Rice Biofortification: High Iron, Zinc, and Vitamin-A to Fight against “Hidden Hunger”
Abstract
1. Introduction
2. Rice Biofortification
- High iron rice;
- High zinc rice;
- Golden rice (high carotenoid rice).
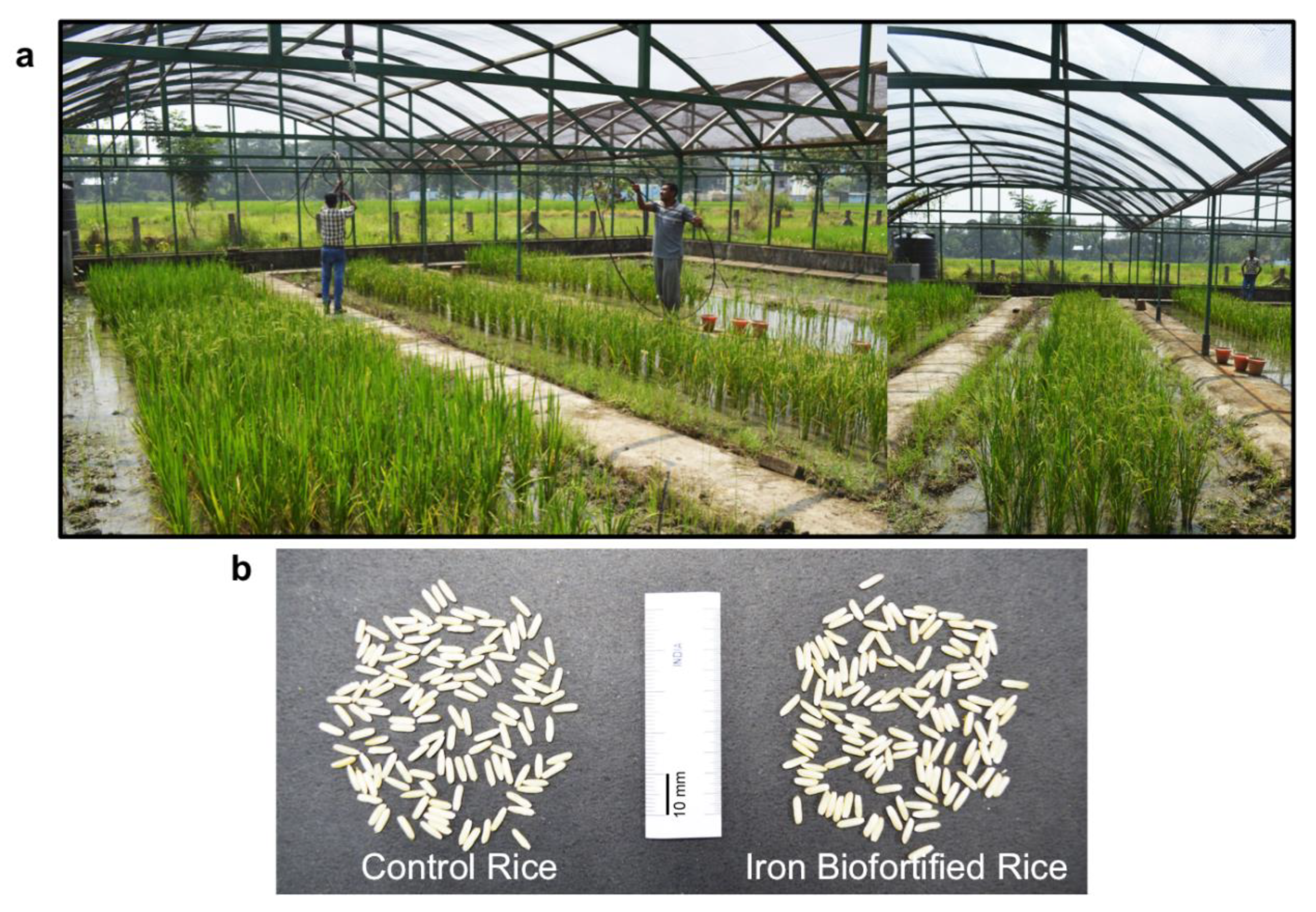
3. High Iron Rice
3.1. Iron Biofortification via Conventional Plant Breeding
3.2. Iron Biofortification via Molecular Plant Breeding
3.2.1. Enhancement of Iron Storage in Rice
3.2.2. Enhancement of Plant Iron Uptake from the Soil via Chelation-Based Strategy
3.2.3. Enhancement of Iron Influx in Seeds
3.2.4. Enhancement of Iron Uptake and Translocation
3.2.5. Enhancement of Iron Translocation
3.2.6. Manipulation of Iron Uptake and Translocation Regulators
3.2.7. Low Phytate Rice by Using RNAi Technology
3.2.8. Release of Phytic Acid Bound Iron
3.2.9. Combination of Multiple Transgenes
4. High Zinc Rice
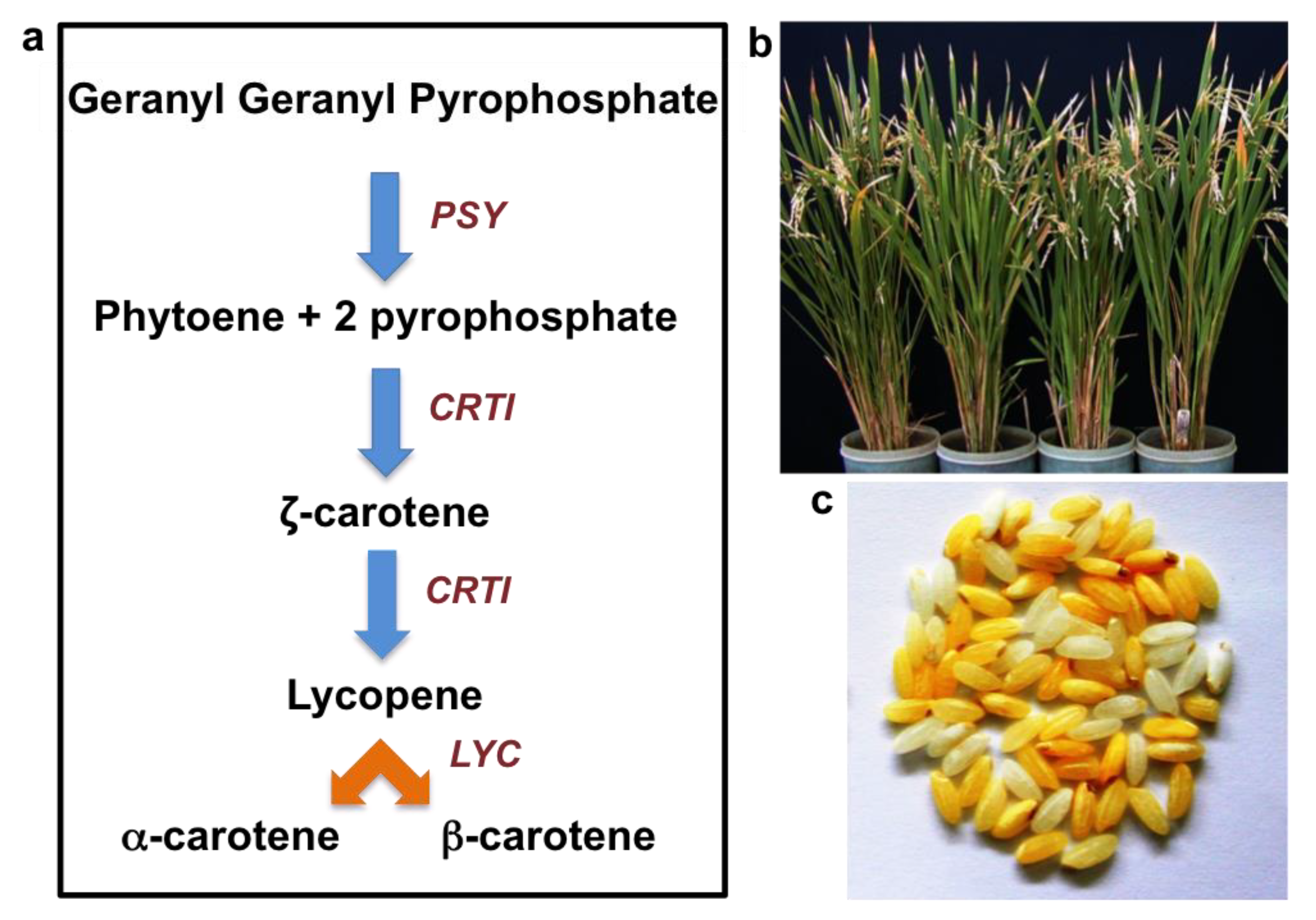
5. Provitamin-A (β-Carotene) Biofortified Rice—‘Golden Rice’
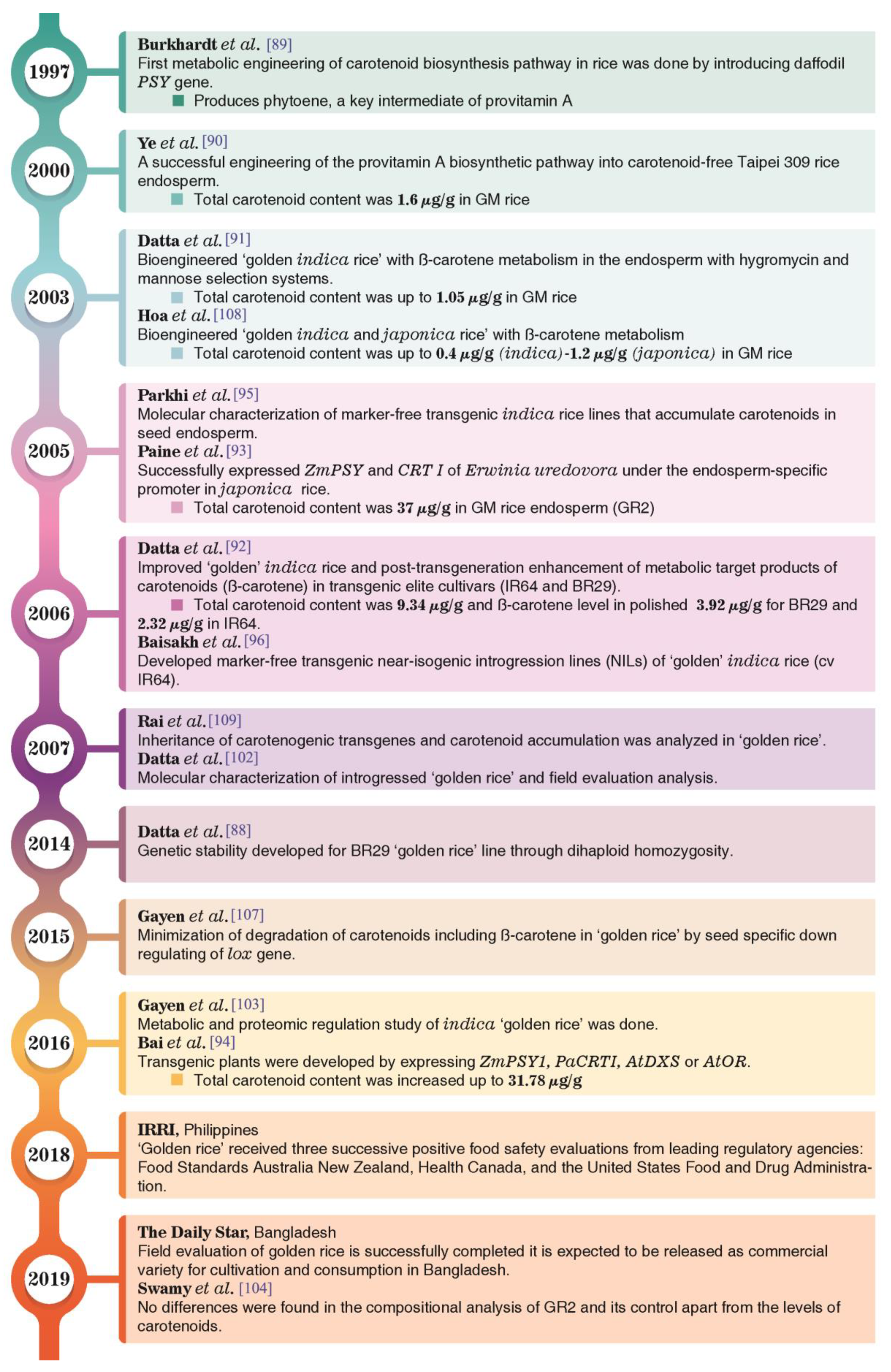
5.1. Development of Golden Rice
5.2. Long Term Storage of ‘Golden Rice’
6. Food Quality and Safety Analysis of Biofortified Rice
7. Regulatory Challenges of Biofortified Rice
8. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization FAO; International Fund for Agricultural Development IFAD; World Food Programme WFP. The State of Food Insecurity in the World 2015; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Datta, K.; Datta, S.K. Rice vitamins. In Rice Chemistry and Technology, 1st ed.; Bao, J., Ed.; Elsevier Inc.: Amsterdam, The Netherlands; AACC International: St. Paul, MN, USA, 2019; Volume 7, pp. 195–220. [Google Scholar]
- Champagne, E.T.; Wood, D.F.; Juliano, B.O.; Bechtel, D.B. The rice grain and its gross composition. In: Champagne ET (ed) Rice Chemistry and Technology. Cereal. Chem. 2004, 3, 93–96. [Google Scholar]
- Luh, B.S. Rice—I: Production, 2nd ed.; Van Nostrand Reinhold: New York, NY, USA, 1991. [Google Scholar]
- Dexter, P.B. Rice Fortification for Developing Countries; OMNI/USAID: Washington, DC, USA, 1998.
