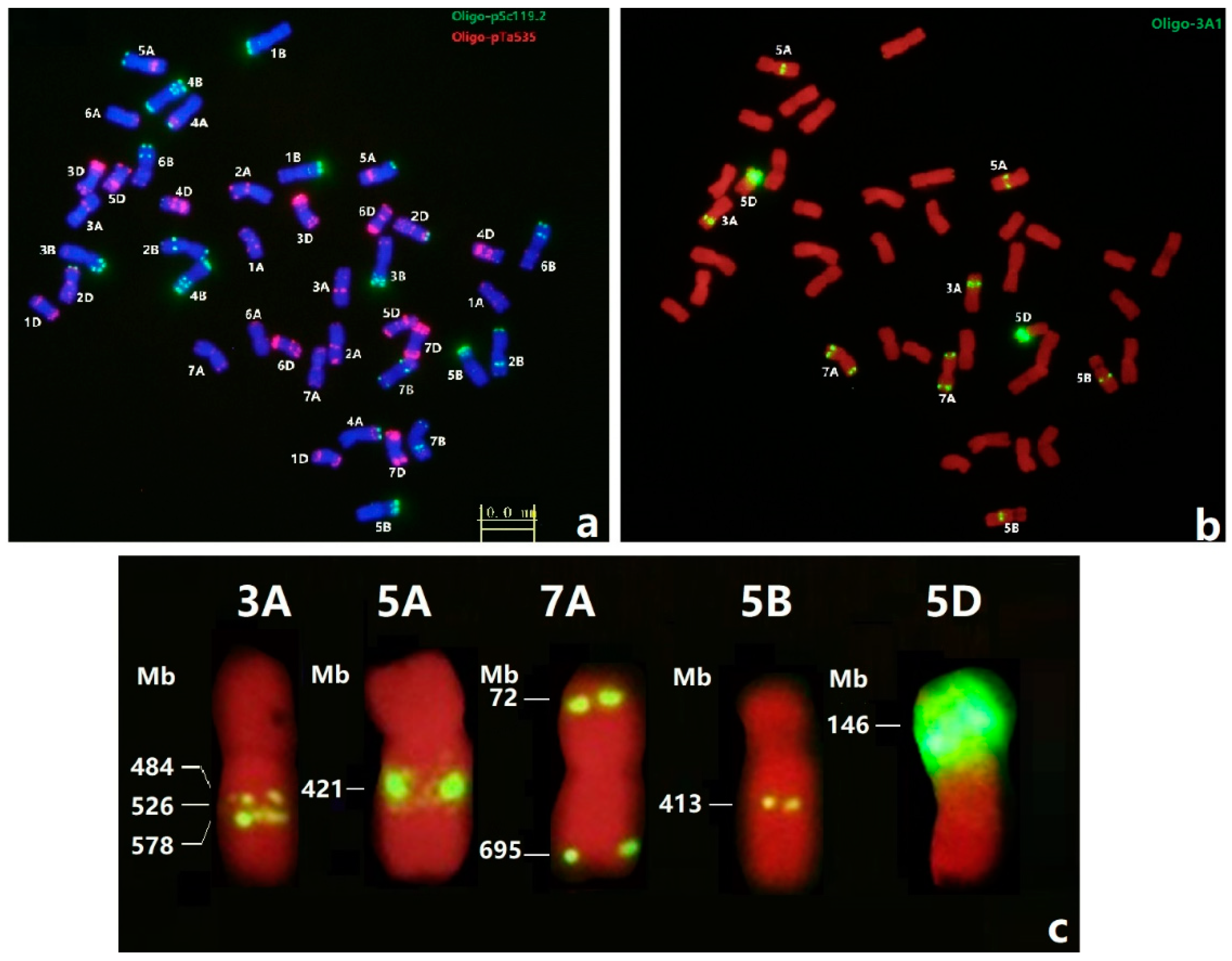
| Species | Chromosome | Copy Number | Species | Chromosome | Copy Number |
|---|---|---|---|---|---|
| 3A | 14,987 | T. urartu | 3A | 2304 | |
| 5A | 11,083 | 5A | 1199 | ||
| T. aestivum | 7A | 7828 | 7A | 835 | |
| 5B | 8444 | T. dicoccoides | 3A | 2765 | |
| 5D | 28,964 | 5A | 16,482 | ||
| 7A | 6449 | ||||
| Ae. tauschii | 5D | 5404 | 5B | 2200 |
| Chromosome | Physical Location (bp) | N | S | π | Hd ± SD | Tajima’s D |
|---|---|---|---|---|---|---|
| 3A | 484,160,160–484,477,359 | 674 | 16 | 0.131 | 0.925 ± 0.007 | −1.865 * |
| 486,643,172–486,682,396 | 370 | 24 | 0.147 | 0.987 ± 0.002 | −1.851 * | |
| 525,926,313–525,999,776 | 356 | 22 | 0.127 | 0.965 ± 0.005 | −2.014 * | |
| 526,002,472–526,436,842 | 1065 | 17 | 0.150 | 0.959 ± 0.004 | −1.648 | |
| 536,013,749–536,065,681 | 280 | 22 | 0.144 | 0.981 ± 0.003 | −1.991 * | |
| 577,811,191–577,988,203 | 685 | 24 | 0.155 | 0.991 ± 0.001 | −1.719 | |
| 578,013,600–578,743,842 | 2821 | 5 | 0.194 | 0.713 ± 0.006 | −0.916 | |
| 5A | 420,664,083–420,976,973 | 4031 | 4 | 0.159 | 0.520 ± 0.010 | −0.998 |
| 5B | 412,801,830–414,820,389 | 2806 | 9 | 0.130 | 0.765 ± 0.007 | −1.456 |
| 441,918,962–442,205,867 | 616 | 22 | 0.127 | 0.973 ± 0.004 | −1.920 * | |
| 5D | 109,229,486–109,417,074 | 908 | 19 | 0.129 | 0.948 ± 0.005 | −1.843 * |
| 139,568,074–144,410,834 | 3529 | 3 | 0.093 | 0.263 ± 0.010 | −1.276 | |
| 146,286,068–146,411,073 | 762 | 21 | 0.149 | 0.980 ± 0.003 | −1.765 | |
| 146,494,968–147,180,961 | 2924 | 9 | 0.119 | 0.693 ± 0.010 | −1.519 | |
| 147,436,033–147,525,419 | 570 | 22 | 0.143 | 0.983 ± 0.003 | −1.883 * | |
| 7A | 72,124,011–72,576,573 | 1466 | 14 | 0.120 | 0.866 ± 0.008 | −1.743 |
| 695,384,113–695,559,280 | 621 | 22 | 0.128 | 0.970 ± 0.004 | −1.942 * |
| Species | Origin | Accessions | 5B | 5A | 2BL |
|---|---|---|---|---|---|
| T. dicoccoides | Japan | H137 | ++ | ++ | |
