Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. RNA Extraction and Library Preparation for Illumina Sequencing
2.3. Construction of Library
2.4. Sequencing Quality Control and Reads Mapping to the Reference Genome
2.5. Differentially Expressed Gene Detection
2.6. Gene Enrichment Analysis
2.7. Co-Expression Gene Models
2.8. Validation of RNA-seq Data
3. Results
3.1. RNA Sequencing of Different Samples
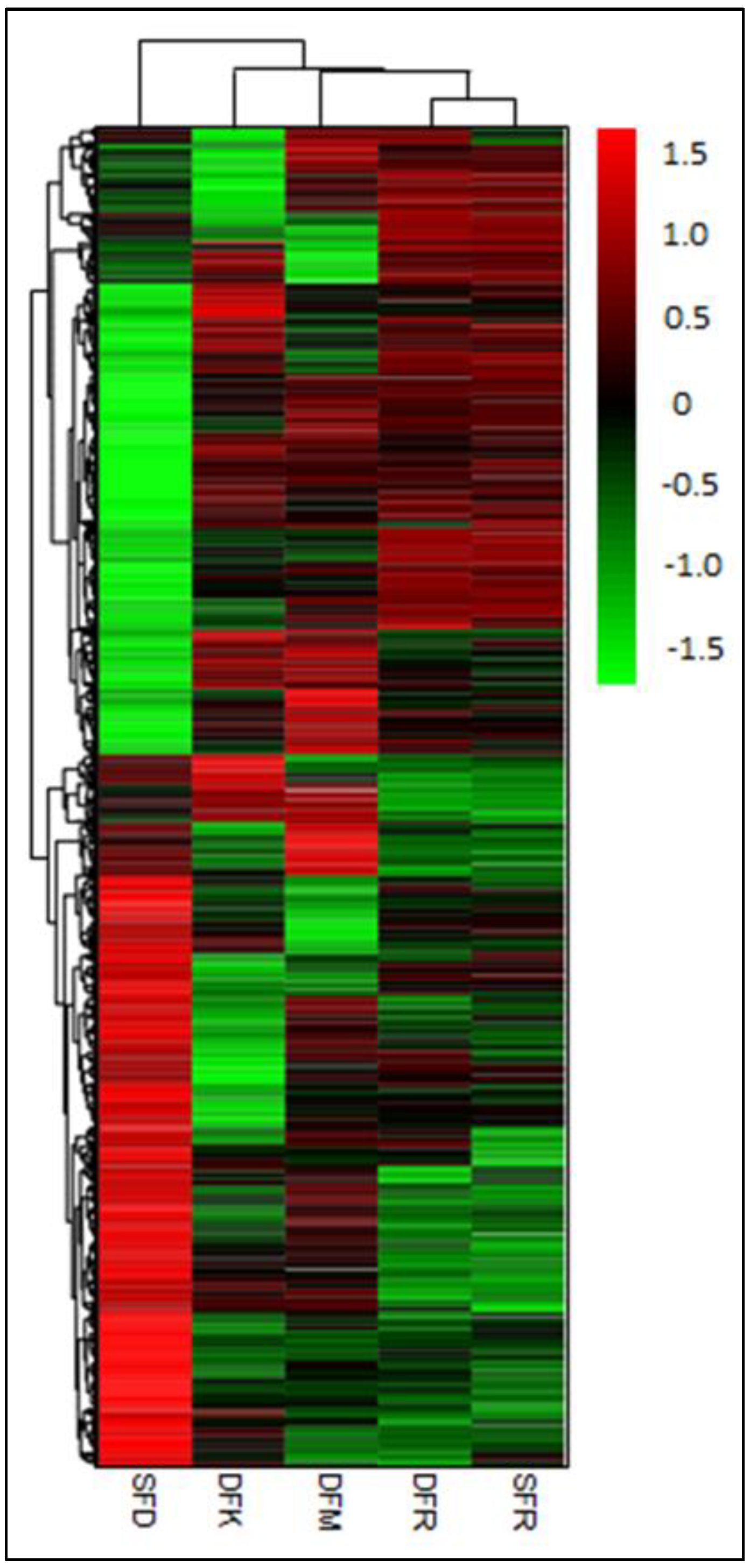
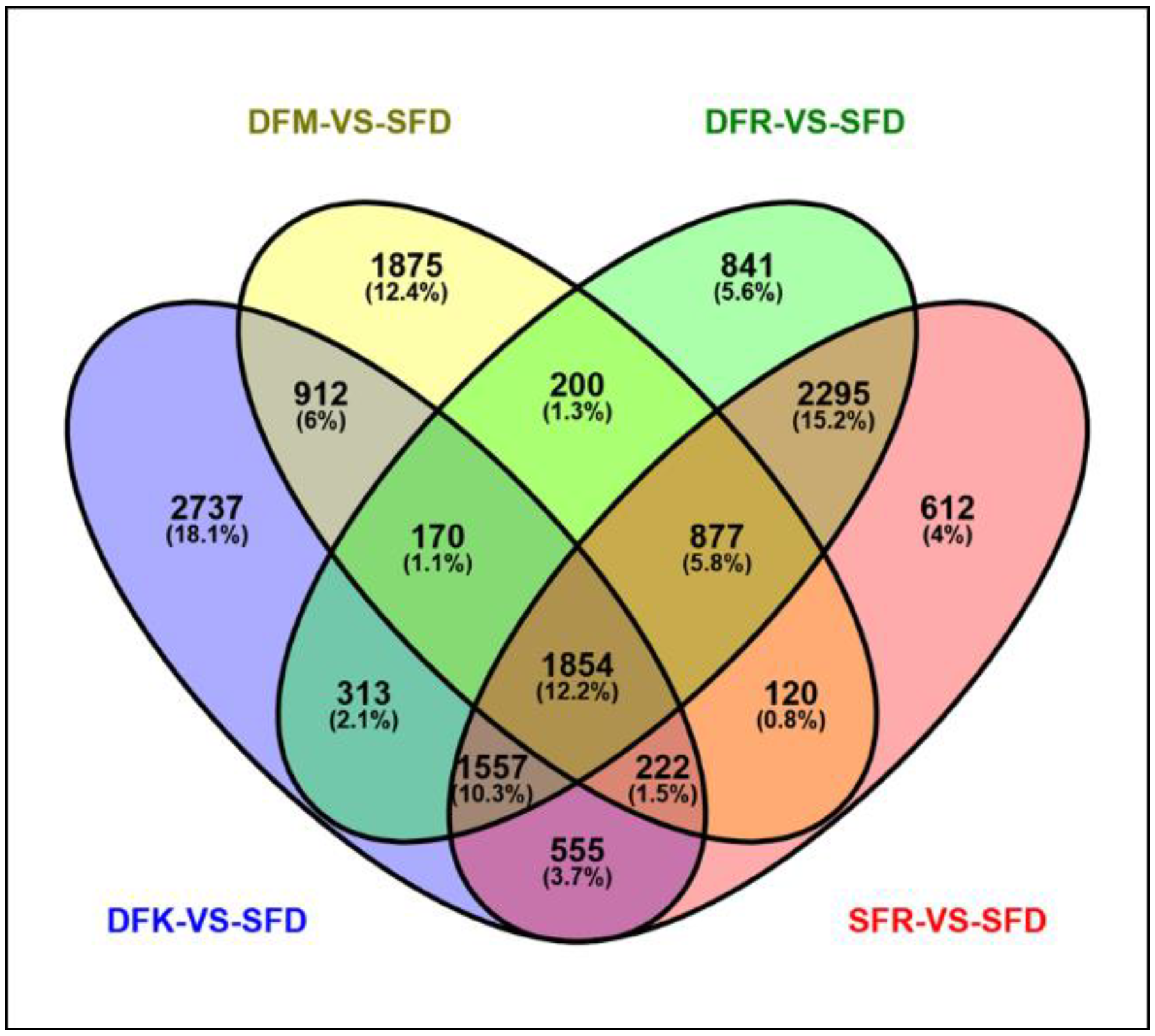
3.2. Transcriptome Dynamics of Double Flowering
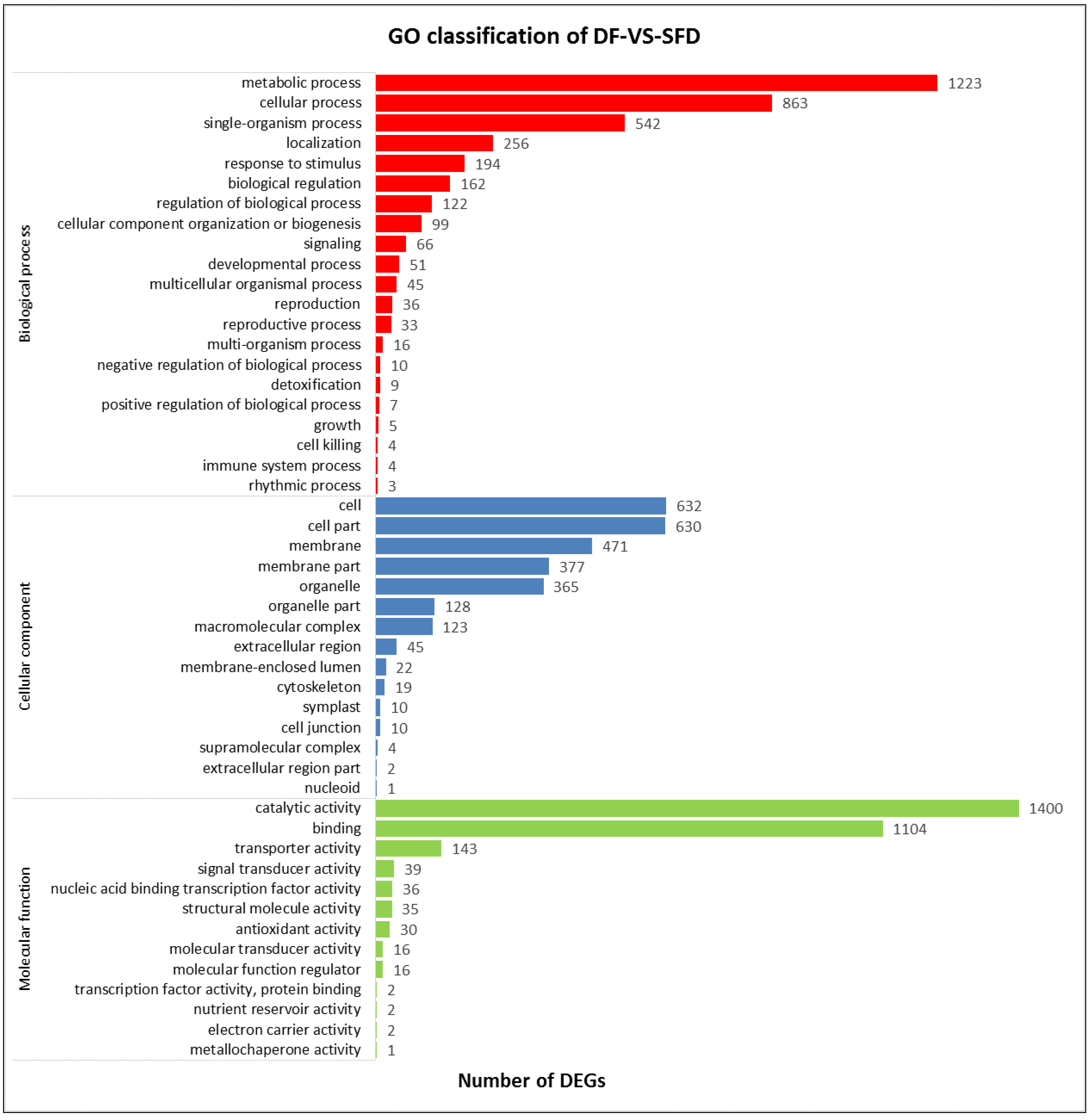
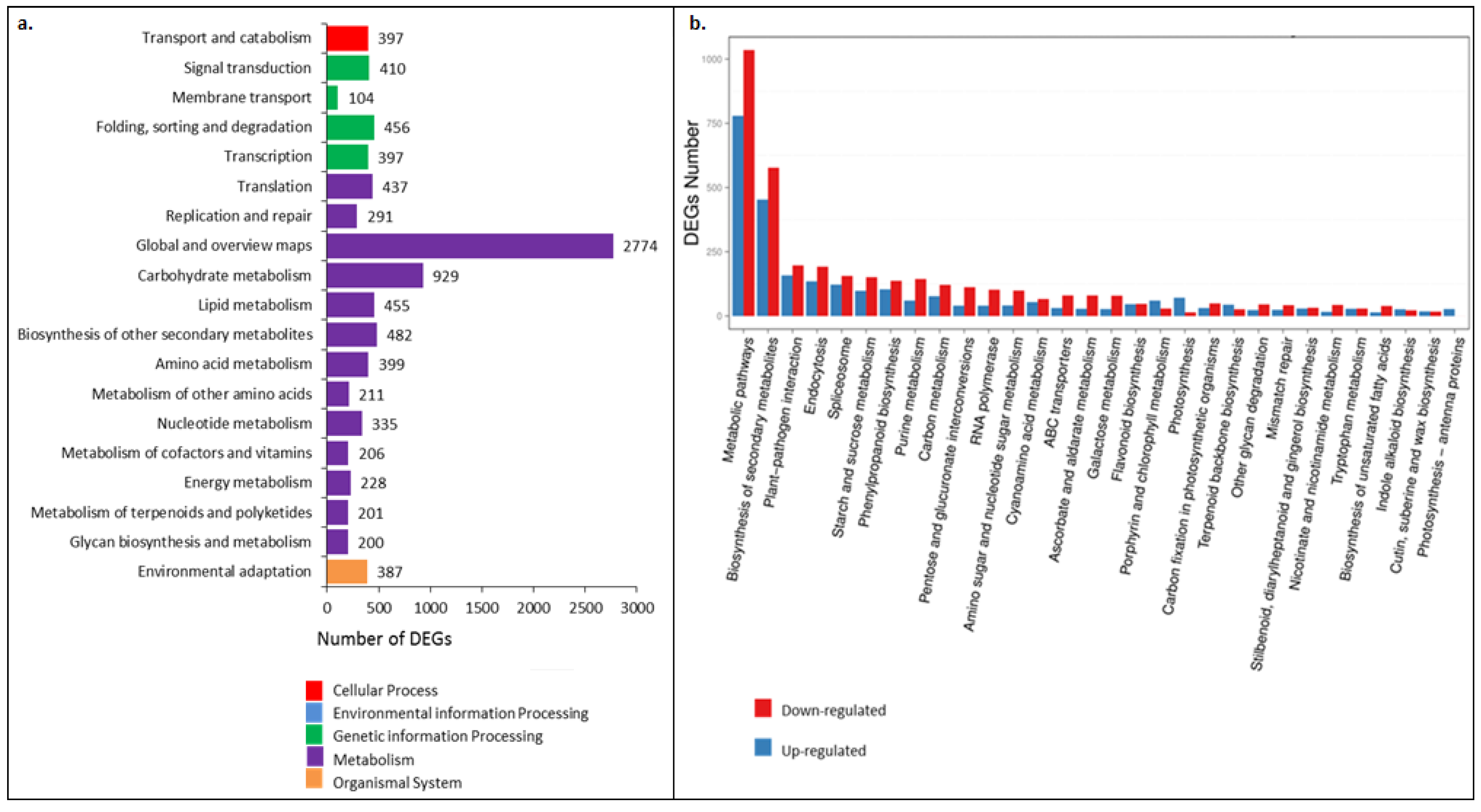
3.3. Gene Ontology and Kyoto Encyclopedia of Genes and Genome (KEGG) analysis
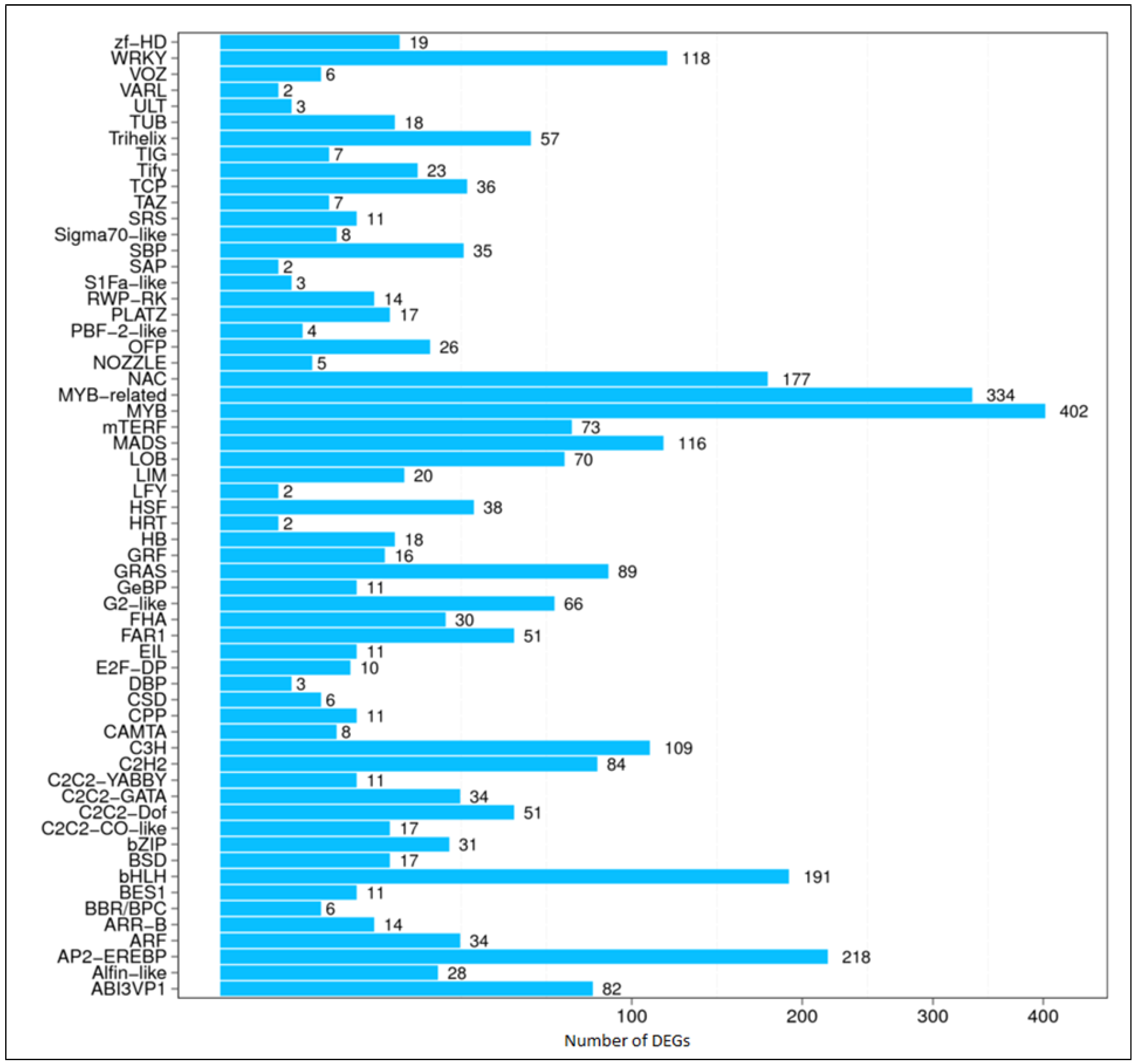
3.4. Recognizing TF-Encoding Genes Associated with Floral Organ Development
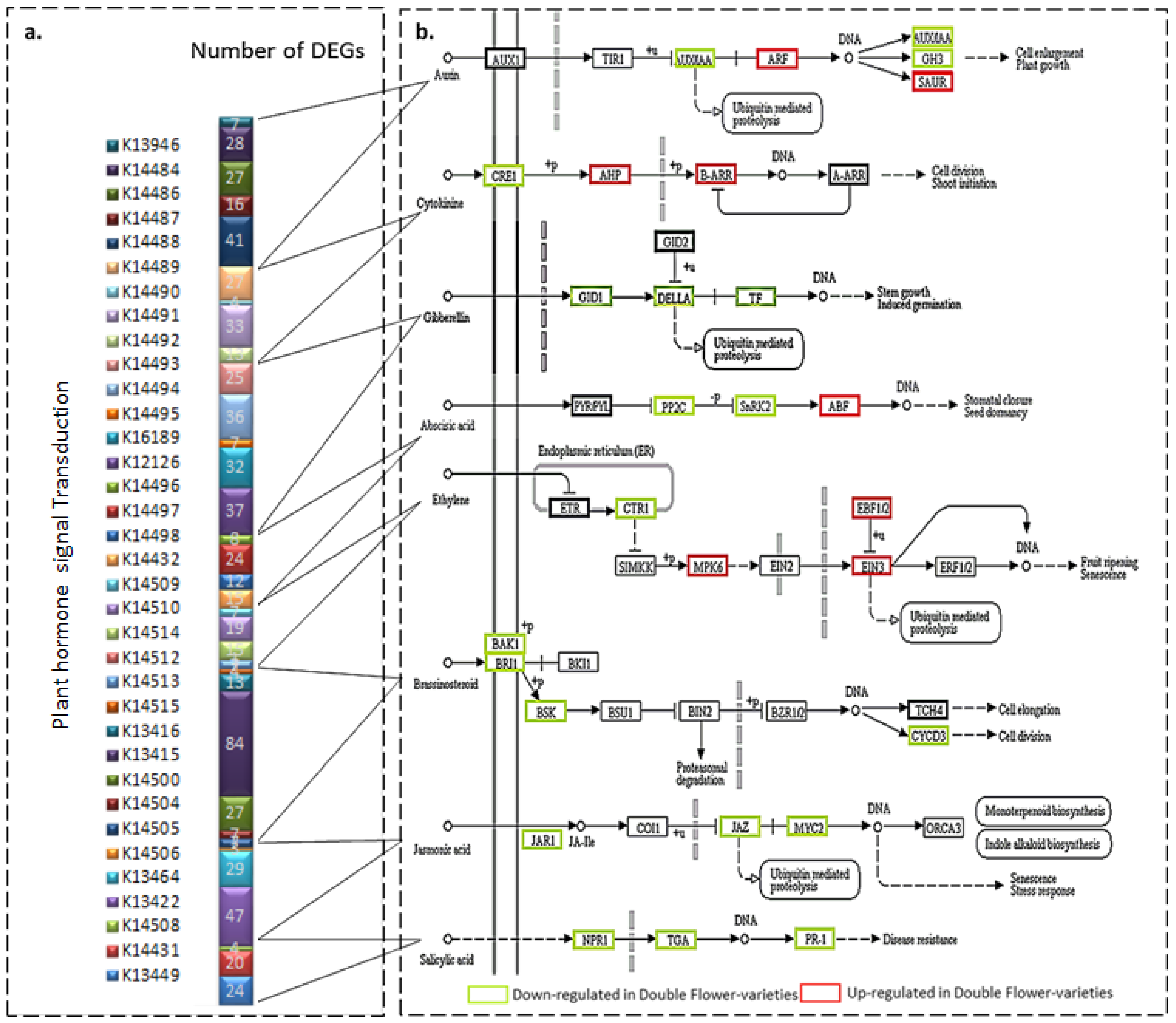
3.5. Recognizing DEGs Related to Hormones
3.6. Weighted Gene co-Expression Analysis (WGCNA)
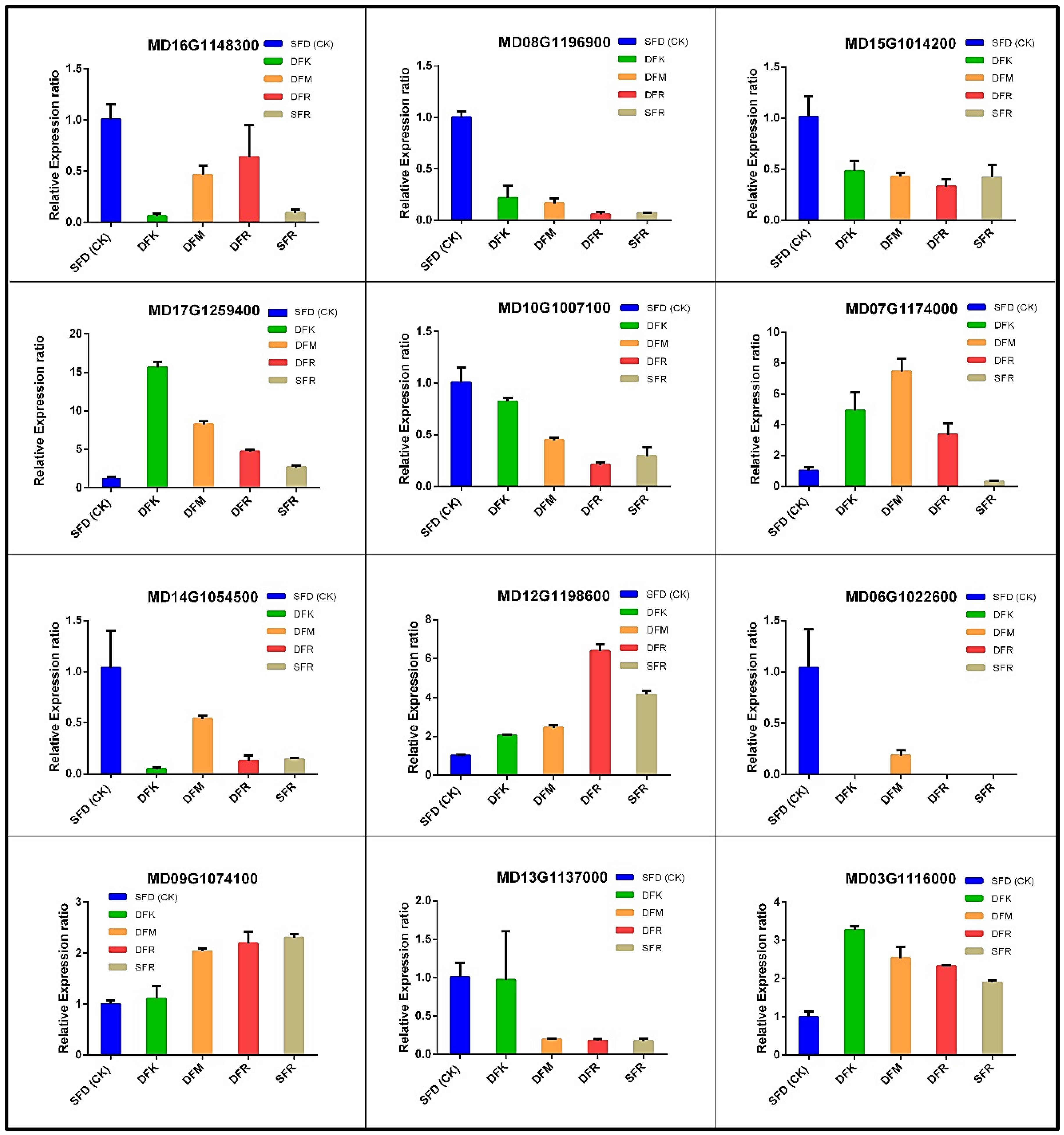
3.7. Validation of the Randomly Selected Genes Expression
