Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops
Abstract
1. Introduction
2. Pattern-Triggered Immunity Forms a Robust Host Barrier to Invaders
2.1. Microbe-Associated Molecular Patterns (MAMPs)
2.2. Damage-Associated Molecular Patterns (DAMPs)
2.3. Pattern Recognition Receptors (PRRs)
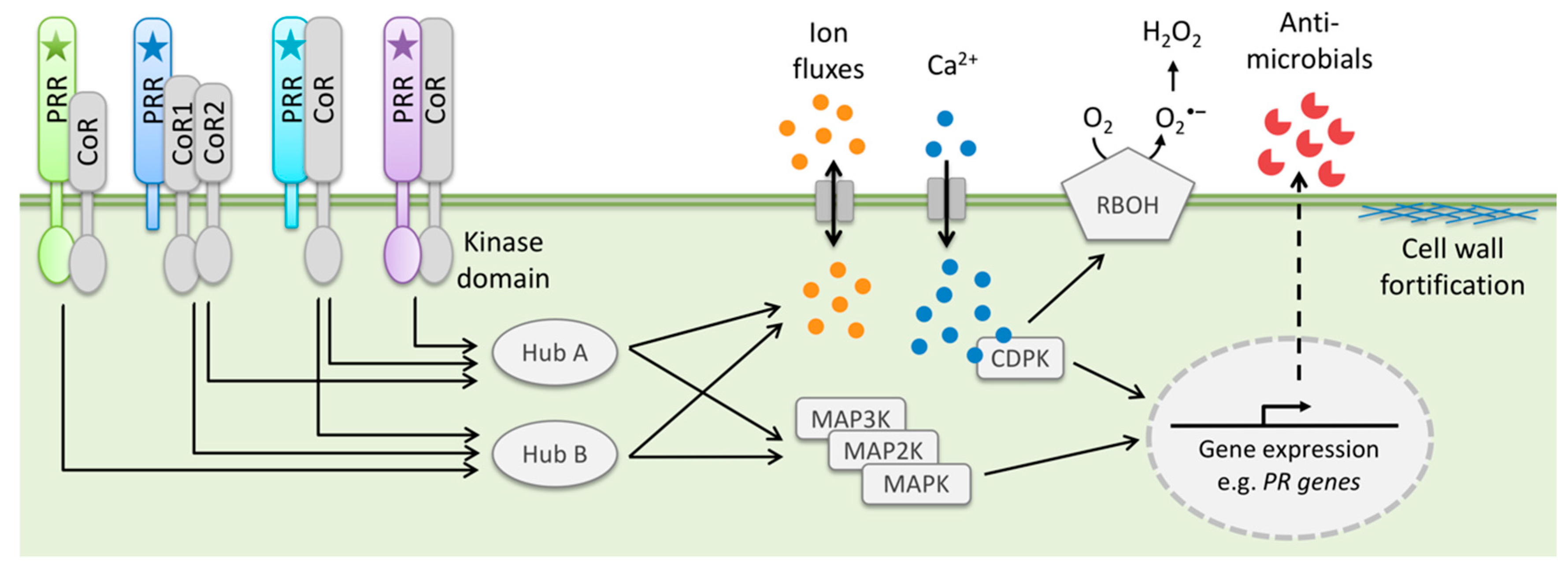
2.4. Pattern-Triggered Immunity (PTI)
3. Prospects of Deploying PTI for Broad-Spectrum Disease Resistance Engineering in Crops
3.1. Pattern- versus Effector-Triggered Immunity
3.2. PRR Transfer Between Plants Species
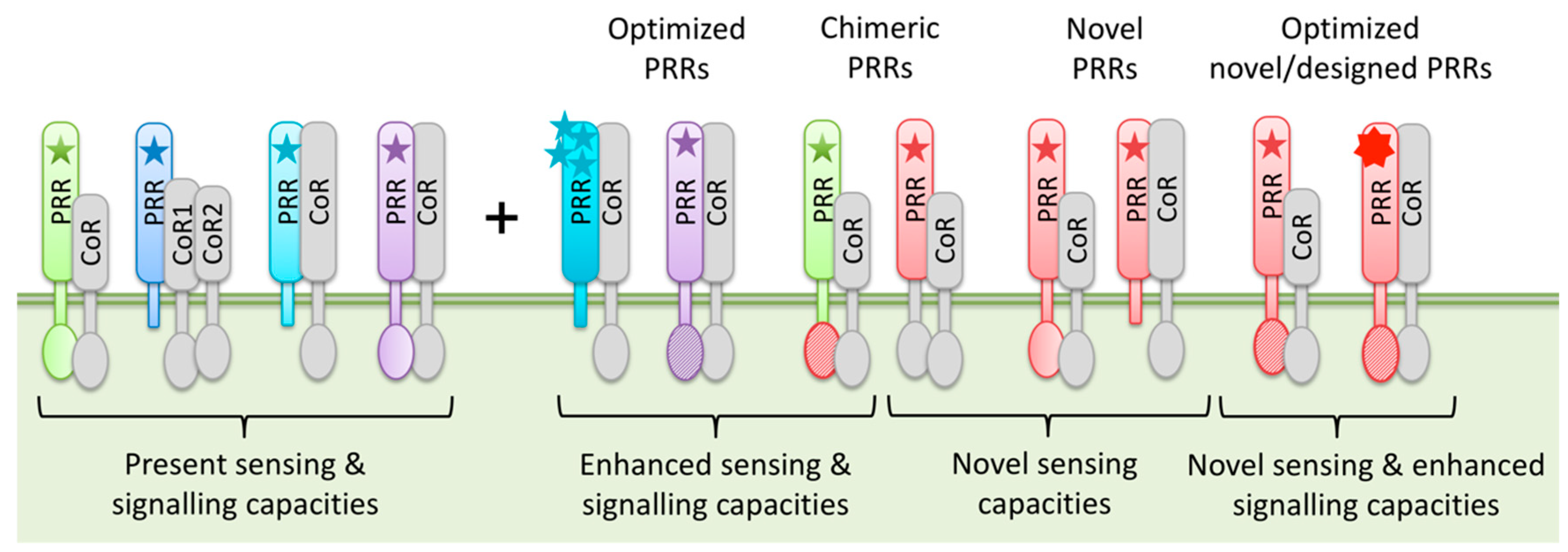
3.3. PRR Engineering
3.4. Exploiting DAMP Signalling
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Oerke, E. Crop Losses to Animal Pests, Plant Pathogens, and Weeds. In Encyclopedia of Pest Management, Volume II; CRC Press: Boca Raton, FL, USA, 2009; pp. 116–120. [Google Scholar]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The Emergence of Ug99 Races of the Stem Rust Fungus is a Threat to World Wheat Production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Scheel, D.; Lee, J. Challenges in the identification of microbe-associated molecular patterns in plant and animal innate immunity: A case study with bacterial lipopolysaccharide. Mol. Plant Pathol. 2016, 17, 1165–1169. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Lewis, J.; Yan, S.; Liu, H.; Clarke, C.R.; Campanile, F.; Almeida, N.F.; Studholme, D.J.; Lindeberg, M.; Schneider, D.; et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 2011, 7, e1002130. [Google Scholar]
- Gómez-Gómez, L.; Felix, G.; Boller, T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999, 18, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Pfund, C.; Tans-Kersten, J.; Dunning, F.M.; Alonso, J.M.; Ecker, J.R.; Allen, C.; Bent, A.F. Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2004, 17, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Sun, W. Within-species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 2006, 18, 764–779. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.R.; Chinchilla, D.; Hind, S.R.; Taguchi, F.; Miki, R.; Ichinose, Y.; Martin, G.B.; Leman, S.; Felix, G.; Vinatzer, B.A. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol. 2013, 200, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, Z.; Wang, H.; Liu, L.; Lu, F.; Yang, J.; Zhang, M.; Zhang, S.; Guo, Z.; Bent, A.F.; et al. Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola. Mol. Plant 2015, 8, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- McCann, H.C.; Nahal, H.; Thakur, S.; Guttman, D.S. Identification of innate immunity elicitors using molecular signatures of natural selection. Proc. Natl. Acad. Sci. USA 2012, 109, 4215–4220. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.-I.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S. Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 2017, 38, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.; Manohar, M.; von Reuss, S.H.; Chen, S.; Koch, A.; Kaplan, F.; Choe, A.; Micikas, R.J.; Wang, X.; Kogel, K.