- Masuda, H.; Usuda, K.; Kobayashi, T.; Ishimaru, Y.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2009, 2, 155–166. [Google Scholar] [CrossRef]
- Datta, S.K.; Bouis, H.E. Application of biotechnology to improving the nutritional quality rice. Food Nutr. Bull. 2000, 21, 451–456. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Jasrasaria, R.; Naghavi, M.; Wulf, S.K.; Johns, N.; Lozano, R.; Regan, M.; Weatherall, D.; Chou, D.P.; Eisele, T.P.; et al. A systematic analysis of global anemia burden from 1990 to 2010. Blood 2014, 123, 615–624. [Google Scholar] [CrossRef]
- Virmani, S.S.; Ilyas-Ahmed, M. Rice breeding for sustainable production. In Breeding Major Food Staples; Blackwell Publishing Ltd.: Oxford, UK, 2008. [Google Scholar]
- Sperotto, R.A.; Ricachenevsky, F.K.; Waldow, V.d.A.; Fett, J.P. Iron biofortification in rice: It’s a long way to the top. Plant Sci. 2012, 190, 24–39. [Google Scholar] [CrossRef]
- Haas, J.D.; Beard, J.L.; Murray-Kolb, L.E.; del Mundo, A.M.; Felix, A.; Gregorio, G.B. Iron biofortified rice improves the iron stores of nonanemic Filipino women. J. Nutr. 2005, 135, 2823–2830. [Google Scholar] [CrossRef]
- Vasconcelos, M.; Datta, K.; Oliva, N.; Khalekuzzaman, M.; Torrizo, L.; Krishnan, S.; Oliveira, M.; Goto, F.; Datta, S.K. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 2003, 164, 371–378. [Google Scholar] [CrossRef]
- Paul, S.; Ali, N.; Datta, S.K.; Datta, K. Development of an iron-enriched high-yieldings Indica rice cultivar by introgression of a high-iron trait from transgenic iron-biofortified rice. Plant. Foods Hum. Nutr. 2014, 69, 203–208. [Google Scholar] [CrossRef]
- Qu, L.Q.; Yoshihara, T.; Ooyama, A.; Goto, F.; Takaiwa, F. Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 2005, 222, 225–233. [Google Scholar] [CrossRef]
- Lucca, P.; Hurrell, R.; Potrykus, I. Fighting iron deficiency anemia with iron-rich rice. J. Am. Coll. Nutr. 2002, 21, 184S–190S. [Google Scholar] [CrossRef] [PubMed]
- Goto, F.; Yoshihara, T.; Shigemoto, N.; Toki, S.; Takaiwa, F. Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 1999, 17, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Khalekuzzaman, M.; Datta, K.; Oliva, N.; Alam, M.; Datta, S. Stable integration, expression and inheritance of the ferritin gene in transgenic elite indica rice cultivar BR29 with enhanced iron level in the endosperm. Indian J. Biotechnol. 2006, 5, 26–31. [Google Scholar]
- Oliva, N.; Chadha-Mohanty, P.; Poletti, S.; Abrigo, E.; Atienza, G.; Torrizo, L.; Garcia, R.; Duenas, C., Jr.; Poncio, M.A.; Balindong, J.; et al. Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol. Breed. 2014, 33, 23–37. [Google Scholar] [CrossRef]
- Paul, S.; Ali, N.; Gayen, D.; Datta, S.K.; Datta, K. Molecular breeding of Osfer2 gene to increase iron nutrition in rice grain. GM Crops Food 2012, 3, 310–316. [Google Scholar] [CrossRef]
- Johnson, A.A.T.; Kyriacou, B.; Callahan, D.L.; Carruthers, L.; Stangoulis, J.; Lombi, E.; Tester, M. Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of Rice endosperm. PLoS ONE 2011, 6, e24476. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.-S.; Jeon, U.S.; Kim, Y.-K.; Schjoerring, J.K.; An, G. Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 2012, 33, 269–275. [Google Scholar] [CrossRef]
- Lee, S.; Jeon, U.S.; Lee, S.J.; Kim, Y.-K.; Persson, D.P.; Husted, S.; Schjorring, J.K.; Kakei, Y.; Masuda, H.; Nishizawa, N.K.; et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. USA 2009, 106, 22014–22019. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Suzuki, M.; Morikawa, K.C.; Nakanishi, H.; Takahashi, M.; Saigusa, M.; Mori, S.; Nishizawa, N.K. Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. J. Soil Sci. Plant. Nutr. 2008, 54, 77–85. [Google Scholar] [CrossRef]
- Masuda, H.; Suzuki, M.; Morikawa, K.C.; Kobayashi, T.; Nakanishi, H.; Takahashi, M.; Saigusa, M.; Mori, S.; Nishizawa, N.K. Increase in iron and zinc concentrations in rice grains via the introduction of barley genes involved in phytosiderophore synthesis. Rice 2008, 1, 100–108. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.H.; Yi, H.Y.; Gong, J.M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Bashir, K.; Takahashi, R.; Akhtar, S.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, A.; Bhattacharya, S.; Sengupta, S.; Ali, N.; Sarkar, S.N.; Datta, K.; Datta, S.K. RNAi-mediated silencing of ITPK gene reduces phytic acid content, alters transcripts of phytic acid biosynthetic genes, and modulates mineral distribution in rice seeds. Rice Sci. 2019. Available online: http://www.ricescience.org/EN/abstract/abstract9861.shtml (accessed on 17 August 2019).
- Ali, N.; Paul, S.; Gayen, D.; Sarkar, S.N.; Datta, S.K.; Datta, K. RNAi mediated down regulation of myo-inositol-3-phosphate synthase to generate low phytate rice. Rice 2013, 6, 12. [Google Scholar] [CrossRef]
- Ali, N.; Paul, S.; Gayen, D.; Sarkar, S.N.; Datta, K.; Datta, S.K. Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS ONE 2013, 8, e68161. [Google Scholar] [CrossRef]
- Lucca, P.; Hurrell, R.; Potrykus, I. Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains. Theor. Appl. Genet. 2001, 102, 392–397. [Google Scholar] [CrossRef]
- Wirth, J.; Poletti, S.; Aeschlimann, B.; Yakandawala, N.; Drosse, B.; Osorio, S.; Tohge, T.; Fernie, A.R.; Gunther, D.; Gruissem, W.; et al. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 2009, 7, 631–644. [Google Scholar] [CrossRef]
- Boonyaves, K.; Gruissem, W.; Bhullar, N.K. NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol. Biol. 2016, 90, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sengupta, S.; Karmakar, A.; Sarkar, S.N.; Gangopadhyay, G.; Datta, K.; Datta, S.K. Genetically engineered rice with appA gene enhanced phosphorus and minerals. J. Plant Biochem. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Masuda, H.; Ishimaru, Y.; Aung, M.S.; Kobayashi, T.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Nishizawa, N.K. Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2012, 2, 543. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Kobayashi, T.; Ishimaru, Y.; Takahashi, M.; Aung, M.S.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Iron biofortification in rice by the introduction of three barley genes participated in mugineic acid biosynthesis with soybean ferritin gene. Front. Plant Sci. 2013, 4, 132. [Google Scholar] [CrossRef]
- Boonyaves, K.; Wu, T.-Y.; Gruissem, W.; Bhullar, N.K. Enhanced grain iron levels in Rice expressing an iron-regulated metal transporter, nicotianamine synthase, and ferritin gene cassette. Front. Plant Sci. 2017, 8, 130. [Google Scholar] [CrossRef]
- Trijatmiko, K.R.; Duenas, C.; Tsakirpaloglou, N.; Torrizo, L.; Arines, F.M.; Adeva, C.; Balindong, J.; Oliva, N.; Sapasap, M.V.; Borrero, J.; et al. Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 2016, 6, 19792. [Google Scholar] [CrossRef]
- Aung, M.S.; Masuda, H.; Kobayashi, T.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. Iron biofortification of Myanmar rice. Front. Plant Sci. 2013, 4, 158. [Google Scholar]