4. Discussion
5. Conclusions
Supplementary Materials
Availability of Supporting Data
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Reynold, J.; Tampion, J. Double Flowers: A Scientific Study; Polytechnic of Central London Press: Pembridge, UK, 1983; p. 41. [Google Scholar]
- Ramirez, F.; Davenport, T.L. Apple pollination: A review. Sci. Hortic. 2013, 162, 188–203. [Google Scholar] [CrossRef]
- Garratt, M.P.D.; Breeze, T.D.; Boreux, V.; Fountain, M.T.; Mckerchar, M.; Webber, S.M.; Coston, D.J.; Jenner, N.; Dean, R.; Westbury, D.B. Apple Pollination: Demand Depends on Variety and Supply Depends on Pollinator Identity. PLoS ONE 2016, 11, e0153889. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. The ABC model of flower development: then and now. Development 2012, 139, 4095–4098. [Google Scholar] [CrossRef] [PubMed]
- Chanderbali, A.S.; Berger, B.A.; Howarth, D.G.; Soltis, P.S.; Soltis, D.E. Evolving ideas on the origin and evolution of flowers: New perspectives in the genomic era. Genetics 2016, 202, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Zik, M.; Irish, V.F. Global identification of target genes regulated by APETALA3 (AP3) and PISTILLATA floral homeotic gene action. Plant Cell 2003, 15, 207. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Hall, J.C. Evolutionary dynamics of genes controlling floral development. Curr. Opin. Plant Biol. 2005, 8, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Sablowski, R. Flowering and determinacy in Arabidopsis. J. Exp. Bot. 2007, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Egea-Cortines, M.; Saedler, H.; Sommer, H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999, 18, 5370–5379. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhao, Y. A role for auxin in flower development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Pfluger, J.; Zambryski, P. The role of SEUSS in auxin response and floral organ patterning. Development 2004, 131, 4697–4707. [Google Scholar] [CrossRef] [PubMed]
