Long Non-Coding RNAs: Rising Regulators of Plant Reproductive Development
Abstract
1. Introduction
2. Identification and Specific Expression Pattern of Plant lncRNAs
3. Regulation Mechanisms of lncRNAs in Plant Reproductive Development
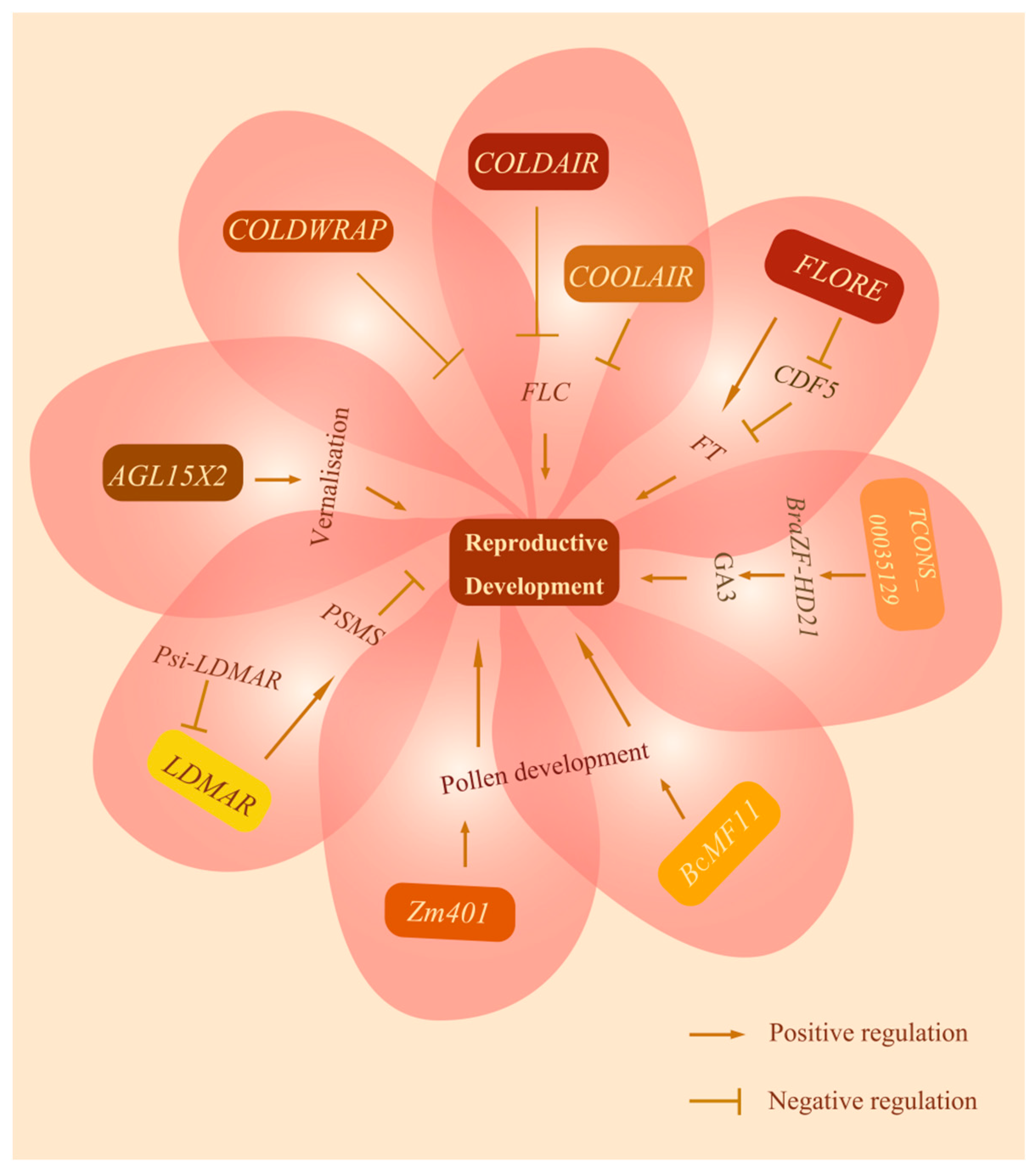
4. Function of lncRNAs in Regulation of Plant Reproductive Development
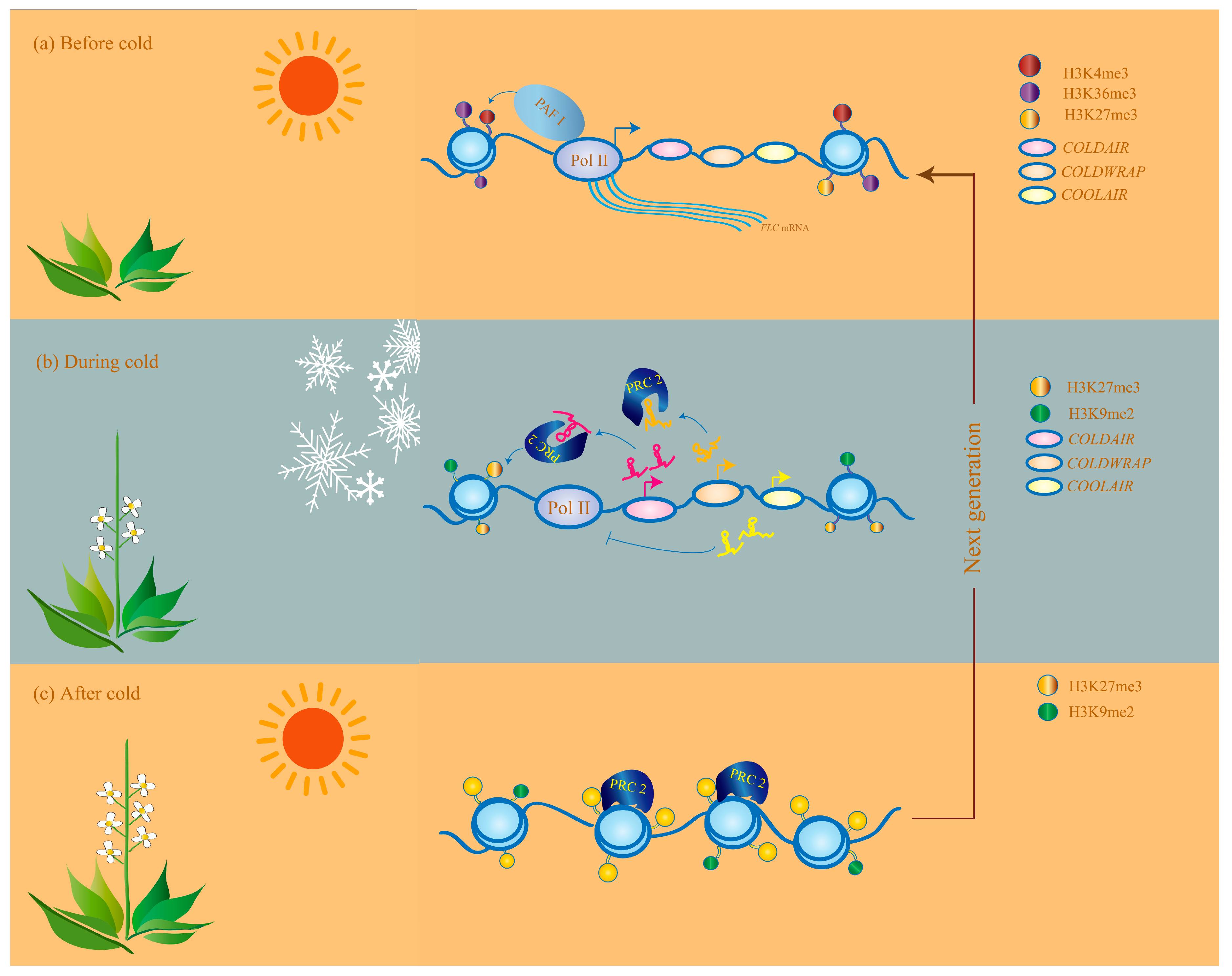
4.1. Vernalization
4.2. Photoperiodic-Sensitive Male Sterility and Pollen Development
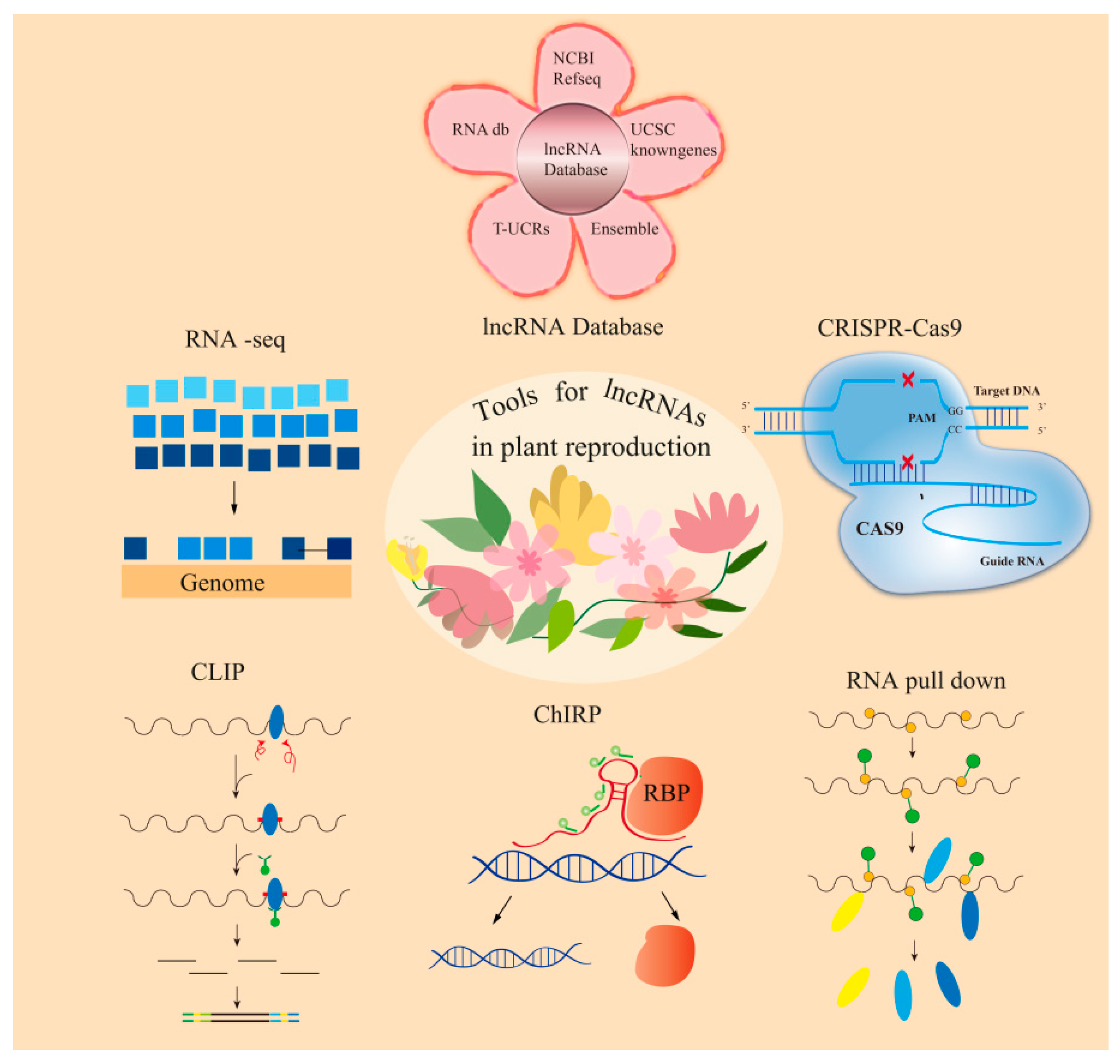
5. Perspectives
5.1. Technical Challenges
5.2. Strategies
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Wilkie, J.D.; Sedgley, M.; Olesen, T. Regulation of floral initiation in horticultural trees. J. Exp. Bot. 2008, 59, 3215–3228. [Google Scholar] [CrossRef]
- Cui, H.; Onyango, P.; Brandenburg, S.; Wu, Y.; Hsieh, C.L.; Feinberg, A.P. Loss of Imprinting in Colorectal Cancer Linked to Hypomethylation of H19 and IGF2. Cancer Res. 2002, 62, 6442–6446. [Google Scholar] [PubMed]
- Ausín, I.; Alonso-Blanco, C.; Martínez-Zapater, J.M. Environmental regulation of flowering. Int. J. Dev. Boil. 2004, 49, 689–705. [Google Scholar] [CrossRef]
- Amasino, R.M. Vernalization and flowering time. Curr. Opin. Biotechnol. 2005, 16, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Crevillen, P.; Liu, F.; Ecker, J.R.; Jerzmanowski, A.; Dean, C. Small RNA-Mediated Chromatin Silencing Directed to the 3′ Region of the Arabidopsis Gene Encoding the Developmental Regulator, FLC. Proc. Natl. Acad. Sci. USA 2007, 104, 3633–3638. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Wirth, S.; Merchan, F.; Laporte, P.; D’Aubenton-Carafa, Y.; Hirsch, J.; Maizel, A.; Mallory, A.; Lucas, A.; Deragon, J.M. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009, 19, 57–69. [Google Scholar] [CrossRef]
- Bai, Y.; Dai, X.; Harrison, A.P.; Chen, M. RNA regulatory networks in animals and plants: A long noncoding RNA perspective. Brief. Funct. Genom. 2015, 14, 91–101. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, X.W.; Shen, Y.H.; Pang, L.X.; Zhang, A.Q.; Fu, Z.Y.; Chen, J.T.; Guo, X.R.; Gan, W.H.; Ji, C.B. Distinct expression profiles of LncRNAs between brown adipose tissue and skeletal muscle. Biochem. Biophys. Res. Commun. 2014, 443, 1028–1034. [Google Scholar] [CrossRef]
- Yuen-Yi, T.; Moriarity, B.S.; Wuming, G.; Ryutaro, A.; Ashutosh, T.; Hiroko, K.; Peter, R.; Brian, R.; Kacey, G.; Beadnell, T.C. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Mchugh, C.A.; Chun-Kan, C.; Amy, C.; Surka, C.F.; Christina, T.; Patrick, M.D.; Amy, P.J.; Mario, B.; Christina, B.; Annie, M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xin, L.; Hui, Z.; Liu, H. Long non-coding RNAs: Potential new biomarkers for predicting tumor invasion and metastasis. Mol. Cancer 2016, 15, 62. [Google Scholar] [CrossRef]
- Lossos, I.S.; Morgensztern, D. Prognostic Biomarkers in Diffuse Large B-Cell Lymphoma. J. Clin. Oncol. 2006, 24, 995–1007. [Google Scholar] [CrossRef]