- Theil, E. Ferritin: Structure, gene regulation, and cellular function in animals, plants and microorganisms. Annu. Rev. Biochem. 1987, 56, 289–315. [Google Scholar] [CrossRef]
- Andrews, S.C.; Arosio, P.; Bottke, W.; Briat, J.F.; Von Darl, M.; Harrison, P.M.; Laulhere, J.P.; Levi, S.; Lobreaux, S.; Yewdall, S.J. Structure, function and evolution of ferritins. J. Inorg. Biochem. 1992, 47, 116–174. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin: At the crossroads of iron and oxygen metabolism. J. Nutr. 2003, 133, 1549–1553. [Google Scholar] [CrossRef]
- Kok, A.D.; Yoon, L.L.; Sekeli, R.; Yeong, W.C.; Yusof, Z.N.B.; Song, L.K. Iron biofortification of rice: Progress and prospects. In Rice Crop—Current Developments; Saha, F., Khan, Z.H., Iqbal, A., Eds.; Intech Open: London, UK, 2018; Volume 3, pp. 25–44. [Google Scholar]
- Theil, E.C. Iron homeostasis and nutritional iron deficiency. J. Nutr. 2011, 141, 724S–728S. [Google Scholar] [CrossRef] [PubMed]
- Romheld, V.; Marschner, H. Genotypical differences among graminaceous species in release of phytosiderphores and uptake of iron phytosiderophores. Plant Soil 1990, 123, 147–153. [Google Scholar] [CrossRef]
- Marschner, H.; Romheld, V. Strategies for plants for acquisition of iron. Plant Soil 1994, 165, 261–274. [Google Scholar] [CrossRef]
- Huguchi, K.; Suzuki, K.; Nakanishi, H.; Yamaguchi, H.; Nishizawa, N.K.; Mori, S. Cloning of nicotamine synthase genes, novel genes involved in the synthesis of phytosiderophores. Plant Physiol. 1999, 119, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Takahashi, M.; Sato, Y.; Uozumi, N.; Nakanlshl, H.; Nlshlzawa, N.K. Phytosiderophore efflux transporters are crucial for iron acquisition in germinaceous plants. J. Biol. Chem. 2011, 286, 5446–5454. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.; Lou, G.M. Iron uptake and transport in plants: The good, the bad, and the ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+ -phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef]
- Bashir, K.; Inoue, H.; Nagasaka, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 2006, 281, 32395–32402. [Google Scholar] [CrossRef]
- Nakanishi, H.; Yamahuchi, H.; Sasakuma, T.; Nishizawa, N.K.; Mori, S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol. Biol. 2000, 44, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nakanish, H.; Takahashi, M.; Kawasaki, S.; Nishizawa, N.; Mori, S. In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2′-deoxymugineic acid to mugineic acid in transgenic rice. Planta 2001, 212, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Von-Wiren, N.; Khodr, H.; Hider, R.C. Hydroxylated phytosiderophore species possess an enhanced chelate stability and affinity for iron(III). Plant Physiol. 2000, 124, 1149–1158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, S.A.; Punshon, T.; Lanzirotti, A.; Li, L.; Alonso, J.M.; Ecker, J.R.; Kaplan, J.; Guerinot, M.L. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 2006, 314, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Itai, R.N.; Nakanishi, H.; Inoue, H.; Kobayashi, T.; Suzuki, M.; Takahashi, M.; Mori, S.; Nishizawa, N.K. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006, 57, 2867–2878. [Google Scholar] [CrossRef]
- Larson, S.R.; Rutger, J.N.; Young, K.A.; Raboy, V. Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid 1 mutation. Crop Sci. 2000, 40, 1397–1405. [Google Scholar] [CrossRef]
- Kim, S.I.; Andaya, C.B.; Newman, J.W.; Goyal, S.S.; Tai, T.H. Isolation and characterization of a low phytic acid rice mutant reveals a mutation in the rice orthologue of maize MIK. Theor. Appl. Genet. 2008, 117, 1291–1301. [Google Scholar] [CrossRef]
- Ali, N.; Datta, S.K.; Datta, K. RNA interference in designing transgenic crops. GM Crops 2010, 1, 207–213. [Google Scholar] [CrossRef]
- Kuwano, M.; Mimura, T.; Takaiwa, F.; Yoshida, K.T. Generation of stable ‘low phytic acid’ transgenic rice through antisense repression of the 1 D -myoinositol 3-phosphate synthase gene using the 18-kDa oleosin promoter. Plant Biotechnol. J. 2009, 7, 96–105. [Google Scholar] [CrossRef]
- Suzuki, M.; Tanaka, K.; Kuwano, M.; Yoshida, K.T. Expression pattern of inositol phosphate-related enzymes in rice (Oryza sativa L.): Implications for the phytic acid biosynthetic pathway. Gene 2007, 405, 55–64. [Google Scholar] [CrossRef]
- Kuwano, M.; Ohyama, A.; Tanaka, Y.; Mimura, T.; Takaiwa, F.; Yoshida, K.T. Molecular breeding for transgenic rice with low-phytic acid phenotype through manipulating myo-inositol 3 phosphate synthase gene. Mol. Breeding 2006, 18, 263–272. [Google Scholar] [CrossRef]
- Feng, X.; Yoshida, K.T. Molecular approaches for producing low-phytic-acid grains in rice. Plant Biotechnol. 2004, 21, 183–189. [Google Scholar] [CrossRef]
- Welch, R.M. Breeding strategies for biofortified staple plant foods to reduce micronutrient malnutrition globally. J. Nutr. 2002, 132, 495S–499S. [Google Scholar] [CrossRef] [PubMed]
- Pasamontes, L.; Haiker, M.; Wyss, M.; Tessier, M.; Van-Loon, A.P.G.M. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 1997, 63, 1696–1700. [Google Scholar] [PubMed]
- Taylor, P.G.; Martinez, T.C.; Romano, E.L.; Layrisse, M. The effect of cysteine-containing peptides released during meat digestion on iron absorption in humans. Am. J. Clin. Nutr. 1986, 43, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Kagi, J.H.R.; Schaffer, A. Biochemistry of metallothionein. Biochemistry 1988, 27, 8509–8515. [Google Scholar] [CrossRef]
- Hsieh, H.; Liu, W.; Huang, P.C. A novel stress-inducible metallothionein-like gene from rice. Plant Mol. Biol. 1995, 28, 381–389. [Google Scholar] [CrossRef]
- Graham, R.D.; Knez, M.; Welch, R.M. How much nutritional iron deficiency in humans globally is due to an underlying zinc deficiency? Adv. Agron. 2012, 115, 1–40. [Google Scholar]
- Levenson, C.W.; Morris, D. Zinc and neurogenesis: Making new neurons from development to adulthood. Adv. Nutr. 2011, 2, 96–100. [Google Scholar] [CrossRef]
- Hefferon, K. Biotechnological approaches for generating zinc-enriched crops to combat malnutrition. Nutrients 2019, 11, 253. [Google Scholar] [CrossRef]
- Prasad, R.; Shivay, Y.S.; Kumar, D. Agronomic biofortification of cereal grains with iron and zinc. Adv. Agro. 2014, 125, 55–91. [Google Scholar]
- Arsenault, J.E.; Yakes, E.A.; Hossain, M.B.; Islam, M.M.; Ahmed, T.; Hotz, C.; Lewis, B.; Rahman, A.S.; Jamil, K.M.; Brown, K.H. The current high prevalence of dietary zinc inadequacy among children and women in rural Bangladesh could be substantially ameliorated by zinc biofortification of rice. J. Nutr. 2010, 140, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, P. HarvestPlus Talks Zinc Rice with Farmers in Southeastern Bangladesh. 2018. Available online: https://www.harvestplus.org/knowledge-market/in-the-news/harvestplus-talks-zinc-rice-farmers-southeastern-bangladesh (accessed on 14 April 2019).
