Towards the Understanding of Important Coconut Endosperm Phenotypes: Is there an Epigenetic Control?
Abstract
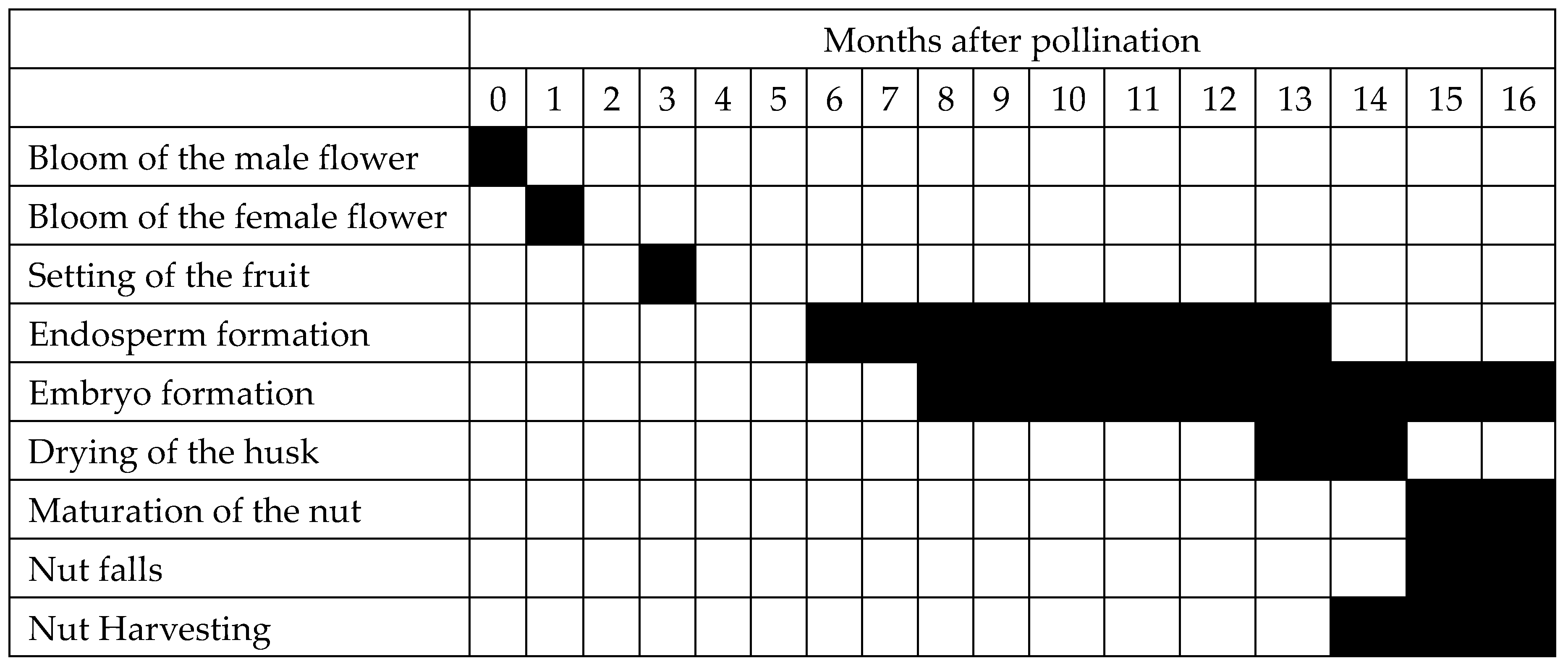
1. The Coconut Endosperm, Its Development and Fatty Acid Composition
2. The Makapuno and Lono Endosperm Phenotypes
2.1. Makapuno
2.2. Lono
3. Molecular Characterization of the Coconut
4. Plant Epigenetics and Endosperm Development
5. Plant Tissue Culture and Epigenetics
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Child, R. Coconuts, 2nd ed.; Longman Group Ltd.: London, UK, 1974; ISBN 0-582-46675-X. [Google Scholar]
- Sugimura, Y.; Itano, M.; Salud, C.D.; Otsuji, K.; Yamaguchi, H. Biometric analysis on diversity of coconut palm: Cultivar classification by botanical and agronomical traits. Euphytica 1997, 98, 29–35. [Google Scholar] [CrossRef]
- Naresh Kumar, S.; Balakrishna, A. Seasonal variations in fatty acid composition of oil in developing coconut. J. Food Qual. 2009, 32, 158–176. [Google Scholar] [CrossRef]
- Gunn, B.F.; Baudouin, L.; Olsen, K.M. Independent origins of cultivated coconut (Cocos nucifera L.) in the Old World tropics. PLoS ONE 2011, 6, e21143. [Google Scholar] [CrossRef] [PubMed]
- Teh, H.F.; Neoh, B.K.; Ithnin, N.; Daim, L.D.J.; Ooi, T.E.K.; Appleton, D.R. Review: Omics and strategic yield improvement in oil crops. JAOCS 2017, 94, 1225–1244. [Google Scholar] [CrossRef]
- Tammes, P. Review of coconut selection in India. Euphytica 1955, 4, 17–24. [Google Scholar] [CrossRef]
- Menon, K.P.; Pandalai, K.M. The Coconut Palm, A Monograph; Indian Central Coconut Committee: Kerala, India, 1958. [Google Scholar]
- Hulme, P.; Benkman, C. Granivory. In Plant-Animal Interactions: An Evolutionary Approach; Blackwell Science Ltd.: Oxford, UK, 2002; pp. 132–156. ISBN 978-0-632-05267-7. [Google Scholar]
- Huang, Y.-Y.; Lee, C.-P.; Fu, J.L.; Chang, B.C.-H.; Matzke, A.J.M.; Matzke, M. De novo transcriptome sequence assembly from coconut leaves and seeds with a focus on factors involved in RNA-directed DNA methylation. G3 2014, 4, 2147. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.B.C.; Sousa, C.N.S.; Meneses, L.N.; Ximenes, N.C.; Santos Junior, M.A.; Vasconcelos, G.S.; Lima, N.B.C.; Patrocinio, M.C.A.; Macedo, D.; Vasconcelos, S.M.M. Cocos nucifera (L.) (Arecaceae): A phytochemical and pharmacological review. Braz. J. Med. Biol. Res. 2015, 48, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Kappally, S.; Shirwaikar, A.; Shirwaikar, A. Coconut oil—A review of potential applications. Hygeia J. Drugs Med. 2015. [Google Scholar] [CrossRef]
- Tangwatcharin, P.; Khopaibool, P. Activity of virgin coconut oil, lauric acid or monolaurin in combination with lactic acid against Staphylococcus aureus. Southeast Asian J. Trop. Med. Public Health 2012, 43, 969–985. [Google Scholar] [PubMed]
- Arlee, R.; Suanphairoch, S.; Pakdeechanuan, P. Differences in chemical components and antioxidant-related substances in virgin coconut oil from coconut hybrids and their parents. Int. Food Res. J. 2013, 20, 2103–2109. [Google Scholar]
- Laureles, L.R.; Rodriguez, F.M.; Reaño, C.E.; Santos, G.A.; Laurena, A.C.; Mendoza, E.M.T. Variability in fatty acid and triacylglycerol composition of the oil of coconut (Cocos nucifera L.) hybrids and their parentals. J. Agric. Food Chem. 2002, 50, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Naresh Kumar, S. Copra, oil and fatty acid database. In Variability in Content and Composition of Fatty Acids in Coconut Oil Due to Genetic and Environmental Factors; ICAR: New Delhi, India, 2005. [Google Scholar]
- Naresh Kumar, S. Capillary gas chromatography method for fatty acid analysis of coconut oil. J. Plant. Crops 2007, 35, 23–27. [Google Scholar]
- Kumar, S.N. Variability in coconut (Cocos nucifera L.) germplasm and hybrids for fatty acid profile of oil. J. Agric. Food Chem. 2011, 59, 13050–13058. [Google Scholar] [CrossRef] [PubMed]
- Zambiazi, R.; Przybylski, R.; Zambiazi, M.; Mendonca, C. Fatty acid composition of vegetable oils and fats. Boletim Centro de Pesquisa de Processamento de Alimentos 2007, 25, 111–120. [Google Scholar]
- Kostik, V.; Memeti, S.; Bauer, B. Fatty acid composition of edible oils and fats. JHED 2013, 4, 112–116. [Google Scholar] [CrossRef]
- Dorni, C.; Sharma, P.; Saikia, G.; Longvah, T. Fatty acid profile of edible oils and fats consumed in India. Food Chem. 2018, 238, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Juliano, J.B. Origin, development and nature of the stony layer of the coconut (Cocos nucifera). Phil. J. Sci. 1926, 30, 187–200. [Google Scholar]