- Matiashernandez, L.; Aguilarjaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Hättasch, C.; Flachowsky, H.; Hanke, M.V.; Lehmann, S.; Gau, A.; Kapturska, D. The switch to flowering: genes involved in floral induction of the apple cultivar ‘Pinova’ and the role of the flowering gene MdFT. In International Symposium on Biotechnology of Fruit Species: BIOTECHFRUIT2008; Hanke, M.-V., Dunemann, F., Flachowsky, H., Eds.; International Society for Horticultural Science: Dresden, Germany, 2009; pp. 701–705. [Google Scholar]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [PubMed]
- Der Linden, C.G.V.; Vosman, B.; Smulders, M.J.M. Cloning and characterization of four apple MADS box genes isolated from vegetative tissue. J. Exp. Bot. 2002, 53, 1025–1036. [Google Scholar] [CrossRef]
- Sung, S.; Yu, G.; Nam, J.; Jeong, D.; An, G. Developmentally regulated expression of two MADS-box genes, MdMADS3 and MdMADS4, in the morphogenesis of flower buds and fruits in apple. Planta 2000, 210, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Klocko, A.L.; Borejsza, W.E.; Brunner, A.M.; Shevchenko, O.; Aldwinckle, H.; Strauss, S.H. Transgenic suppression of AGAMOUS genes in apple reduces fertility and increases floral attractiveness. PLoS ONE 2016, 11, e0159421. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Tanaka-Moriya, Y.; Mimida, N.; Honda, C.; Iwanami, H.; Komori, S.; Wada, M. The analysis of transgenic apples with down-regulated expression of MdPISTILLATA. Plant Biotechnol. 2016, 33, 395–401. [Google Scholar] [CrossRef]
- Di Stilio, V.S.; Kramer, E.M.; Baum, D.A. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae)—A new model for the study of dioecy. Plant J. 2005, 41, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Zheng, B.; Wellmer, F. Floral organogenesis:when knowing your ABCs is not enough. Plant Physiol. 2017, 173, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Cliff, M. Development of a model for prediction of consumer liking from visual attributes of new and established apple cultivars. Am. Pomol. Soc. 2002, 4, 223–229. [Google Scholar]
- Hong, Y.; Jackson, S.D. Floral induction and flower formation—The role and potential applications of miRNAs. Plant Biotechnol. J. 2015, 13, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Chanderbali, A.S.; Yoo, M.-J.; Zahn, L.M.; Brockington, S.F.; Wall, P.K.; Gitzendanner, M.A.; Albert, V.A.; Leebens-Mack, J.; Altman, N.S.; Ma, H. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc. Natl. Acad. Sci. USA 2010, 107, 22570–22575. [Google Scholar] [CrossRef] [PubMed]
- Zahn, L.M.; Ma, X.; Altman, N.S.; Zhang, Q.; Wall, P.K.; Tian, D.; Gibas, C.J.; Gharaibeh, R.; Leebens-Mack, J.H.; Ma, H. Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biol. 2010, 11, R101. [Google Scholar] [CrossRef] [PubMed]
- Romer, J.P. A Srvey of Selection Preferences for Crabapple Cultivars and Species. 2002. Available online: https://lib.dr.iastate.edu/rtd/17486/ (accessed on 26 December 2018).
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.; Antonescu, C.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.A.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. The Apple Genome and Epigenome. Available online: https://iris.angers.inra.fr/gddh13/ (accessed on 26 December 2018).