- Zhou, M.; Zhao, H.; Xu, W.; Bao, S.; Liang, C.; Sun, J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017, 16, 16. [Google Scholar] [CrossRef]
- Ariel, F.; Romerobarrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Bazin, J.; Baileyserres, J. Emerging roles of long non-coding RNA in root developmental plasticity and regulation of phosphate homeostasis. Front. Plant Sci. 2015, 6, 400. [Google Scholar] [CrossRef]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. BBA Gene Regul. Mech. 2016, 1859, 155–162. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Wang, M.B. Molecular Functions of Long Non-Coding RNAs in Plants. Genes 2012, 3, 176–190. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Chen, Y.Q. Long noncoding RNAs: New regulators in plant development. Biochem. Biophys. Res. Commun. 2013, 436, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Gaasterland, T.; Chua, N.H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005, 6. [Google Scholar] [CrossRef]
- Wang, H.; Chua, N.-H.; Wang, X.-J. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006, 7. [Google Scholar] [CrossRef]
- Song, D.; Yang, Y.; Yu, B.; Zheng, B.; Deng, Z.; Lu, B.-L.; Chen, X.; Jiang, T. Computational prediction of novel non-coding RNAs in Arabidopsis thaliana. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Hotto, A.M.; Schmitz, R.J.; Fei, Z.; Ecker, J.R.; Stern, D.B. Unexpected Diversity of Chloroplast Noncoding RNAs as Revealed by Deep Sequencing of the Arabidopsis Transcriptome. G3 2011, 1, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Jun, L.; Choonkyun, J.; Jun, X.; Huan, W.; Shulin, D.; Lucia, B.; Catalina, A.H.; Nam-Hai, C. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar]
- Li, S.; Liberman, L.M.; Mukherjee, N.; Benfey, P.N.; Ohler, U. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res. 2013, 23, 1730–1739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Deng, W.; Fan, X.; Liu, T.-T.; He, G.; Chen, R.; Terzaghi, W.; Zhu, D.; Deng, X.W. Genomic Features and Regulatory Roles of Intermediate-Sized Non-Coding RNAs in Arabidopsis. Mol. Plant 2014, 7, 514–527. [Google Scholar] [CrossRef]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.-C.; Kean, M.J.; Xu, J.; Chua, N.-H. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014, 24, 444–453. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Osato, N.; Yamada, H.; Satoh, K.; Ooka, H.; Yamamoto, M.; Suzuki, K.; Kawai, J.; Carninci, P.; Ohtomo, Y.; Murakami, K.; et al. Antisense transcripts with rice full-length cDNAs. Genome Biol. 2003, 5, R5. [Google Scholar] [CrossRef] [PubMed]
- Markov, A.G.; Falchuk, E.L.; Kruglova, N.M.; Rybalchenko, O.V.; Fromm, M.; Amasheh, S. Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res. 2009, 19, 70–78. [Google Scholar]