- Padua-Resurreccion, A.N.; Banzon, J.A. Fatty acid composition of the oil from progressively maturing bunches of coconut. Philipp. J. Coconut Stud. 1979, 4, 1–16. [Google Scholar]
- Oo, K.C.; Stumpf, P.K. Fatty acid biosynthesis in the developing endosperm of Cocos nucifera. Lipids 1979, 14, 132–143. [Google Scholar] [CrossRef]
- Nevin, K.G.; Rajamohan, T. Effect of topical application of virgin coconut oil on skin components and antioxidant status during dermal wound healing in young rats. Skin Pharmacol. Physiol. 2010, 23, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Intahphuak, S.; Khonsung, P.; Panthong, A. Anti-inflammatory, analgesic, and antipyretic activities of virgin coconut oil. Pharm. Biol. 2010, 48, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Hayatullina, Z.; Muhammad, N.; Mohamed, N.; Soelaiman, I.-N. Virgin coconut oil supplementation prevents bone loss in osteoporosis rat model. Evid.-Based Complement. Altern. Med. ECAM 2012, 2012, 237236. [Google Scholar] [CrossRef] [PubMed]
- Nurul-Iman, B.; Kamisah, Y.; Jaarin, K.; Qodriyah, H. Virgin coconut oil prevents blood pressure elevation and improves endothelial functions in rats fed with repeatedly heated palm oil. Evid. Based Complement. Altern. Med. 2013, 5, 629329. [Google Scholar] [CrossRef] [PubMed]
- Nevin, K.G.; Rajamohan, T. Virgin coconut oil supplemented diet increases the antioxidant status in rats. Food Chem. 2006, 99, 260–266. [Google Scholar] [CrossRef]
- Zakaria, Z.A.; Rofiee, M.S.; Somchit, M.N.; Zuraini, A.; Sulaiman, M.R.; Teh, L.K.; Salleh, M.Z.; Long, K. Hepatoprotective activity of dried- and fermented-processed virgin coconut oil. Evid. Based Complement. Altern. Med. 2011, 2011, 142739. [Google Scholar] [CrossRef] [PubMed]
- Arunima, S.; Rajamohan, T. Effect of virgin coconut oil enriched diet on the antioxidant status and paraoxonase 1 activity in ameliorating the oxidative stress in rats—A comparative study. Food Funct. 2013, 4, 1402–1409. [Google Scholar] [CrossRef] [PubMed]
- Yeap, S.K.; Beh, B.K.; Ali, N.M.; Yusof, H.M.; Ho, W.Y.; Koh, S.P.; Alitheen, N.B.; Long, K. Antistress and antioxidant effects of virgin coconut oil in vivo. Exp. Ther. Med. 2015, 9, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Thoenes, P. Recent trends and medium-term prospects in the global vegetable oil market. In Proceedings of the 10th International Conference: Fat and Oil Industry, Kiev, Ukraine, 23–25 November 2011. [Google Scholar]
- Balachandran, C.; Arumughan, C.; Mathew, A.G. Distribution of major chemical constituents and fatty acids in different regions of coconut endosperm. JAOCS 1985, 62, 1583–1586. [Google Scholar] [CrossRef]
- Kartha, A.R.S.; Narayanan, R. Development of oil in the ripening coconut. Ind. J. Agric. Sci. 1956, 26, 319–327. [Google Scholar]
- Nathanael, W.R.N. Changes in the coconut endosperm during development. Ceylon Coconut Quart. 1959, 10, 35–36. [Google Scholar]
- Balleza, C.F.; Sierra, Z.N. Proximate analysis of the coconut endosperm in progressive stages of development. Philipp. J. Coconut Stud. 1972, 16, 37–43. [Google Scholar]
- Huang, A.H.C. Oil bodies and oleosins in seeds. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1992, 43, 177–200. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Dussert, S.; Guerin, C.; Andersson, M.; Joët, T.; Tranbarger, T.J.; Pizot, M.; Sarah, G.; Omore, A.; Durand-Gasselin, T.; Morcillo, F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 2013, 162, 1337–1358. [Google Scholar] [CrossRef] [PubMed]
- Copeland, E.B. The Coconut; Macmillian and Company, Ltd.: Basingstoke, UK, 1931. [Google Scholar]
- Gonzales, B. The Makapuno coconut. Philipp. Agric. For. 1914, 3, 31–32. [Google Scholar]
- Ramirez, D.A.; Mendoza, E.M.T. The Makapuno Coconut; The National Academy of Science and Technology: Taguig, Philippines, 1998. [Google Scholar]
- De Guzman, E.V.; del Rosario, D.A. The growth and development of Cocos nucifera L. ‘Makapuno’ embryo In-Vitro. Philipp. Agric. 1964, 48, 82–94. [Google Scholar]
- De Guzman, E.V.; Manuel, G.C. Improved root growth in embryo and seedlings culture of coconut ‘Makapuno’ by the incorporation of charcoal in the growth medium. PJCS 1977, 11, 35–39. [Google Scholar]
- Rillo, E.P. Coconut embryo culture. In Proceedings of the International Coconut Biotechnology Symposium, CICY, Merida, Mexico, 1–5 December 1997. [Google Scholar]
- Rillo, E.P. Coconut embryo culture. In Current Advances in Coconut Biotechnology; Oropeza, C., Verdeil, J.L., Ashburner, G.R., Cardeña, R., Santamaría, J.M., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 279–288. ISBN 978-94-015-9283-3. [Google Scholar]