- Kong, L.; Zhang, Y.; Ye, Z.; Liu, X.; Zhao, S.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 28, 27–30. [Google Scholar] [CrossRef]
- Rice, P.M.; Longden, I.; Bleasby, A.J. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; et al. 15 Years of GDR: New Data and Functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Xie, M.; Vasseur, L.; You, M. Selecting and validating reference genes for quantitative real-time PCR in Plutella xylostella (L.). Genome 2018, 61, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sui, S.; Luo, J.; Ma, J.; Zhu, Q.; Lei, X.; Li, M. Generation and analysis of expressed sequence tags from chimonanthus praecox (wintersweet) flowers for discovering stress-responsive and floral development-related genes. Int. J. Genom. 2012. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Nawaz, M.A.; Acet, T.; Abbas, A.; Imran, Q.M.; Shah, K.H.; Rehman, H.M.; Chung, G.; Yang, S.H.; et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 2018, 226, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Malcomber, S.T.; Kellogg, E.A. SEPALLATA gene diversification: Brave new whorls. Trends Plant Sci. 2005, 10, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.F.; Yadav, V.; Tholl, D.; Chetelat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C. A Regulatory network for coordinated flower maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Irish, V.F. Gene networks controlling petal organogenesis. J. Exp. Bot. 2016, 67, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.; Park, C. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Ma, Z.; Huang, H.; Mo, H.; Zhao, T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Galimba, K.D.; Tolkin, T.R.; Sullivan, A.M.; Melzer, R.; Theißen, G.; Di Stilio, V.S. Loss of deeply conserved C-class floral homeotic gene function and C- and E-class protein interaction in a double-flowered ranunculid mutant. Proc. Natl. Acad. Sci. USA 2012, 109, E2267–E2275. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Dorcafornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Ma, H. Regulatory genes in plant development: MADS. Essent. Life Sci. 2009. [Google Scholar] [CrossRef]
- Mondragonpalomino, M. Perspectives on MADS-box expression during orchid flower evolution and development. Front. Plant Sci. 2013, 4, 377. [Google Scholar]
- Chang, Y.; Kao, N.; Li, J.; Hsu, W.; Liang, Y.; Wu, J.; Yang, C. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 2010, 152, 837–853. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Pan, Z.J.; Hsiao, Y.Y.; Chen, L.; Liu, Z. Evolution and function of MADS-box genes involved in orchid floral development. J. Syst. Evol. 2014, 52, 397–410. [Google Scholar] [CrossRef]
- Chang, Y.; Chiu, Y.; Wu, J.; Yang, C. Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.J.; Chen, Y.Y.; Du, J.S.; Chen, Y.Y.; Chung, M.C.; Tsai, W.C.; Wang, C.; Chen, H. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 2014, 202, 1024–1042. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Goto, N.; Pharis, R.P. Role of gibberellins in the development of floral organs of the gibberellin-deficient mutant, GA1-1, of Arabidopsis thaliana. Botany 1999, 77, 944–954. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Chmelnitsky, I.; Colauzzi, M.; Algom, R.; Zieslin, N. Effects of temperature on phyllody expression and cytokinin content in floral organs of rose flowers. Plant Growth Regul. 