- Lu, T.; Zhu, C.; Lu, G.; Guo, Y.; Zhou, Y.; Zhang, Z.; Zhao, Y.; Li, W.; Lu, Y.; Tang, W. Strand-specific RNA-seq reveals widespread occurrence of novel cis- natural antisense transcripts in rice. BMC Genomics 2012, 13, 721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [PubMed]
- Boerner, S.; McGinnis, K.M. Computational Identification and Functional Predictions of Long Noncoding RNA in Zea mays. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Eichten, S.R.; Shimizu, R.; Petsch, K.; Yeh, C.-T.; Wu, W.; Chettoor, A.M.; Givan, S.A.; Cole, R.A.; Fowler, J.E.; et al. Genome-wide discovery and characterization of maize long non-coding RNAs (vol 15, R40, 2014). Genome Biol. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Han, Z.; Guo, Q.; Liu, Y.; Zheng, Y.; Wu, F.; Jin, W. Identification of Maize Long Non-Coding RNAs Responsive to Drought Stress. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Wang, Y.; Yao, Y.; Song, N.; Hu, Z.; Qin, D.; Xie, C.; Peng, H.; Ni, Z.; Sun, Q. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 2011, 11. [Google Scholar] [CrossRef]
- Lu, X.; Chen, X.; Mu, M.; Wang, J.; Wang, X.; Wang, D.; Yin, Z.; Fan, W.; Wang, S.; Guo, L. Genome-wide analysis of long noncoding rnas and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar]
- Zou, C.; Wang, Q.; Lu, C.; Yang, W.; Zhang, Y.; Cheng, H.; Feng, X.; Prosper, M.A.; Song, G. Transcriptome analysis reveals long noncoding RNAs involved in fiber development in cotton (Gossypium arboreum). Sci. China-Life Sci. 2016, 59, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.-Y.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef]
- Celton, J.M.; Gaillard, S.; Bruneau, M.; Pelletier, S.; Aubourg, S.; Martin-Magniette, M.L.; Navarro, L.; Laurens, F.; Renou, J.P. Widespread anti-sense transcription in apple is correlated with siRNA production and indicates a large potential for transcriptional and/or post-transcriptional control. New Phytol. 2014, 203, 287–299. [Google Scholar] [CrossRef]
- Wen, J.; Parker, B.J.; Weiller, G.F. In silico identification and characterization of mRNA-Like Noncoding transcripts in Medicago truncatula. In Silico Biol. 2007, 7, 485–505. [Google Scholar] [PubMed]
- Wang, J.; Jing, L.; Kan, J.; Hong, W.; Li, X.; Yang, Q.; Hui, L.; Chang, Y. Genome-Wide Identification and Functional Prediction of Novel Drought-Responsive lncRNAs inPyrus betulifolia. Genes 2018, 9, 311. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, S.; Gu, C.; Zhou, Y.; Zhou, H. Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol. Biol. 2013, 83, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yang, J.; Li, X.; Liu, X.; Sun, C.; Wu, F.; He, Y. Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 2013, 13, 208. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zheng, Y.; Dong, J.; Yu, J.; Yue, J.; Liu, F.; Guo, X.; Huang, S.; Wisniewski, M.; Sun, J.; et al. Comprehensive transcriptome profiling reveals long noncoding rna expression and alternative splicing regulation during fruit development and ripening in kiwifruit (Actinidia chinensis). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Hao, Z.; Fan, C.; Cheng, T.; Su, Y.; Wei, Q.; Li, G. Genome-Wide Identification, Characterization and Evolutionary Analysis of Long Intergenic Noncoding RNAs in Cucumber. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Flórez-Zapata, N.M.V.; Reyes-Valdés, M.H.; Martínez, O. Long non-coding RNAs are major contributors to transcriptome changes in sunflower meiocytes with different recombination rates. BMC Genomics 2016, 17, 490. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, L.; Zhang, C.; Hao, P.; Jing, X.; Li, X. Global transcriptome analysis reveals extensive gene remodeling, alternative splicing and differential transcription profiles in non-seed vascular plant Selaginella moellendorffii. BMC Genomics 2017, 18, 1042. [Google Scholar] [CrossRef]
- Zhang, G.; Duan, A.; Zhang, J.; He, C. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit. Gene 2017, 596, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yang, Y.; Li, R.; Fu, D.; Wen, L.; Luo, Y.; Zhu, H. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. J. Exp. Bot. 2015, 66, 4483–4495. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, D.D.; Schadt, E.E.; Armour, C.D.; He, Y.D.; Garrettengele, P.; Mcdonagh, P.D.; Loerch, P.M.; Leonardson, A.; Lum, P.Y.; Cavet, G. Experimental annotation of the human genome using microarray technology. Nature 2001, 409, 922–927. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, H.; Xie, S.; Chen, J.; Xu, Y.; Wang, K.; Zhao, H.; Guan, H.; Hu, X.; Jiao, Y. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 20042–20047. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Chao, D.; Jiapei, Y.; Yue, W.; Jingrui, L.; Huixin, L.; Long, H.; Ting, Z.; Yijun, Q.; Gerstein, M.B.; Yan, G. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2015, 80, 848–861. [Google Scholar]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of noncoding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, P.; Rodriguezescudero, A.I. The Solanum commersonii Genome Sequence Provides Insights into Adaptation to Stress Conditions and Genome Evolution of Wild Potato Relatives. Plant Cell 2015, 27, 954–968. [Google Scholar]
- Liu, S.; Sun, Z.; Xu, M. Identification and characterization of long non-coding RNAs involved in the formation and development of poplar adventitious roots. Ind. Crops Prod. 2018, 118, 334–346. [Google Scholar] [CrossRef]
- Kiegle, E.A.; Garden, A.; Lacchini, E.; Kater, M.M. A Genomic View of Alternative Splicing of Long Non-coding RNAs during Rice Seed Development Reveals Extensive Splicing and lncRNA Gene Families. Front. Plant Sci. 2018, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, P.; Wang, Q.; Wang, W.; Zhang, C.; Sun, F.; Liu, Z.; Li, Y.; Hou, X. Comparative transcriptome discovery and elucidation of the mechanism of long noncoding RNAs during vernalization in Brassica rapa. Plant Growth Regul. 2018, 85, 27–39. [Google Scholar] [CrossRef]