- Ishimaru, Y.; Suzuki, M.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005, 56, 3207–3214. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Masuda, H.; Suzuki, M.; Bashir, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J. Exp. Bot. 2007, 58, 2909–2915. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Jiang, Y.; Zhang, H.S. Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol. Biol. Rep. 2007, 36, 281–287. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Shin, R.; Eide, D.J.; Schachtman, P. Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 2003, 133, 126–134. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef]
- Tan, S.; Han, R.; Li, P.; Yang, G.; Li, S.; Zhang, P.; Wang, W.B.; Zhao, W.Z.; Yin, L.P. Over-expression of the MxIRT1 gene increases iron and zinc content in rice seeds. Transgenic Res. 2015, 24, 109–122. [Google Scholar] [CrossRef]
- Johnson, E.J. The role of carotenoids in human health. Nutr. Clin. Care 2002, 5, 56–65. [Google Scholar] [CrossRef]
- Tan, J.; Baisakh, N.; Oliva, N.; Torrizo, L.; Abrigo, E.; Datta, K.; Datta, S.K. HPLC carotenoid profiles of rice germplasm. Int. J. Food. Sci. Technol. 2005, 40, 563–569. [Google Scholar] [CrossRef]
- Datta, K.; Sahoo, G.; Krishnan, S.; Ganguly, M.; Datta, S.K. Genetic stability developed for β-carotene synthesis in BR29 rice line using dihaploid homozygosity. PLoS ONE 2014, 9, e100212. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, P.K.; Beyer, P.; Wunn, J.; Kloti, A.; Armstrong, G.A.; Schledz, M.; von-Lintig, J.; Potrykus, I. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin a biosynthesis. Plant J. 1997, 11, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Al-Babili, S.; Kloti, A.; Zhang, J.; Lucca, P.; Beyer, P.; Potrykus, I. Engineering the provitamin a (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 2000, 287, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Baisakh, N.; Oliva, N.; Torrizo, L.; Abrigo, E.; Tan, J.; Rai, M.; Rehana, S.; Al-Babili, S.; Beyer, P.; et al. Bioengineered ‘golden’ indica rice cultivar with β-carotene metabolism in the endosperm with hygromycin and mannose selection systems. Plant Biotechnol. J. 2003, 1, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Rai, M.; Parkhi, V.; Oliva, N.; Tan, J.; Datta, S.K. Improved ‘golden’ indica rice and post-transgeneration enhancement of metabolic target products of carotenoids (β-carotene) in transgenic elite cultivars (IR64 and BR29). Curr. Sci. 2006, 91, 935–939. [Google Scholar]
- Paine, J.A.; Shipton, C.A.; Chagger, S.; Howles, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hincliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Capell, T.; Berman, J.; Medina, V.; Sandmann, G.; Christou, P.; Zhu, C. Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor-product balance. Plant Biotechnol. J. 2016, 14, 195–205. [Google Scholar] [CrossRef]