2001, 35, 207–214. [Google Scholar] [CrossRef]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 2015, 15, 237. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.E.; Sakai, H.; Finnegan, E.J.; Cao, X.; Meyerowitz, E.M. Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr. Biol. 2000, 10, 179–186. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, B.; Chen, G.; Shi, N.; Zhao, Y.; Yin, Q.; Liu, J. Application of Arabidopsis AGAMOUS second intron for the engineered ablation of flower development in transgenic tobacco. Plant Cell Rep. 2008, 27, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.-B.; Zhang, D.; Li, Y.-M.; Shen, Y.-W.; Zhao, C.-P.; Ma, J.-J.; An, N.; Han, M.-Y. Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 2015, 56, 2052–2068. [Google Scholar] [CrossRef] [PubMed]
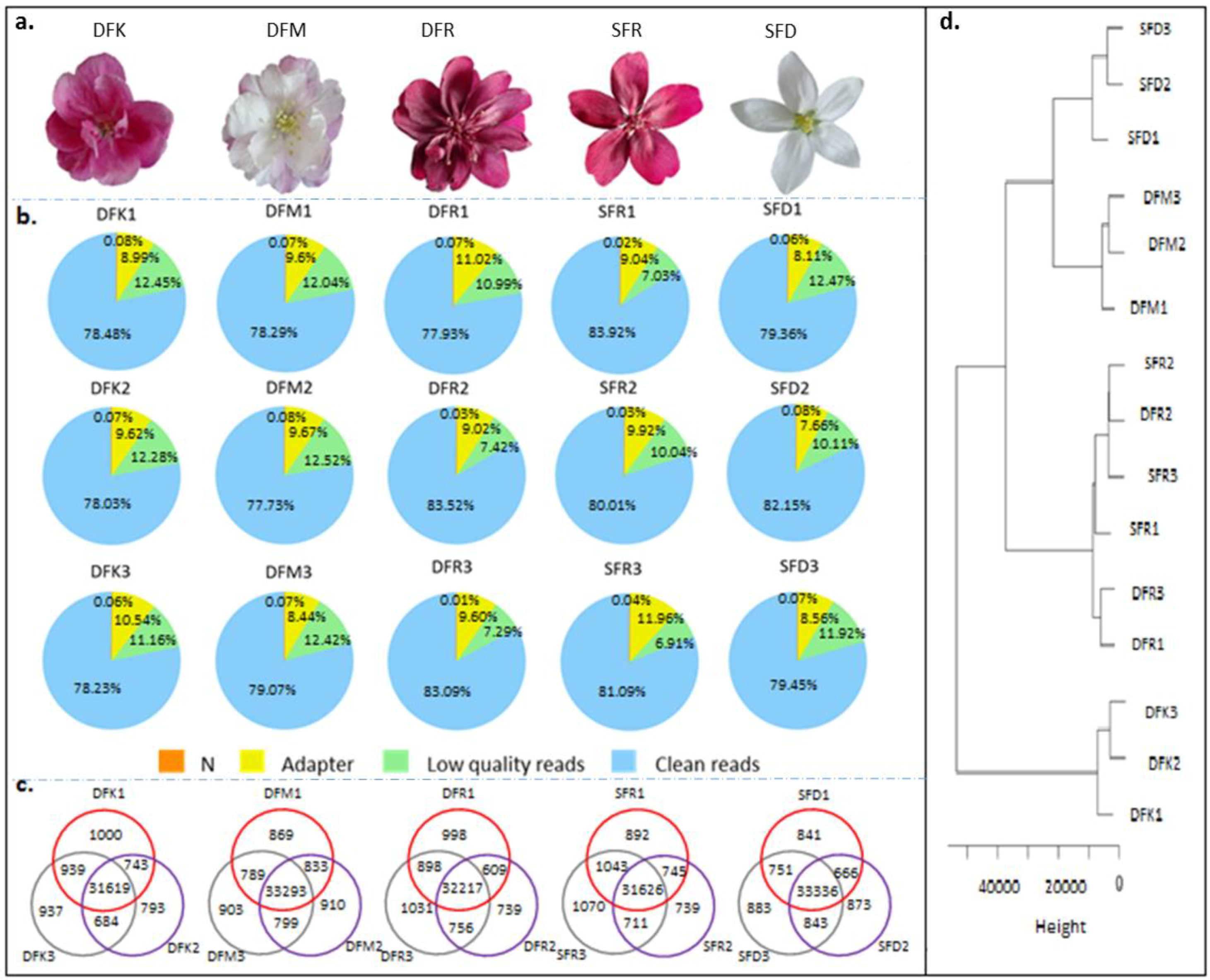

| Sample ID | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Total Clean Bases (Gb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Clean Reads Ratio (%) | Total Mapping Ratio (%) | Uniquely Mapping Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| SFD1 | 55.52 | 44.06 | 6.61 | 97.91 | 93.36 | 79.36 | 76.62 | 37.36 |
| SFD2 | 53.89 | 44.27 | 6.64 | 98.12 | 93.86 | 82.15 | 76.79 | 37.04 |
| SFD3 | 55.53 | 44.12 | 6.62 | 97.94 | 93.44 | 79.45 | 75.93 | 37.16 |
| DFK1 | 53.56 | 42.04 | 6.31 | 97.84 | 93.18 | 78.48 | 72.53 | 34.98 |
| DFK2 | 57.16 | 44.60 | 6.69 | 97.85 | 93.22 | 78.03 | 71.89 | 34.2 |
| DFK3 | 57.16 | 44.72 | 6.71 | 97.97 | 93.48 | 78.23 | 73.03 | 34.46 |
| DFM1 | 57.16 | 44.75 | 6.71 | 97.89 | 93.31 | 78.29 | 73.76 | 34.76 |
| DFM2 | 55.90 | 43.45 | 6.52 | 97.82 | 93.15 | 77.73 | 72.37 | 34.8 |
| DFM3 | 57.16 | 45.20 | 6.78 | 97.9 | 93.36 | 79.07 | 73.56 | 35.32 |
| DFR1 | 57.16 | 44.55 | 6.68 | 98 | 93.6 | 77.93 | 71.5 | 34.17 |
| DFR2 | 53.90 | 45.02 | 6.75 | 98.64 | 95.67 | 83.52 | 72.11 | 34.82 |
| DFR3 | 53.90 | 44.78 | 6.72 | 98.66 | 95.73 | 83.09 | 71.07 | 34.61 |
| SFR1 | 53.90 | 45.23 | 6.78 | 98.69 | 95.84 | 83.92 | 72.06 | 34.74 |
| SFR2 | 55.53 | 44.43 | 6.66 | 98.09 | 94.09 | 80.01 | 72.36 | 34.93 |
| SFR3 | 55.53 | 45.03 | 6.75 | 98.64 | 95.7 | 81.09 | 69.09 | 33.51 |
| Average | 55.53 | 44.42 | 6.66 | 98.13 | 94.07 | 80.02 | 72.98 | 35.13 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gul, H.; Tong, Z.; Han, X.; Nawaz, I.; Wahocho, S.A.; Khan, S.; Zhang, C.; Tian, Y.; Cong, P.; Zhang, L. Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering. Agronomy 2019, 9, 112. https://doi.org/10.3390/agronomy9030112
Gul H, Tong Z, Han X, Nawaz I, Wahocho SA, Khan S, Zhang C, Tian Y, Cong P, Zhang L. Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering. Agronomy. 2019; 9(3):112. https://doi.org/10.3390/agronomy9030112
Chicago/Turabian StyleGul, Hera, Zhaoguo Tong, Xiaolei Han, Iqra Nawaz, Safdar Ali Wahocho, Shumaila Khan, Caixia Zhang, Yi Tian, Peihua Cong, and Liyi Zhang. 2019. "Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering" Agronomy 9, no. 3: 112. https://doi.org/10.3390/agronomy9030112
APA StyleGul, H., Tong, Z., Han, X., Nawaz, I., Wahocho, S. A., Khan, S., Zhang, C., Tian, Y., Cong, P., & Zhang, L. (2019). Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering. Agronomy, 9(3), 112. https://doi.org/10.3390/agronomy9030112