-H.; et al. Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 2015, 6, 7795. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.G.; Darvill, A.G.; Albersheim, P. Host-Pathogen Interactions: XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeans. Plant Physiol. 1981, 68, 1161–1169. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.; Felix, G.; Buchala, A.; Muller, C.; Métraux, J.-P. Perception of free cutin monomers by plant cells. Plant J. 1996, 10, 331–341. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, M.; Zhang, X.; Yao, J.; Zhang, Y.; Mou, Z. A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 2017, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Manohar, M.; Manosalva, P.; Tian, M.; Moreau, M.; Klessig, D.F. Activation of Plant Innate Immunity by Extracellular High Mobility Group Box 3 and Its Inhibition by Salicylic Acid. PLoS Pathog. 2016, 12, e1005518. [Google Scholar] [CrossRef] [PubMed]
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turrà, D.; Di Pietro, A.; Zhang, W. The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7. PLoS Pathog. 2014, 10, e1004331-15. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 10732–10736. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yamashita-Yamada, M.; Hirase, T.; Fujiwara, T.; Tsuda, K.; Hiruma, K.; Saijo, Y. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 2016, 35, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.J.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef] [PubMed]
- Lori, M.; van Verk, M.C.; Hander, T.; Schatowitz, H.; Klauser, D.; Flury, P.; Gehring, C.A.; Boller, T.; Bartels, S. Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: Interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 2015, 66, 5315–5325. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Oome, S.; Raaymakers, T.M.; Cabral, A.; Samwel, S.; Böhm, H.; Albert, I.; Nürnberger, T.; Van den Ackerveken, G. Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 16955–16960. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.; Albert, I.; Oome, S.; Raaymakers, T.M.; Van den Ackerveken, G.; Nürnberger, T. A Conserved Peptide Pattern from a Widespread Microbial Virulence Factor Triggers Pattern-Induced Immunity in Arabidopsis. PLoS Pathog. 2014, 10, e1004491. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Shan, L.; He, P.; de Vries, S.; Kemmerling, B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009, 14, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Brunner, F.; Nürnberger, T. Identification of immunogenic microbial patterns takes the fast lane. Proc. Natl. Acad. Sci. USA 2012, 109, 4029–4030. [Google Scholar] [CrossRef] [PubMed]