| T. dicoccoides | Israel | H141 | +++ | ++ | |
| T. dicoccoides | Syria | H177 | ++ | +++ | |
| T. dicoccoides | Syria | H179 | + | ++ | |
| T. dicoccoides | China | H171 | + | ++ | |
| T. dicoccoides | Israel | H283 | + | +++ | |
| T. carthlicum | Turkey | H149 | + | + | |
| T. carthlicum | Turkey | H94 | + | + | |
| T. carthlicum | USA | AS311 | + | +++ | |
| T. abyssinicum | Ethiopia | H129 | + | + | |
| T. abyssinicum | Ethiopia | H131 | + | + | |
| T. durum | CIMMYT | H281 H322 H268 H242 | + | +++ | + |
| T. durum | CIMMYT | H246 | ++ | +++ | + |
| T. durum | CIMMYT | H261 | ++ | +++ | ++ |
| T. durum | CIMMYT | H242 | + | ++ | + |
| T. durum | CIMMYT | H259 | +++ | +++ | ++ |
| T. durum | Israel | H319 H305 H299 | + | +++ | |
| T. durum | Israel | Langdon | + | ++ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lang, T.; Li, G.; Yu, Z.; Ma, J.; Chen, Q.; Yang, E.; Yang, Z. Genome-Wide Distribution of Novel Ta-3A1 Mini-Satellite Repeats and Its Use for Chromosome Identification in Wheat and Related Species. Agronomy 2019, 9, 60. https://doi.org/10.3390/agronomy9020060
Lang T, Li G, Yu Z, Ma J, Chen Q, Yang E, Yang Z. Genome-Wide Distribution of Novel Ta-3A1 Mini-Satellite Repeats and Its Use for Chromosome Identification in Wheat and Related Species. Agronomy. 2019; 9(2):60. https://doi.org/10.3390/agronomy9020060
Chicago/Turabian StyleLang, Tao, Guangrong Li, Zhihui Yu, Jiwei Ma, Qiheng Chen, Ennian Yang, and Zujun Yang. 2019. "Genome-Wide Distribution of Novel Ta-3A1 Mini-Satellite Repeats and Its Use for Chromosome Identification in Wheat and Related Species" Agronomy 9, no. 2: 60. https://doi.org/10.3390/agronomy9020060
APA StyleLang, T., Li, G., Yu, Z., Ma, J., Chen, Q., Yang, E., & Yang, Z. (2019). Genome-Wide Distribution of Novel Ta-3A1 Mini-Satellite Repeats and Its Use for Chromosome Identification in Wheat and Related Species. Agronomy, 9(2), 60. https://doi.org/10.3390/agronomy9020060