- Islam, M.N.; Azad, A.K.; Namuco, L.O.; Borromeo, T.; Cedo, M.L.O.; Aguilar, E.A. Morphometric characterization and diversity analysis of a Makapuno coconut population in U.P. Los Banos. Pak. J. Agric. Res. 2013, 26, 254–264. [Google Scholar]
- Torres, J. Some notes on Makapuno coconut and its inheritance. Philipp. J. Agric. 1937, 8, 27–37. [Google Scholar]
- Zuniga, L.C. The probable inheritance of the Makapuno character of the coconut. Philipp. Agric. 1953, 36, 402–413. [Google Scholar]
- Cedo, M.L.O.; de Guzman, E.V.; Rimando, T.J. Controlled pollination of embryo-culture “Makapuno” coconut. Philipp. Agric. 1984, 67, 100–104. [Google Scholar]
- Maskromo, I.; Larekeng, S.; Novarianto, H.; Sudarsono, S. Xenia negatively affecting kopyor nut yield in Kalianda Tall kopyor and Pati Dwarf kopyor coconuts. EJFA 2017, 28. [Google Scholar] [CrossRef]
- Satyabalan, K. A note on the occurrence of “buttery” kernel, in coconut. Indian Coconut J. 1953, 6, 152–154. [Google Scholar]
- Adriano, F.T.; Manahan, M. The nutritive value of green, ripe and sport coconuts (buko, niyog and Makapuno). Philipp. Agric. 1931, 20, 195–198. [Google Scholar]
- Deva Kumar, K.; Gautam, R.K.; Ahmad, I.; Dam Roy, S.; Sharma, A. Biochemical, genetic and molecular basis of the novel and commercially important soft endosperm Makapuno coconut—A review. JFAE 2015, 13, 61–65. [Google Scholar]
- Samonte, J.L.; Ramirez, D.A.; Mendoza, E.M.T. Galactomannan in developing normal and Makapuno coconut endosperm. Bull. Philipp. Biochem. Soc. 1987, 7, 15–19. [Google Scholar]
- Mendoza, E.M.T.; Mujer, C.V.; Rodriguez, F.M.; Ramirez, D.A. Ontogeny of some chemical constituents in developing Makapuno and normal coconut endosperms. Kalikasan Philipp. J. Biol. 1982, 11, 293–301. [Google Scholar]
- De la Cruz, R.Y.; Laude, R.P.; Diaz, M.G.Q.; Laurena, A.C.; Mendioro, M.S.; Mendoza, E.M.T. Polymerase chain reaction (PCR)-based cloning of partial cDNAs of selected genes in normal and mutant “Makapuno” endosperms of coconut (Cocos nucifera L.). Philipp. Agric. Sci. 2013, 96, 60–71. [Google Scholar]
- Mujer, C.V.; Arambulo, A.S.; Mendoza, E.M.T.; Ramirez, D.A. The viscous components of the mutant (Makapuno) endoosperm I. Isolation and characterization. Kalikasan Philipp. J. Biol. 1983, 12, 42–50. [Google Scholar]
- Balasubramaniam, K. Polysaccharides of the kernel of maturing and matured coconuts. J. Food Sci. 1976, 41, 1370–1373. [Google Scholar] [CrossRef]
- Mujer, C.V.; Ramirez, D.A.; Mendoza, E.M.T. α-d-galactosidase deficiency in coconut endosperm: Its possible pleiotropic effects in makapuno. Phytochemistry 1984, 23, 893–894. [Google Scholar] [CrossRef]
- Mujer, C.V.; Ramirez, D.A.; Mendoza, E.M.T. Coconut α-d-galactosidase isoenzymes: Isolation, purification and characterization. Phytochemistry 1984, 23, 1251–1254. [Google Scholar] [CrossRef]
- Samonte, J.L.; Mendoza, E.M.T.; Ilag, L.L.; De La Cruz, N.B.; Ramirez, D.A. Galactomannan degrading enzymes in maturing normal and makapuno and germinating normal coconut endosperm. Phytochemistry 1989, 28, 2269–2273. [Google Scholar] [CrossRef]
- Diaz, M.G.Q. Differential Gene Expression of the Enzymes of Galactomannan Degradation in the Developing Endosperms of Normal and “Makapuno” Coconut (Cocos nucifera Linn.). Ph.D. Thesis, University of the Philippines Los Banos, Los Banos, Laguna, Philippines, 2002. [Google Scholar]
- De la Cruz, R.Y.; Bugayong, V.J.D. Mutations in the alpha-D-galactosidase gene suggest molecular basis of the mutant “Makapuno” coconut (Cocos nucifera L.) phenomenon. Philipp. Agric. Sci. 2016, 99, 321–325. [Google Scholar]
- Carpio, C.B.; Santos, G.A.; Emmanuel, E.E.; Novarianto, H. Research on coconut genetic resources in Southeast and East Asia. In Coconut Genetic Resources; International Plant Genetic Resources Institute—Regional Office for Asia, the Pacific and Oceania (IPGRI-APO): Serdang, Malaysia, 2005; pp. 533–545. ISBN 92-9043-629-8. [Google Scholar]
- Niral, V.; Samsudeen, K.; Nair, R.V. Genetic resources of coconut. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute, Kasaragod, Kerala, India, 25–28 October 2010; Thomas, G.V., Krishnakumar, V., Augustine Jerard, B.A., Eds.; pp. 22–28. [Google Scholar]
- Padolina, W.G. Contribution of biological research to the development of the coconut industry. JAOCS 1985, 62, 206–210. [Google Scholar] [CrossRef]
- Central Plantation Crops Research Institute Kasaragod (CPCRI). Annual Report 2003–2004; CPCRI: Kerala, India, 2004; p. 152.