- Hind, S.R.; Strickler, S.R.; Boyle, P.C.; Dunham, D.M.; Bao, Z.; O’Doherty, I.M.; Baccile, J.A.; Hoki, J.S.; Viox, E.G.; Clarke, C.R.; et al. Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2016, 2, 16128. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Inagaki, H.; Takai, R.; Hirai, H.; Che, F.-S. Two distinct EF-Tu epitopes induce immune responses in rice and Arabidopsis. Mol. Plant-Microbe Interact. 2014, 27, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.G.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Gisch, N.; Schäffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Sánchez-Carballo, P.M.; Zähringer, U.; Hückelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between Innate Immunity and the Plant Microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nat. Publ. Group 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.-F.; Nomura, K.; Aung, K.; Velásquez, A.C.; Yao, J.; Boutrot, F.; Chang, J.H.; Zipfel, C.; He, S.Y. Bacteria establish an aqueous living space in plants crucial for virulence. Nature 2016, 539, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Halane, M.K.; Gassmann, W.; Stacey, G. The Role of Plant Innate Immunity in the Legume-Rhizobium Symbiosis. Annu. Rev. Plant Biol. 2017, 68, 535–561. [Google Scholar] [CrossRef] [PubMed]
- Albersheim, P.; Valent, B.S. Host-pathogen interactions in plants. Plants, when exposed to oligosaccharides of fungal origin, defend themselves by accumulating antibiotics. J. Cell Biol. 1978, 78, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Somssich, I.E.; Science, K. Pathogen defence in plants—A paradigm of biological complexity. Trends Plant Sci. 1998, 3, 86–90. [Google Scholar] [CrossRef]
- Ayers, A.R.; Valent, B.; Ebel, J.; Albersheim, P. Host-Pathogen Interactions: XI. Composition and Structure of Wall-released Elicitor Fractions. Plant Physiol. 1976, 57, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Fuller, K.W. Characterization of components from culture filtrates of Botrytis cinerea which stimulate phaseollin biosynthesis in Phaseolus vulgaris cell suspension cultures. Physiol. Plant Pathol. 1977, 11, 287–296. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, S.E.V.; Rougon-Cardoso, A.; Sherwood, E.; Peeters, N.; Dahlbeck, D.; Van Esse, H.P.; Smoker, M.; Rallapalli, G.; Thomma, B.P.H.J.; Staskawicz, B.; et al. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 2010, 28, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Holton, N.; Nekrasov, V.; Ronald, P.C.; Zipfel, C. The Phylogenetically-Related Pattern Recognition Receptors EFR and XA21 Recruit Similar Immune Signaling Components in Monocots and Dicots. PLoS Pathog. 2015, 11, e1004602-22. [Google Scholar] [CrossRef] [PubMed]
- Schwessinger, B.; Bahar, O.; Thomas, N.; Holton, N.; Nekrasov, V.; Ruan, D.; Canlas, P.E.; Daudi, A.; Petzold, C.J.; Singan, V.R.; et al. Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses. PLoS Pathog. 2015, 11, e1004809. [Google Scholar]