- Parkhi, V.; Rai, M.; Tan, J.; Oliva, N.; Rehana, S.; Bandyopadhyay, A.; Torrizo, L.; Ghole, V.; Datta, K.; Datta, S.K. Molecular characterization of marker free transgenic indica rice lines that accumulate carotenoids in seed endosperm. Mol. Genet. Genomics 2005, 274, 325–336. [Google Scholar] [CrossRef]
- Baisakh, N.; Rehana, S.; Rai, M.; Oliva, N.; Tan, J.; Mackill, D.; Khush, G.S.; Datta, K.; Datta, S.K. Marker-free transgenic (MFT) near-isogenic introgression lines (NILs) of ‘golden’ indica rice (cv IR64) with accumulation of provitamin A in the endosperm tissue. Plant Biotechnol. J. 2006, 4, 467–475. [Google Scholar] [CrossRef]
- Cotsaftis, O.; Sallaud, C.; Breitler, J.C.; Meynard, D.; Greco, R.; Pereira, A.; Guiderdoni, E. Transposon-mediated generation of T-DNA and marker free rice plants expressing a Bt endotoxin gene. Mol. Breed. 2002, 10, 165–180. [Google Scholar] [CrossRef]
- Zubko, E.; Scutt, C.; Meyer, P. Intrachromosomal recombination between aatP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol. 2000, 18, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Odell, J.; Caimi, P.; Sauer, B.; Russell, S. Site directed recombination in the genome of transgenic tobacco. Mol. Gen. Genet. 1990, 223, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.M.; Davis, R.W. Functional expression of the yeast FLP/FRT site specific recombination system in Nicotiana tabacum. Mol. Gen. Genet. 1994, 242, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Depicker, A.; Herman, L.; Jacobs, S.; Schell, J.; van Montagu, M. Frequency of simultaneous transformation with different T-DNAs and their relevance to the Agrobacterium-plant cell interaction. Mol. Gen. Genet. 1985, 201, 477–484. [Google Scholar] [CrossRef]
- Datta, S.K.; Datta, K.; Parkhi, V.; Rai, M.; Baisakh, N.; Sahoo, G.; Rehana, S.; Bandyopadhyay, A.; Alamgir, M.; Ali, M.S.; et al. Golden rice: Introgression, breeding, and field evaluation. Euphytica 2007, 154, 271–278. [Google Scholar] [CrossRef]
- Gayen, D.; Ghosh, S.; Paul, S.; Sarkar, S.N.; Datta, S.K.; Datta, K. Metabolic regulation of carotenoid-enriched golden rice line. Front. Plant Sci. 2016, 7, 1622. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.M.; Samia, M.; Boncodin, R.; Marundan, S.; Rebong, D.B.; Ordonio, R.L.; Miranda, R.T.; Rebong, A.T.O.; Alibuyog, A.Y.; Adeva, C.C.; et al. Compositional analysis of genetically engineered GR2E “Golden Rice” in comparison to that of conventional rice. J. Agric. Food Chem. 2019. [Google Scholar] [CrossRef]
- Carrera, A.; Echenique, V.; Zhang, W.; Helguera, M.; Manthey, F.; Schrager, A.; Picca, A.; Cervigni, G.; Dubcovsky, J. A deletion at the Lpx-B1 locus is associated with low lipoxygenase activity and improved pasta color in durum wheat (Triticum turgidum ssp. durum). J. Cereal. Sci. 2007, 45, 67–77. [Google Scholar] [CrossRef]
- Gayen, D.; Ali, N.; Ganguly, M.; Paul, S.; Datta, K.; Datta, S.K. RNAi mediated silencing of lipoxygenase gene to maintain rice grain quality and viability during storage. Plant Cell Tiss. Org. Cul. 2014, 118, 229–243. [Google Scholar] [CrossRef]