- Niral, V.; Devakumar, K.; Umamaheswari, T.S.; Naganeeswaran, S.; Nair, R.V.; Jerard, B.A. Morphological and molecular characterization of a large fruited unique coconut accession from Vaibhavwadi, Maharashtra, India. Indian J. Genet. 2013, 73, 220–224. [Google Scholar] [CrossRef]
- Devakumar, K.; Jerard, B.A.; Dhanapal, R.; Damodaran, V.; Niral, V.; Naganeeswaran, S. Genetic relatedness of coconut populations from Andaman and Nicobar Islands traced to the Indian sub-continent using microsatellite markers. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute, Kasaragod, Kerala, India, 25–28 October 2010; Thomas, G.V., Krishnakumar, V., Augustine Jerard, B.A., Eds.; pp. 67–75. [Google Scholar]
- Rognon, F. Biologie florale du cocotier, Duree et succession des phases males et femelles chez divers types de cocotiers. Oleagineux 1976, 31, 13–18. [Google Scholar]
- Regi, T.; Josephrajkumar, A. Flowering and pollination biology in coconut. J. Plant. Crops 2013, 41, 109–117. [Google Scholar]
- Ratnambal, M.J.; Nair, M.K.; Muralidharan, K.; Kumaran, P.M.; Rao, E.V.V.B.; Pillai, R.V. Coconut Descriptors: Part I; Central Plantation Crops Research Institute Kasaragod: Kerala, India, 1995. [Google Scholar]
- Carpio, C.; Manohar, E.; Rillo, E.P.; Cueto, C.; Orense, O.; Areza-Ubaldo, M.B.; Alfiler, A.R. Status of coconut genetic resources research in the Philippines. In Coconut Genetic Resources; International Plant Genetic Resources Institute—Regional Office for Asia, the Pacific and Oceania (IPGRI-APO): Serdang, Malaysia, 2005; pp. 630–647. ISBN 92-9043-629-8. [Google Scholar]
- Rajagopai, V.; Kumaran, P.M.; Arulraj, S.; Arunachalam, V. Research on coconut genetic resources in Southeast and East Asia. In Coconut Genetic Resources; International Plant Genetic Resources Institute—Regional Office for Asia, the Pacific and Oceania (IPGRI-APO): Serdang, Malaysia, 2005; pp. 573–580. ISBN 92-9043-629-8. [Google Scholar]
- Niral, V.; Jerard, B.A.; Samsudeen, K.; Patil, P.D.; Ananda, K.S.; Nair, R.V.; Thomas, G.V. International coconut gene bank for South Asia—Conservation and Characterization. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute, Kasaragod, Kerala, India, 25–28 October 2010; Thomas, G.V., Krishnakumar, V., Augustine Jerard, B.A., Eds.; pp. 35–40. [Google Scholar]
- Thomas, G.V.; Niral, V.; Jerard, B.A. Current status of coconut diversity conservation and utilization in India. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute Kasaragod, Kerala, India, 25–28 October 2010; Central Plantation Crops Research Institute: Kerala, India, 2010; pp. 10–21. [Google Scholar]
- Aala, W.F.J.; Custodio, C.J.C.; Lado, J.P.; Berdos, M.L.G.; Cueto, C.A.; Diaz, M.G.Q. Oil body enlargement and not number is responsible for increased oil content in solid endosperm of Philippine Lono tall coconut (Cocos nucifera L.). In Proceedings of the 25th International Conference and 43rd PSBMB Annual Convention, Manila, Philippines, 5–7 December 2016; p. 152. [Google Scholar]
- Castillo, E.M. Chemical Characterization of Makapuno and Normal Coconut Endosperms. Ph.D. Thesis, University of the Philippines Los Banos, Los Banos, Laguna, Philippines, 1988. [Google Scholar]
- Bagde, A.S.; Patil, P.D.; Pashte, V.V. Screening of coconut genotypes against coconut eriophyid mite (Aceria guerreronis Keifer.). IJBSM 2016, 7, 18–23. [Google Scholar] [CrossRef]
- Gurav, S.S.; Desai, V.S.; Nagwekar, D.D.; Narangakal, A.L. Reaction of different coconut genotypes against eriophyrid mite, Aceria guerreronis Keifer in Konkan region of Maharashta. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute, Kasaragod, Kerala, India, 25–28 October 2010; Thomas, G.V., Krishnakumar, V., Augustine Jerard, B.A., Eds.; pp. 375–377. [Google Scholar]
- Jacob, P.M.; Thomas, R.J.; Nair, R.V. Achievements of breeding for resistance to root (wilt) disease in coconut. In Proceedings of the International Conference on Coconut Biodiversity for Prosperity, Central Plantation Crops Research Institute, Kasaragod, Kerala, India, 25–28 October 2010; Thomas, G.V., Krishnakumar, V., Augustine Jerard, B.A., Eds.; pp. 41–48. [Google Scholar]
- Jerard, B.A.; Damodaran, V.; Niral, V.; Samsudeen, K.; Rajesh, M.K.; Sankaran, M. Conservation and utilization of soft endosperm coconut accession from Andaman Islands. J. Plant. Crops 2013, 41, 14–21. [Google Scholar] [CrossRef]