- Rodriguez-Moreno, L.; Song, Y.; Thomma, B.P. Transfer and engineering of immune receptors to improve recognition capacities in crops. Curr. Opin. Plant Biol. 2017, 38, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Schoonbeek, H.-J.; Wang, H.-H.; Stefanato, F.L.; Craze, M.; Bowden, S.; Wallington, E.; Zipfel, C.; Ridout, C.J. Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 2015, 206, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Lorenzen, J.; Bahar, O.; Ronald, P.; Tripathi, L. Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol. J. 2014, 12, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.A.; Murata, M.M.; El-Shamy, H.A.; Graham, J.H.; Grosser, J.W. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018, 27, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Verzaux, E.; Chaparro-Garcia, A.; Bijsterbosch, G.; Keizer, L.C.P.; Zhou, J.; Liebrand, T.W.H.; Xie, C.; Govers, F.; Robatzek, S.; et al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 2015, 1, 15034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fraiture, M.; Kolb, D.; Loffelhardt, B.; Desaki, Y.; Boutrot, F.F.G.; Tor, M.; Zipfel, C.; Gust, A.A.; Brunner, F. Arabidopsis RECEPTOR-LIKE PROTEIN30 and Receptor-Like Kinase SUPPRESSOR OF BIR1-1/EVERSHED Mediate Innate Immunity to Necrotrophic Fungi. Plant Cell 2013, 25, 4227–4241. [Google Scholar] [CrossRef] [PubMed]
- Jehle, A.K.; Lipschis, M.; Albert, M.; Fallahzadeh-Mamaghani, V.; Fürst, U.; Mueller, K.; Felix, G. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 2013, 25, 2330–2340. [Google Scholar] [CrossRef] [PubMed]
- Hegenauer, V.; Fürst, U.; Kaiser, B.; Smoker, M.; Zipfel, C.; Felix, G.; Stahl, M.; Albert, M. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 2016, 353, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Mott, G.A.; Thakur, S.; Smakowska, E.; Wang, P.W.; Belkhadir, Y.; Desveaux, D.; Guttman, D.S. Genomic screens identify a new phytobacterial microbe-associated molecular pattern and the cognate Arabidopsis receptor-like kinase that mediates its immune elicitation. Genome Biol. 2016, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nsibo, D.L.; Juhar, H.M.; Govers, F.; Bouwmeester, K. Ectopic expression of Arabidopsis L-type lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 in Nicotiana benthamiana confers Phytophthora resistance. Plant Cell Rep. 2016, 35, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.-B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2015, 33, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Weide, R.; Govers, F.; Bouwmeester, K. L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J. Exp. Bot. 2015, 66, 6731–6743. [Google Scholar] [CrossRef] [PubMed]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojärvi, J.; Rayapuram, C.; Idänheimo, N.; Hunter, K.; Kimura, S.; Merilo, E.; et al. On behalf of the CRK Consortium. Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress. PLoS Genet. 2015, 11, e1005373. [Google Scholar] [CrossRef] [PubMed]
- Pfeilmeier, S.; George, J.; Morel, A.; Roy, S.; Smoker, M.; Stansfeld, L.; Downie, A.; Peeters, N.; Malone, J.; Zipfel, C. Heterologous expression of the immune receptor EFR in Medicago truncatula reduces pathogenic infection, but not rhizobial symbiosis. bioRxiv 2017. [Google Scholar] [CrossRef]