- Gayen, D.; Ali, N.; Sarkar, S.N.; Datta, S.K.; Datta, K. Down-regulation of lipoxygenase gene reduces degradation of carotenoids of golden rice during storage. Planta 2015, 242, 353–363. [Google Scholar] [CrossRef]
- Hoa, T.T.C.; Al-Babili, S.; Schaub, P.; Potrykus, I.; Beyer, P. Golden indica and japonica rice lines amenable to deregulation. Plant Physiol. 2003, 133, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Datta, K.; Parkhi, V.; Tan, J.; Oliva, N.; Chawla, H.S.; Datta, S.K. Variable T-DNA linkage configuration affects inheritance of carotenogenic transgenes and carotenoid accumulation in transgenic indica rice. Plant Cell Rep. 2007, 26, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Gayen, D.; Sarkar, S.N.; Datta, S.K.; Datta, K. Comparative analysis of nutritional compositions of transgenic high iron rice with its non-transgenic counterpart. Food Chem. 2013, 138, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Slamet-Loedin, I.H.; Johnson-Beebout, S.E.; Impa, S.; Tsakirpaloglou, N. Enriching rice with Zn and Fe while minimizing Cd risk. Front. Plant Sci. 2015, 6, 121. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome Editing in Rice: Recent Advances, Challenges, and Future Implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
- Napier, J.A.; Haslam, R.P.; Tsalavouta, M.; Sayanova, O. The challenges of delivering genetically modified crops with nutritional enhancement traits. Nat. Plant. 2019, 5, 563–567. [Google Scholar] [CrossRef]
- Dubock, A. The present status of Golden Rice. J. Huazhong Agric. Univ. 2014, 33, 69–84. [Google Scholar]
- Bollinedi, H.; Krishnan, G.; Prabhu, K.V.; Singh, N.K.; Mishra, S.; Khurana, J.P.; Singh, A.K. Molecular and functional characterization of GR2-R1 event based backcross derived lines of Golden Rice in the genetic background of a mega rice variety Swarna. PLoS ONE 2017, 12, 0169600. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Lang, Z.; Ramon, J.B.; Zhu, J.K. Genome editing-principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 2017, 36, 291–309. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genomics 2016, 43, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Xiao, L.; Hua, K.; Zoua, C.; Zhao, Y.; Bressanb, R.A.; Zhu, J.-K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Biswal, A.K.; Dionora, J.; Perdigon, K.M.; Balahadia, C.P.; Mazumdar, S.; Chater, C.; Lin, H.C.; Coe, R.A.; Kretzschmar, T.; et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017, 36, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zeng, D.; Yu, S.; Cui, C.; Li, J.; Li, H.; Chen, J.; Zhang, R.; Zhao, X.; Chen, L.; et al. From Golden Rice to aSTARice: Bioengineering astaxanthin biosynthesis in rice endosperm. Mol. Plant 2018, 11, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Descalsota, G.I.L.; Swamy, B.P.M.; Zaw, H.; Inabangan-Asilo, M.A.; Amparado, A.; Mauleon, R.; Chadha-Mohanty, P.; Arocena, E.C.; Raghavan, C.; Leung, H.; et al. Genome-wide association mapping in a rice MAGIC plus population detects QTLs and genes useful for Biofortification. Front. Plant Sci. 2018, 9, 1347. [Google Scholar] [CrossRef]