- Yu, T.; Tzeng, D.; Li, R.; Chen, J.; Zhong, S.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Genome-wide identification of long non-coding RNA targets of the tomato MADS box transcription factor RIN and function analysis. Ann. Bot. 2018. [Google Scholar] [CrossRef] [PubMed]
- Juan, W.; Toshihiro, O.; Toru, F.; Takahiko, T.; Masahiro, S.; Yasushi, Y. A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Boil. 2012, 9, 302–313. [Google Scholar]
- Cho, J.; Koo, D.H.; Nam, Y.W.; Han, C.T.; Lim, H.T.; Bang, J.W.; Hur, Y. Isolation and characterization of cDNA clones expressed under male sex expression conditions in a monoecious cucumber plant (Cucumis sativus L. cv. Winter Long). Euphytica 2006, 146, 271–281. [Google Scholar] [CrossRef]
- Markus, W.; Rita, G.H.; Friedrich, S.F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Boil. 2014, 85, 541–550. [Google Scholar]
- Miao-Chih, T.; Ohad, M.; Yue, W.; Nima, M.; Wang, J.K.; Fei, L.; Yang, S.; Eran, S.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar]
- Nakamoto, S.; Tashiro, K.; Matsumoto, A. Rice ENOD40: Isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 2010, 18, 121–129. [Google Scholar]
- Dai, X.Y.; Yu, J.J.; Zhao, Q.; Zhu, D.Y.; Ao, G.M. Non-coding RNA for ZM401, a pollen-specific gene of Zea mays. Acta Bot. Sin. 2004, 46, 497–504. [Google Scholar]
- Jack, T. Molecular and Genetic Mechanisms of Floral Control. Plant Cell 2004, 16, S1–S17. [Google Scholar] [CrossRef]
- Filipe, B.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Boil. 2015, 16, 727–741. [Google Scholar]
- Zhao, J.; Liu, Y.; Huang, G.; Cui, P.; Zhang, W.; Zhang, Y. Long non-coding RNAs in gastric cancer: Versatile mechanisms and potential for clinical translation. Am. J. Cancer Res. 2015, 5, 907–927. [Google Scholar]
- Wang, P.; Xu, J.; Wang, Y.; Cao, X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 2017, 358, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Yvonne, T.; Lev, K.; Leonardo, S.; Dror, W.; Mynn, T.S.; Ugo, A.; Florian, K.; Laura, P.; Paolo, P.; Ferdinando, D.C. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar]
- Ding, J.; Shen, J.; Mao, H.; Xie, W.; Li, X.; Zhang, Q. RNA-directed dna methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 2012, 5, 1210–1216. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, Q.; Li, J.; Jiang, D.; Zhou, L.; Wu, P.; Lu, S.; Li, F.; Zhu, L.; Liu, Z. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012, 22, 649–660. [Google Scholar] [CrossRef]
- Komiya, R.; Ohyanagi, H.; Niihama, M.; Watanabe, T.; Nakano, M.; Kurata, N.; Nonomura, K. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014, 78, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Jing, Z.; Bryan, K.S.; Jennifer, A.E.; Ji-Joon, S.; Jeannie, T.L. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar]