- Kim, S.Y.; Shang, Y.; Joo, S.-H.; Kim, S.-K.; Nam, K.H. Overexpression of BAK1 causes salicylic acid accumulation and deregulation of cell death control genes. Biochem. Biophys. Res. Commun. 2017, 484, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Wyrsch, I.; Strutt, J.; Wimalasekera, R.; Webb, A.; Boller, T.; Robatzek, S. Expression patterns of FLAGELLIN SENSING 2 map to bacterial entry sites in plant shoots and roots. J. Exp. Bot. 2014, 65, 6487–6498. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, C.; Robatzek, S. Plants and pathogens: Putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 2012, 15, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Brunner, F.D.R.; Nürnberger, T. Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 2010, 21, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Trdá, L.; Fernandez, O.; Boutrot, F.; Héloir, M.-C.; Kelloniemi, J.; Daire, X.; Adrian, M.; Clément, C.; Zipfel, C.; Dorey, S.; et al. The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 2013, 201, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Febres, V.J.; Jones, J.B.; Moore, G.A. A survey of FLS2 genes from multiple citrus species identifies candidates for enhancing disease resistance to Xanthomonas citri ssp. citri. Hortic. Res. 2016, 3, 80. [Google Scholar] [CrossRef] [PubMed]
- Helft, L.; Thompson, M.; Bent, A.F. Directed Evolution of FLS2 towards Novel Flagellin Peptide Recognition. PLoS ONE 2016, 11, e0157155. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H.; et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Kouzai, Y.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 2010, 64, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Kouzai, Y.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. Enhancement of MAMP signaling by chimeric receptors improves disease resistance in plants. Plant Signal. Behav. 2011, 6, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Han, M.; Blanco-Portales, R.; Song, W.; Weide, R.; Guo, L.-Y.; van der Vossen, E.A.G.; Govers, F. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol. J. 2014, 12, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Pontiggia, D.; Raggi, S.; Cheng, Z.; Scaloni, F.; Ferrari, S.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2015, 112, 5533–5538. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranf, S. Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops. Agronomy 2018, 8, 134. https://doi.org/10.3390/agronomy8080134
Ranf S. Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops. Agronomy. 2018; 8(8):134. https://doi.org/10.3390/agronomy8080134
Chicago/Turabian StyleRanf, Stefanie. 2018. "Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops" Agronomy 8, no. 8: 134. https://doi.org/10.3390/agronomy8080134
APA StyleRanf, S. (2018). Pattern Recognition Receptors—Versatile Genetic Tools for Engineering Broad-Spectrum Disease Resistance in Crops. Agronomy, 8(8), 134. https://doi.org/10.3390/agronomy8080134