- Xiao, Y.; Xu, P.; Fan, H.; Baudouin, L.; Xia, W.; Bocs, S.; Xu, J.; Li, Q.; Guo, A.; Zhou, L.; et al. The genome draft of coconut (Cocos nucifera). Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lantican, D.; Strickler, S.; Canama, A.; Gardoce, R.; Mueller, L.; Galvez, H. The coconut genome: Providing a reference sequence towards coconut varietal improvement. In Proceedings of the Plant and Animal Genome XXVI Conference, San Diego, CA, USA, 13–17 January 2018. [Google Scholar]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.; Chan, K.-L.; Halim, M.A.; et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335. [Google Scholar] [CrossRef] [PubMed]
- Al-Dous, E.K.; George, B.; Al-Mahmoud, M.E.; Al-Jaber, M.Y.; Wang, H.; Salameh, Y.M.; Al-Azwani, E.K.; Chaluvadi, S.; Pontaroli, A.C.; DeBarry, J.; et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 2011, 29, 521. [Google Scholar] [CrossRef] [PubMed]
- Al-Mssallem, I.S.; Hu, S.; Zhang, X.; Lin, Q.; Liu, W.; Tan, J.; Yu, X.; Liu, J.; Pan, L.; Zhang, T.; et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013, 4, 2274. [Google Scholar] [CrossRef] [PubMed]
- Masoudi-Nejad, A.; Narimani, Z.; Hosseinkhan, N. De novo assembly algorithms. In Next Generation Sequencing and Sequence Assembly: Methodologies and Algorithms; Springer Briefs in Systems Biology; Springer: New York, NY, USA, 2013; Volume 4, p. 86. ISBN 978-1-4614-7725-9. [Google Scholar]
- Huang, Y.-Y.; Matzke, A.J.M.; Matzke, M. Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera). PLoS ONE 2013, 8, e74736. [Google Scholar] [CrossRef] [PubMed]
- Aljohi, H.A.; Liu, W.; Lin, Q.; Zhao, Y.; Zeng, J.; Alamer, A.; Alanazi, I.O.; Alawad, A.O.; Al-Sadi, A.M.; Hu, S.; et al. Complete sequence and analysis of coconut palm (Cocos nucifera) mitochondrial genome. PLoS ONE 2016, 11, e0163990. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Xiao, Y.; Yang, Y.; Xia, W.; Mason, A.S.; Xia, Z.; Qiao, F.; Zhao, S.; Tang, H. RNA-Seq analysis of Cocos nucifera: Transcriptome sequencing and de novo assembly for subsequent functional genomics approaches. PLoS ONE 2013, 8, e59997. [Google Scholar] [CrossRef] [PubMed]
- Saensuk, C.; Wanchana, S.; Choowongkomon, K.; Wongpornchai, S.; Kraithong, T.; Imsabai, W.; Chaichoompu, E.; Ruanjaichon, V.; Toojinda, T.; Vanavichit, A.; et al. De novo transcriptome assembly and identification of the gene conferring a “pandan-like” aroma in coconut (Cocos nucifera L.). Plant Sci. 2016, 252, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, M.K.; Fayas, T.P.; Naganeeswaran, S.; Rachana, K.E.; Bhavyashree, U.; Sajini, K.K.; Karun, A. De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing. Protoplasma 2016, 253, 913–928. [Google Scholar] [CrossRef] [PubMed]
- Armero, A.; Baudouin, L.; Bocs, S.; This, D. Improving transcriptome de novo assembly by using a reference genome of a related species: Translational genomics from oil palm to coconut. PLoS ONE 2017, 12, e0173300. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Sircar, G.; Pandey, N.; Gupta Bhattacharya, S. Mining novel allergens from coconut pollen employing manual de novo sequencing and homology-driven proteomics. J. Proteome Res. 2015, 14, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Ong-Abdullah, M.; Ordway, J.M.; Jiang, N.; Ooi, S.-E.; Kok, S.-Y.; Sarpan, N.; Azimi, N.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 2015, 525, 533. [Google Scholar] [CrossRef] [PubMed]
- Alwee, S.S.; Van der Linden, C.G.; Van der Schoot, J.; de Folter, S.; Angenent, G.C.; Cheah, S.-C.; Smulders, M.J.M. Characterization of oil palm MADS box genes in relation to the mantled flower abnormality. Plant Cell Tissue Organ Cult. 2006, 85, 331–344. [Google Scholar] [CrossRef]
- Jaligot, E.; Beulé, T.; Rival, A. Methylation-sensitive RFLPs: Characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor. Appl. Genet. 2002, 104, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Matthes, M.; Singh, R.; Cheah, S.-C.; Karp, A. Variation in oil palm (Elaeis guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor. Appl. Genet. 2001, 102, 971–979. [Google Scholar] [CrossRef]
- Jaligot, E.; Rival, A.; Beulé, T.; Dussert, S.; Verdeil, J.-L. Somaclonal variation in oil palm (Elaeis guineensis Jacq.): The DNA methylation hypothesis. Plant Cell Rep. 2000, 19, 684–690. [Google Scholar] [CrossRef]
- Corley, R.H.V.; Lee, C.H.; Law, L.H.; Wong, C.Y. Abnormal flower development in oil palm clones. Plant. (Kuala Lumpur) 1986, 62, 233–240. [Google Scholar]
- Rival, A.; Jaligot, E.; Beulé, T.; Tregear, J.W.; Finnegan, J. The oil palm “mantled” somaclonal variation: A model for epigenetic studies in higher plants. In Acta Horticulturae; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2009; pp. 177–182. [Google Scholar]