- Lipshitz, H.D.; Peattie, D.A.; Hogness, D.S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987, 1, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xie, S.; Liu, Y.; Yi, F.; Yu, J. Genome-wide annotation of genes and noncoding RNAs of foxtail millet in;response to simulated drought stress by deep sequencing. Plant Mol. Boil. 2013, 83, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ci, D.; Tian, M.; Zhang, D. Stable methylation of a non-coding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. J. Exp. Bot. 2015, 67, 1477–1492. [Google Scholar] [CrossRef]
- Ietswaart, R.; Wu, Z.; Dean, C. Flowering time control: Another window to the connection between antisense RNA and chromatin. Trends Genet. 2012, 28, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James, P.W.; Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef] [PubMed]
- He, Y. Control of the Transition to Flowering by Chromatin Modifications. Mol. Plant 2009, 2, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Richard, A. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Csorba, T.; Skourtistathaki, K.; Proudfoot, N.J.; Dean, C. R-Loop Stabilization Represses Antisense Transcription at the Arabidopsis FLC Locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Rouse, D.T.; Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 2000, 97, 3753–3758. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Lim, J.; Dale, J.M.; Chen, H.; Shinn, P.; Palm, C.J.; Southwick, A.M.; Wu, H.C. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 2003, 302, 842–846. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Masumi, R.; Jean, F.E.; Buzas, D.M.; Dennis, E.S. Vernalization-Repression of Arabidopsis FLC Requires Promoter Sequences but Not Antisense Transcripts. PLoS ONE 2011, 6, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Crevillén, P.; Yang, H.; Cui, X.; Greeff, C.; Trick, M.; Qiu, Q.; Cao, X.; Dean, C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 2014, 515, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Kim, D.H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.F.; Chua, N.H. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Cheng, D.; Cui, J.; Dai, C.; Luo, C.; Liu, T.; Li, J. Vernalisation mediated LncRNA-like gene expression in Beta vulgaris. Funct. Plant Boil. 2017, 44, 720–726. [Google Scholar] [CrossRef]
- Loraine, A.E.; Mccormick, S.; Estrada, A.; Patel, K.; Peng, Q. RNA-Seq of Arabidopsis Pollen Uncovers Novel Transcription and Alternative Splicing. Plant Physiol. 2013, 162, 1092–1109. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.Z.; Ou, L.J.; Deng, L.X.; Luan, S.; Chen, L.B. Fertility response to photoperiod and temperature in indica photoperiod-sensitive male sterile rice. Russ. J. Plant Physiol. 2008, 55, 694–698. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef]