- Christian, P.; West, K.P., Jr. Interaction between zinc and vitamin A: An update. Am. J. Clin. Nutr. 1998, 68, 435–441. [Google Scholar] [CrossRef]
- Graham, R.D.; Rosser, J.M. Carotenoids in staple foods: Their potential to improve human nutrition. Food Nutr. Bull. 2000, 21, 404–409. [Google Scholar] [CrossRef]
- King, J.C.; Donangelo, C.M.; Woodhouse, L.R.; Mertz, S.D.; Shames, D.M.; Viteri, F.E.; Cheng, Z.; Welch, R.M. Measuring iron and zinc bioavailability in humans. Food Nutr. Bull. 2000, 21, 434–439. [Google Scholar] [CrossRef]
- Singh, S.P.; Gruissem, W.; Bhullar, N.K. Single genetic locus improvement of iron, zinc and β-carotene content in rice grains. Sci. Rep. 2017, 7, 6883. [Google Scholar] [CrossRef]
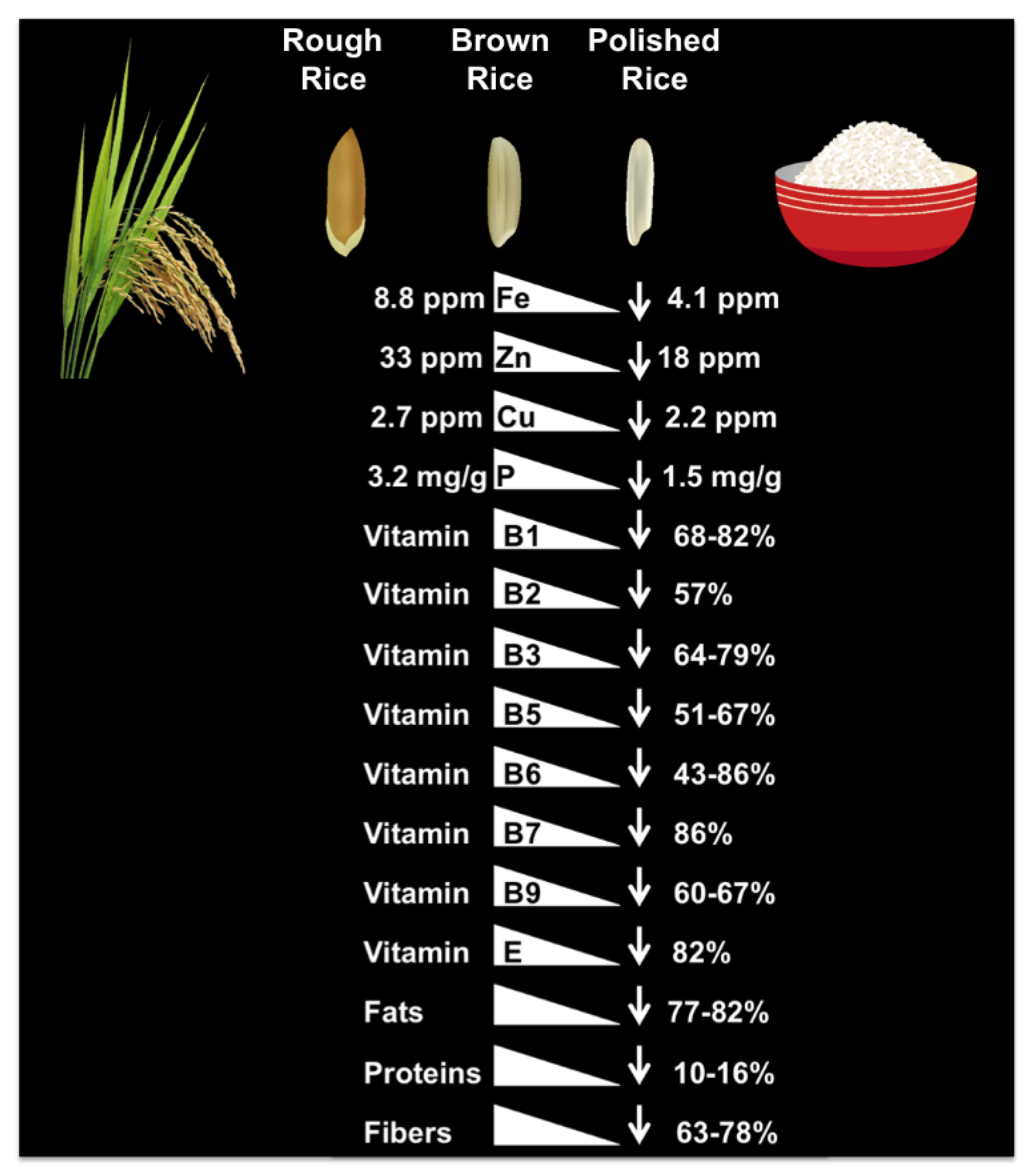
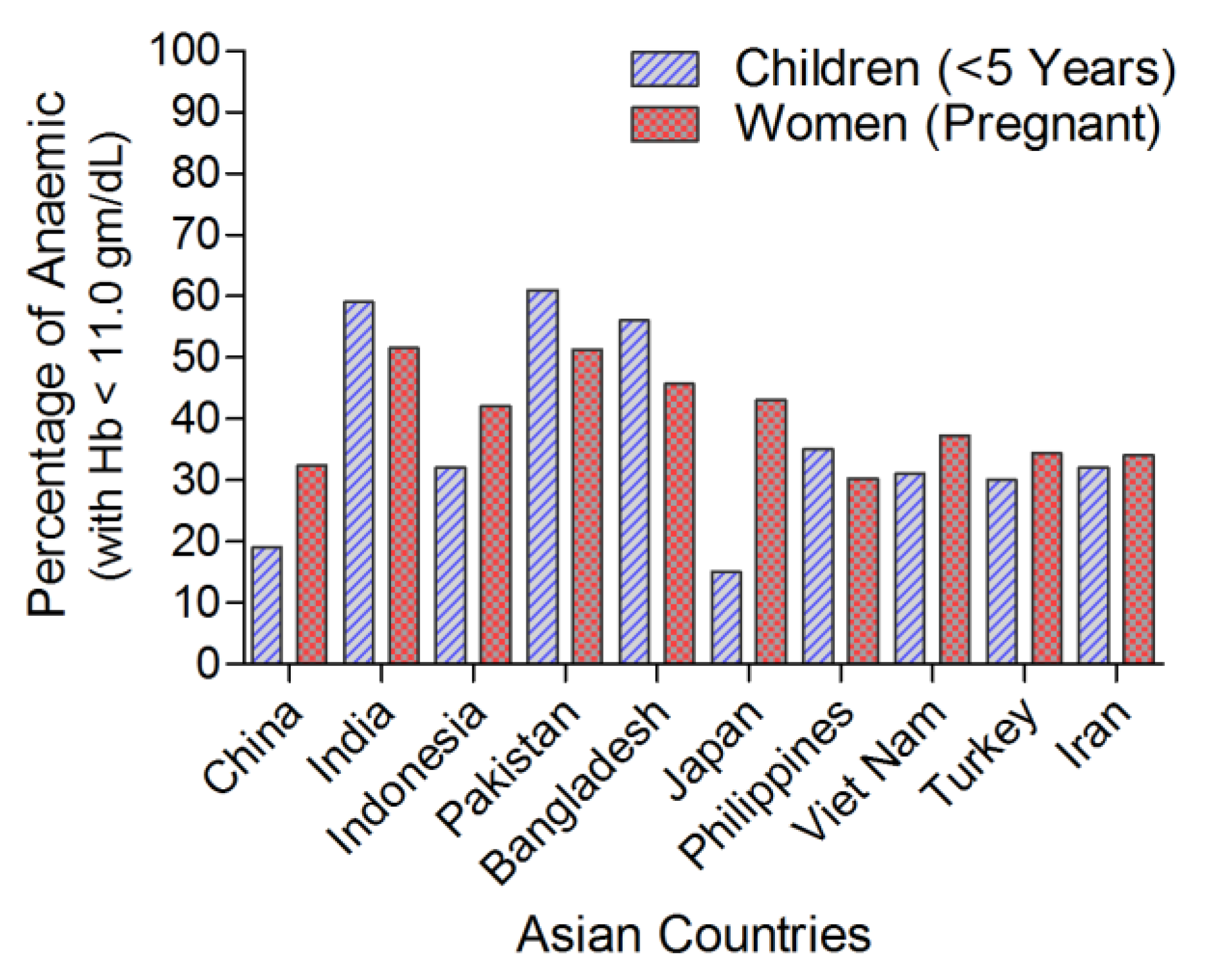
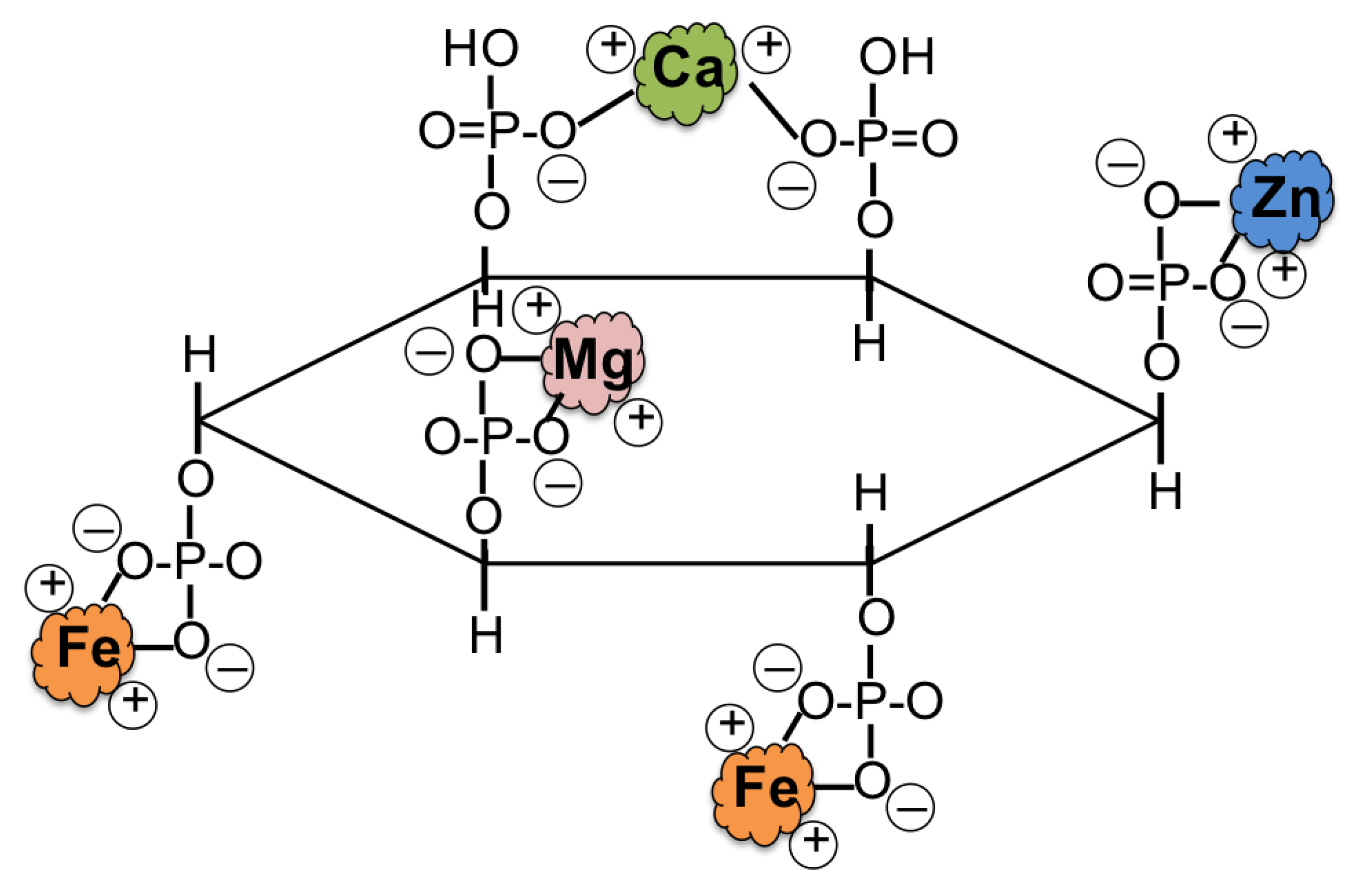
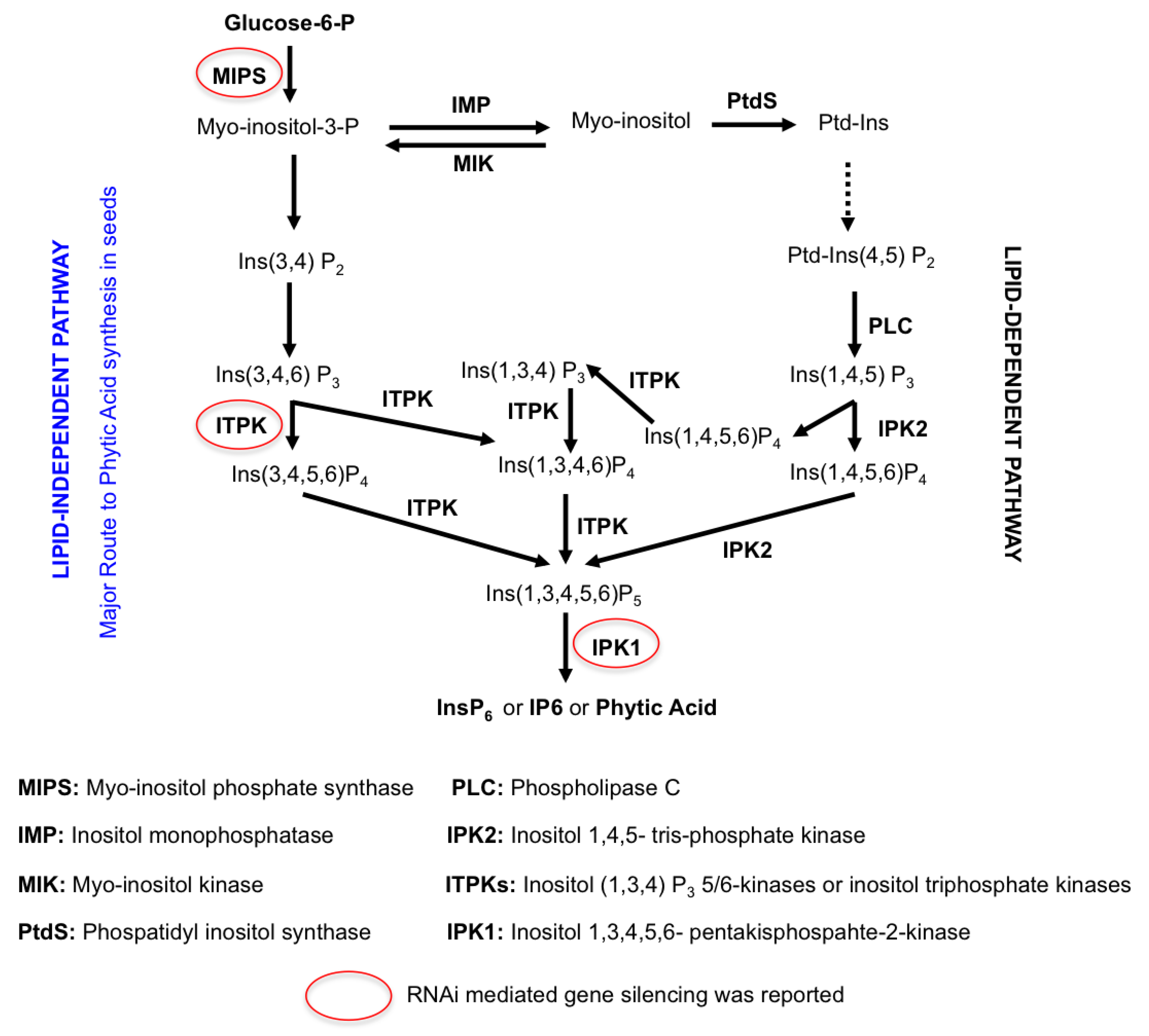
| Rice Iron Biofortification Strategies | Iron Increased in Biofortified Rice Seed | Reference |
|---|---|---|
| Improving iron storage via ferritin | 1.5–2.2-fold in brown rice (BR) 2.0–3.7-fold in polished rice (PR) | [15,16] [13,17,18,19,20] |
| Chelation based strategy (via NAS gene) | 2.3–4.0-fold in PR | [7,21,22,23] |
| Enhancing iron influx (via OsYSL2 gene) | 4.4-fold in PR | [24] |
| Enhancing iron uptake and translocation (via IDS3 gene) | 1.3-fold in BR 1.4-fold in PR | [25] [26] |
| Enhancing iron translocation (silencing OsVITs genes) | 1.4-fold in BR 1.8-fold in PR | [27] [28] |
| Manipulation of iron uptake and translocation regulators | 2.0–3.8-fold in BR 2.9-fold in PR | [29,30] [29] |
| Low phytate rice (RNAi silencing of phytic acid) | 1.3–1.8-fold in PR | [31,32,33] |
| Release of phytic acid bound iron (by phytase gene) | 2.0-fold in BR 2.0–6.3-fold in PR | [34] [35,36,37] |
| Multiple transgenes combination | 6.0-fold in BR 3.4–6.0-fold in PR | [38] [39,40,41,42] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumder, S.; Datta, K.; Datta, S.K. Rice Biofortification: High Iron, Zinc, and Vitamin-A to Fight against “Hidden Hunger”. Agronomy 2019, 9, 803. https://doi.org/10.3390/agronomy9120803
Majumder S, Datta K, Datta SK. Rice Biofortification: High Iron, Zinc, and Vitamin-A to Fight against “Hidden Hunger”. Agronomy. 2019; 9(12):803. https://doi.org/10.3390/agronomy9120803
Chicago/Turabian StyleMajumder, Shuvobrata, Karabi Datta, and Swapan Kumar Datta. 2019. "Rice Biofortification: High Iron, Zinc, and Vitamin-A to Fight against “Hidden Hunger”" Agronomy 9, no. 12: 803. https://doi.org/10.3390/agronomy9120803
APA StyleMajumder, S., Datta, K., & Datta, S. K. (2019). Rice Biofortification: High Iron, Zinc, and Vitamin-A to Fight against “Hidden Hunger”. Agronomy, 9(12), 803. https://doi.org/10.3390/agronomy9120803