- Eeuwens, C.J.; Lord, S.; Donough, C.R.; Rao, V.; Vallejo, G.; Nelson, S. Effects of tissue culture conditions during embryoid multiplication on the incidence of “mantled” flowering in clonally propagated oil palm. Plant Cell Tissue Organ Cult. 2002, 70, 311–323. [Google Scholar] [CrossRef]
- Triantaphyllopoulos, K.A.; Ikonomopoulos, I.; Bannister, A.J. Epigenetics and inheritance of phenotype variation in livestock. Epigenet. Chromatin 2016, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Springer, N.M.; Schmitz, R.J. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 2017, 18, 563. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jacobsen, S.E.; Reik, W. Epigenetic reprogramming in plant and animal development. Science 2010, 330, 622. [Google Scholar] [CrossRef] [PubMed]
- Maleszka, R.; Mason, P.H.; Barron, A.B. Epigenomics and the concept of degeneracy in biological systems. Brief. Funct. Genomics 2014, 13, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Zhao, X. Epigenetic marks: Regulators of livestock phenotypes and conceivable sources of missing variation in livestock improvement programs. Front. Genet. 2015, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.R.; Shah, S.M.; Irshad, M. Immediate and transgenerational regulation of plant stress response through DNA methylation. J. Agric. Sci. 2015, 7, 144–151. [Google Scholar] [CrossRef]
- Alonso, C.; Pérez, R.; Bazaga, P.; Herrera, C.M. Global DNA cytosine methylation as an evolving trait: Phylogenetic signal and correlated evolution with genome size in angiosperms. Front. Genet. 2015, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Lauria, M.; Rossi, V. Epigenetic control of gene regulation in plants. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2011, 1809, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Kimatu, J.N.; Xu, K.; Liu, B. DNA cytosine methylation in plant development. J. Genet. Genomics 2010, 37, 1–12. [Google Scholar] [CrossRef]
- Langevin Scott, M.; Kelsey Karl, T. The fate is not always written in the genes: Epigenomics in epidemiologic studies. Environ. Mol. Mutagen. 2013, 54, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Naveh-Many, T.; Cedar, H.; Razin, A. Sequence specificity of methylation in higher plant DNA. Nature 1981, 292, 860. [Google Scholar] [CrossRef] [PubMed]
- King, G.J.; Amoah, S.; Kurup, S. Exploring and exploiting epigenetic variation in crops. Genome 2010, 53, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Greenberg, M.V.C.; Feng, S.; Bernatavichute, Y.V.; Jacobsen, S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013, 152, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Custard, K.D.; Brown, R.C.; Lemmon, B.E.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 2006, 18, 805. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Comai, L. A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 1998, 149, 307. [Google Scholar] [PubMed]
- Bender, J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004, 55, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Gouil, Q.; Baulcombe, D.C. DNA Methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016, 12, e1006526. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.E.; Bewick, A.J.; Ji, L.; Alabady, M.S.; Kim, K.D.; Li, Q.; Rohr, N.A.; Rambani, A.; Burke, J.M.; Udall, J.A.; et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.-L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, M.W.; Tanurdžić, M.; Lippman, Z.; Jiang, H.; Carrasquillo, R.; Rabinowicz, P.D.; Dedhia, N.; McCombie, W.R.; Agier, N.; Bulski, A.; et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007, 5, e174. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.; Poole, M.; Manning, K.; King, G.J. Genetics and epigenetics of fruit development and ripening. Curr. Opin. Plant Biol. 2008, 11, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009, 5, e1000530. [Google Scholar] [CrossRef] [PubMed]
- Stam, M.; Belele, C.; Dorweiler, J.E.; Chandler, V.L. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002, 16, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, N.; Xu, C.; Zhong, S.; Lin, X.; Yang, J.; Zhou, T.; Yuliang, A.; Wu, Y.; Chen, Y.-R.; et al. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. PNAS 2014, 111, 10642. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L. Paramutation: From maize to mice. Cell 2007, 128, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Missirian, V.; Henikoff, S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 2011, 6, e23687. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Han, S.S.; Wang, Y.; Zhang, X.Z.; Han, Z.H. Variations in leaf morphology and DNA methylation following in vitro culture of Malus xiaojinensis. Plant Cell Tissue Organ Cult. 2012, 111, 153–161. [Google Scholar] [CrossRef]