- Ma, J.; Yan, B.Y.; Qin, F.; Yang, Y.; Hao, X.; Yu, J.; Zhao, Q.; Zhu, D.; Ao, G. Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell. Biochem. 2010, 105, 136–146. [Google Scholar] [CrossRef]
- Song, J.H.; Cao, J.S.; Yu, X.L.; Xiang, X. BcMF11, a putative pollen-specific non-coding RNA from Brassica campestris ssp. chinensis. J. Plant Physiol. 2007, 164, 1097–1100. [Google Scholar] [CrossRef]
- Song, J.H.; Cao, J.S.; Wang, C.G. BcMF11, a novel non-coding RNA gene from Brassica campestris, is required for pollen development and male fertility. Plant Cell Rep. 2013, 32, 21–30. [Google Scholar] [CrossRef]
- Ilott, N.E.; Ponting, C.P. Predicting long non-coding RNAs using RNA sequencing. Methods 2013, 63, 50–59. [Google Scholar] [CrossRef]
- Fan, C.; Hao, Z.; Yan, J.; Li, G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genomics 2015, 16, 793. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Van, A.O. Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. Wormbook 2012, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Sorensen, E.B.; Lynch, T.R.; Kimble, J. C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. eLife 2016, 5, e18370. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.; Duncan, S.; Dean, C. Mutually exclusive sense-antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 2016, 7, 13031. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Van den Bogaard, P.; Rifkin, S.A.; Van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Shechner, D.M.; Hacisuleyman, E.; Younger, S.T.; Rinn, J.L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 2015, 12, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wen, L.; Zhu, H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015, 5, 59. [Google Scholar] [CrossRef]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of\r, lncRNA1459\r, alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Liu, J.; Chen, C.; Liao, Q.; Xu, P.; Xu, H.; Xiao, T.; Cao, Z.; Peng, J. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR library. Nat. Biotechnol. 2016, 34, 1279–1286. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2016, 355, eaah7111. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; Mcdonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, J.; Fang, J.Y. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Clin. Gastroenterol. Hepatol. 2015, 13, e100–e101. [Google Scholar] [CrossRef]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2013, 21, 685–693. [Google Scholar] [CrossRef]
- Yang, L.; Froberg, J.E.; Lee, J.T. Long noncoding RNAs: Fresh perspectives into the RNA world. Trends Biochem. Sci. 2014, 39, 35–43. [Google Scholar] [CrossRef]
| Plant Species | Numbers of lncRNAs | Approaches | Type(s) of lncRNAs | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | 1340 1320 76 179 107 6480 2418 838 37,238 955 | In silico-EST In silico-EST In silico-EST Tiling array RNA-seq Tiling array RNA-seq RNA-seq Tiling array RNA-seq | cis-NATs trans-NATs lncRNAs lncRNAs lncRNAs lincRNAs cis-NATs Intermediate ncRNAs sense/antisense lncRNAs lncRNAs | [22] [23] [8] [24] [25] [26] [27] [28] [29] [30] |