- Schwabe, W.W.; Blake, J.; Fulford, R.M. Application of Tissue Culture Techniques for Vegetative Propagation; FAO Plant News Special Issue (AGP:NL/13); Plant Production and Protection Division, Food and Agriculture Organization: Rome, Italy, 1970; pp. 34–35. [Google Scholar]
- Berdasco, M.; Alcázar, R.; García-Ortiz, M.V.; Ballestar, E.; Fernández, A.F.; Roldán-Arjona, T.; Tiburcio, A.F.; Altabella, T.; Buisine, N.; Quesneville, H.; et al. Promoter DNA hypermethylation and gene repression in undifferentiated Arabidopsis cells. PLoS ONE 2008, 3, e3306. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, C.; Nic-Can, G.; Ojeda, G.; Herrera-Herrera, J.L.; López-Torres, A.; Wrobel, K.; Robert-Díaz, M.L. KNOX1 is expressed and epigenetically regulated during in vitro conditions in Agave spp. BMC Plant Biol. 2012, 12, 203. [Google Scholar] [CrossRef] [PubMed]
- Moricova, P.; Ondrej, V.; Navratilova, B.; Luhova, L. Changes of DNA methylation and hydroxymethylation in plant protoplast cultures. Acta Biochim. Pol. 2013, 60, 33–36. [Google Scholar] [PubMed]
- Valledor, L.; Hasbún, R.; Meijón, M.; Rodríguez, J.L.; Santamaría, E.; Viejo, M.; Berdasco, M.; Feito, I.; Fraga, M.F.; Cañal, M.J.; Rodríguez, R. Involvement of DNA methylation in tree development and micropropagation. Plant Cell Tissue Organ Cult. 2007, 91, 75–86. [Google Scholar] [CrossRef]
- Zakrzewski, F.; Schmidt, M.; Van Lijsebettens, M.; Schmidt, T. DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.). Plant J. 2017, 90, 1156–1175. [Google Scholar] [CrossRef] [PubMed]
- Rival, A.; Jaligot, E.; Beulé, T.; Finnegan, E.J. Isolation and expression analysis of genes encoding MET, CMT, and DRM methyltransferases in oil palm (Elaeis guineensis Jacq.) in relation to the ‘mantled’ somaclonal variation. J. Exp. Bot. 2008, 59, 3271–3281. [Google Scholar] [CrossRef] [PubMed]
- LoSchiavo, F.; Pitto, L.; Giuliano, G.; Torti, G.; Nuti-Ronchi, V.; Marazziti, D.; Vergara, R.; Orselli, S.; Terzi, M. DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones and hypomethylating drugs. Theor. Appl. Genet. 1989, 77, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Smulders, M.J.M.; de Klerk, G.J. Epigenetics in plant tissue culture. Plant Growth Regul. 2011, 63, 137–146. [Google Scholar] [CrossRef]
- Shimron-Abarbanell, D.; Breiman, A. Comprehensive molecular characterization of tissue-culture-derived Hordeum marinum plants. Theor. Appl. Genet. 1991, 83, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Devaux, P.; Kilian, A.; Kleinhofs, A. Anther culture and Hordeum bulbosum-derived barley doubled haploids mutations and methylation. Mol. Gen. Genet. 1993, 241, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Pila Quinga, L.A.; Pacheco de Freitas Fraga, H.; do Nascimento Vieira, L.; Guerra, M.P. Epigenetics of long-term somatic embryogenesis in Theobroma cacao L.: DNA methylation and recovery of embryogenic potential. Plant Cell Tissue Organ Cult. 2017, 131, 295–305. [Google Scholar] [CrossRef]
- Jaligot, E.; Adler, S.; Debladis, É.; Beulé, T.; Richaud, F.; Ilbert, P.; Finnegan, E.J.; Rival, A. Epigenetic imbalance and the floral developmental abnormality of the in vitro-regenerated oil palm Elaeis guineensis. Ann. Bot. 2011, 108, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Aké, F.; Castillo-Castro, E.; Pool, F.B.; Espadas, F.; Santamaría, J.M.; Robert, M.L.; De-la-Peña, C. Physiological differences and changes in global DNA methylation levels in Agave angustifolia Haw. albino variant somaclones during the micropropagation process. Plant Cell Rep. 2016, 35, 2489–2502. [Google Scholar] [CrossRef] [PubMed]
- Rival, A.; Ilbert, P.; Labeyrie, A.; Torres, E.; Doulbeau, S.; Personne, A.; Dussert, S.; Beulé, T.; Durand-Gasselin, T.; Tregear, J.W.; Jaligot, E. Variations in genomic DNA methylation during the long-term in vitro proliferation of oil palm embryogenic suspension cultures. Plant Cell Rep. 2013, 32, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Peredo, E.L.; Ángeles Revilla, M.; Arroyo-García, R. Assessment of genetic and epigenetic variation in hop plants regenerated from sequential subcultures of organogenic calli. J. Plant Physiol. 2006, 163, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Rhee, Y.; Sekhon, R.S.; Chopra, S.; Kaeppler, S. Tissue culture-induced novel epialleles of a Myb transcription factor encoded by pericarp color1 in maize. Genetics 2010, 186, 843. [Google Scholar] [CrossRef] [PubMed]