| Oryza Sativa Linanaeus | 945 7486 3819 2224 | In silico-EST and Microarray In silico-genome annotation RNA-seq RNA-seq | sense/antisense lncRNAs cis-NATs and trans-NATs cis-NATs lincRNAs and NATs | [31] [32] [33] [34] |
| Zea mays L. | 1802 20,163 1724 | In silico-EST In silico-EST and RNA-seq RNA-seq | lncRNAs lncRNAs lncRNAs | [35] [36] [37] |
| Triticumaestivum L. | 125 | Microarray and RNA-seq | lncRNAs | [38] |
| Gossypium | 10,820 5996 | reproducibility-based RNA-seq strand-specific RNA sequencing | lincRNAs, inronic and antisense lncRNAs. lincRNAs and NATs | [39] [40] |
| Populus L. | 2542 17 | RNA-seq stress-specific differentially methylated regions sequencing | lincRNAs lncRNAs | [41] [40] |
| Malus pumila Mill. | 54,975 | Microarray | sense/antisense lncRNAs | [42] |
| Setaria italica | 584 | RNA-seq | lincRNAs and NATs | |
| Medicago truncatula | 503 | In silico-genome annotation | lncRNAs | [43] |
| Pyrus betulifolia | 14,478 | RNA-seq | lncRNAs | [44] |
| Prunuspersica | 1417 | Transcriptome sequencing | lncRNAs | [45] |
| Brassica rapa L. | 1301 | RNA-seq | cis-NATs | [46] |
| Actinidia chinensis Planch | 7051 | RNA-seq | lncRNAs | [47] |
| Cucumis sativus L. | 3274 | RNA-seq | lincRNAs | [48] |
| Helianthus annuus | 6895 | RNA-seq | lncRNAs | [49] |
| Selaginella | 230 | RNA-seq | lncRNAs | [50] |
| Sea buckthorn | 3428 | RNA-seq | lincRNAs anti-sense lncRNAs intronic lncRNAs | [51] |
| Solanum lycopersicum | 3679 | RNA-seq | lincRNAs anti-sense lncRNAs intronic lncRNAs | [52] |
| Name | Species | Tissue Specificity | Biological Function | Refs. |
|---|---|---|---|---|
| AtR8 | Arabidopsis thaliana | Root | Hypoxic stress | [63] |
| CsM10 | Cucumis sativus L. | Apices of seedlings | Sex differentiation | [64] |
| GmENOD40 | Giycine max | Nodule | Nodule formation | [66] |
| OsENOD40 | Oryza sativa L. | Stem | Nodule formation | [67] |
| Zm401 | Zea mays | Pollen | Fertility | [68] |
| asHSFB2a | Arabidopsis thaliana | Female gametophyte | Vegetative and gametophytic development | [65] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Zhu, H. Long Non-Coding RNAs: Rising Regulators of Plant Reproductive Development. Agronomy 2019, 9, 53. https://doi.org/10.3390/agronomy9020053
Yu T, Zhu H. Long Non-Coding RNAs: Rising Regulators of Plant Reproductive Development. Agronomy. 2019; 9(2):53. https://doi.org/10.3390/agronomy9020053
Chicago/Turabian StyleYu, Tongtong, and Hongliang Zhu. 2019. "Long Non-Coding RNAs: Rising Regulators of Plant Reproductive Development" Agronomy 9, no. 2: 53. https://doi.org/10.3390/agronomy9020053
APA StyleYu, T., & Zhu, H. (2019). Long Non-Coding RNAs: Rising Regulators of Plant Reproductive Development. Agronomy, 9(2), 53. https://doi.org/10.3390/agronomy9020053