- Adam, H.; Jouannic, S.; Orieux, Y.; Morcillo, F.; Richaud, F.; Duval, Y.; Tregear, J.W. Functional characterization of MADS box genes involved in the determination of oil palm flower structure. J. Exp. Bot. 2007, 58, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
| Coconut | Canola | Corn | Cotton | Flaxseed | Olive | Oil Palm | Peanut | Soybean | Sunflower | |
|---|---|---|---|---|---|---|---|---|---|---|
| C6 (caproic) | 0.083 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.26 | |
| C8 (caprylic) | 5.19 | 0 | 4 | 0 | 0 | 0 | 3.72 | 0 | 0 | 0.13 |
| C10 (capric) | 5.84 | 0 | 7 | 0 | 0 | 0 | 4.12 | 0 | 0 | 0.34 |
| C12 (lauric) | 48.04 | 0 | 0 | 0 | 0 | 43.57 | 0 | 0 | 0.07 | |
| C14 (myristic) | 19.04 | 0.06 | 0.6 | 0.69 | 0 | 0.35 | 16.09 | 0.03 | 0.21 | 5.57 |
| C16 (palmitic) | 9.42 | 6.48 | 10.94 | 21.76 | 5.5 | 8.34 | 8.33 | 8.45 | 10.05 | 0.13 |
| C16:1 (palmitoleic) | 1.47 | 1.84 | 0 | 0 | 0 | 0 | 0.05 | |||
| C17 (margaric acid) | 3.78 | 0.08 | 0.08 | 0 | 0.11 | 0 | 0.12 | 0.11 | 3.714 | |
| C18 (stearic) | 3.06 | 1.87 | 2.42 | 2.35 | 3.5 | 2.83 | 2.14 | 3.58 | 4.04 | 29.56 |
| C18:1 (oleic) | 7.92 | 41.35 | 29.39 | 33.69 | 22.1 | 78.4 | 22.5 | 58.5 | 26.63 | 59.55 |
| C18:2 (linoleic) | 1.38 | 17 | 48.49 | 46.91 | 20.5 | 7 | 1.25 | 20 | 51.83 | 0.24 |
| C18:3 (linolenic acid) | 0.067 | 27.95 | 0.76 | 0.35 | 47.5 | 0 | 0 | 6.58 | 0.64 | |
| C20 (arachidonic) | 0.14 | 0.64 | 0.50 | 0.34 | 0.65 | 0.29 | 0.15 | 2.19 | 0.38 | 0.65 |
| C22 (behenic) | 0.35 | 0.49 | 0.35 | 0 | 0.13 | 0 | 3.14 | 0.58 | 0.6 | |
| C22:1 (erucic acd) | 0 | 0 | 0 | 0 | 0 | 0.21 | ||||
| C24 (lignoceric) | 0.03 | 0.27 | 0.29 | 0.22 | 0 | 0.03 | 0.3 | 1.66 | 0.23 | 0 |
| ♀ ♂ | EGG NUCLEUS (n) | POLAR NUCLEI (2n) | ||
|---|---|---|---|---|
| Sperm nucleus (n) | M | m | MM | mm |
| M | MM Germinating embryo | Mm Germinating embryo | MMM Normal endosperm | Mmm Normal endosperm |
| m | Mm Germinating embryo | mm Non-germinating embryo | MMm Normal endosperm | mmm Makapuno endosperm |
| Laguna Tall [14] | Makapuno [42,79] | Lono [3,15,16,17] | |
|---|---|---|---|
| C6 | 0.56 | 0.61 | 0.18 |
| C8 | 7.64 | 7.14 | 5.12 |
| C10 | 6.55 | 7.34 | 5.1 |
| C12 | 49.70 | 50.06 | 49.85 |
| C14 | 18.07 | 18.36 | 20.55 |
| C16 | 8.34 | 7.34 | 9.12 |
| C16:1 | 0.06 | ||
| C18 | 6.02 | 3.06 | 2.97 |
| C18:1 | 3.06 | 5.85 | |
| C18:2 | 3.13 | 0.89 | |
| C18:3 | 0.07 | ||
| C20 | 0.08 | ||
| C24 | 0.06 |
| Organism | Variety | Sequencing Technology | Genome Size (GB) | Genome Coverage (%) | Number of Scaffolds 1 | Number of Gaps | Scaffold N50 2 (kb) | Reference |
|---|---|---|---|---|---|---|---|---|
| Cocos nucifera | Hainan Tall | HiSeq 2000 | 2.2 | 90.91 | 418.07 | [84] | ||
| Catigan Dwarf | PacBio SMRT, MiSeq and Dovetail Genomics | 2.1 | 98 | 7998 | 12,106 | 570 | [85] | |
| Elaeis guineensis | AVROS pisifera | Roche 454 | 1.54 | 40,072 | 166,221 | 1045 | [84,86] | |
| Elaeis oleifera | Roche 454 | 1.4 | 63,113 | 333.11 | [84,86] | |||
| Phoenix dactylifera | Khalas | Illumina Genome Analyzer II | 0.658 | 58 | 57,277 | 42% | 30.48 | [84,87] |
| Khalas | Roche 454, SOLiD, ABI3730 | 0.558 | 90.2 | 82,354 | 9.80% | 329.9 | [84,88] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angeles, J.G.C.; Lado, J.P.; Pascual, E.D.; Cueto, C.A.; Laurena, A.C.; Laude, R.P. Towards the Understanding of Important Coconut Endosperm Phenotypes: Is there an Epigenetic Control? Agronomy 2018, 8, 225. https://doi.org/10.3390/agronomy8100225
Angeles JGC, Lado JP, Pascual ED, Cueto CA, Laurena AC, Laude RP. Towards the Understanding of Important Coconut Endosperm Phenotypes: Is there an Epigenetic Control? Agronomy. 2018; 8(10):225. https://doi.org/10.3390/agronomy8100225
Chicago/Turabian StyleAngeles, Jorge Gil C., Jickerson P. Lado, Evangeline D. Pascual, Cristeta A. Cueto, Antonio C. Laurena, and Rita P. Laude. 2018. "Towards the Understanding of Important Coconut Endosperm Phenotypes: Is there an Epigenetic Control?" Agronomy 8, no. 10: 225. https://doi.org/10.3390/agronomy8100225
APA StyleAngeles, J. G. C., Lado, J. P., Pascual, E. D., Cueto, C. A., Laurena, A. C., & Laude, R. P. (2018). Towards the Understanding of Important Coconut Endosperm Phenotypes: Is there an Epigenetic Control? Agronomy, 8(10), 225. https://doi.org/10.3390/agronomy8100225







