Shaping Plant Adaptability, Genome Structure and Gene Expression through Transposable Element Epigenetic Control: Focus on Methylation
Abstract
1. Introduction
1.1. Epigenetic Modifications
1.2. Plant Transposable Elements
1.3. TE Epigenetic Regulation Mechanisms
1.4. Linking Plant Responses to Transposable Element-Derived Epigenetic Changes
2. Stress, Development and Genome Size: How TE Epigenetic Changes Alter the Genomic Landscape and Influence Plant Adaptability
2.1. Prominent Examples of Abiotic Driven Changes
2.2. TEs and Plant Defense Responses
2.3. Benefits of Activating/Deactivating TEs during Development
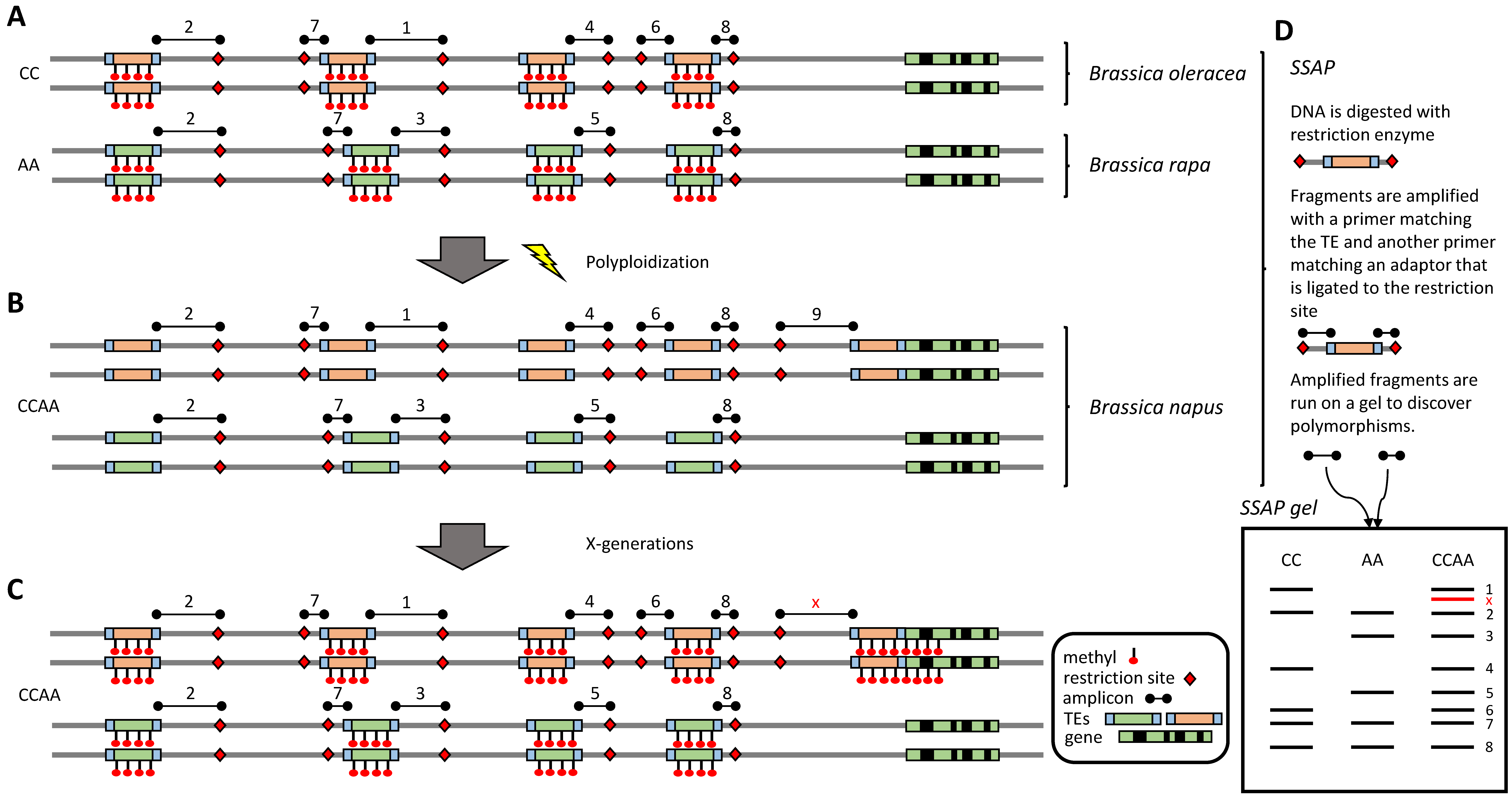
2.4. Can Polyploidy Transiently Affect TE Activity?
2.5. TEs Can Boost Genome Size Divergence among Related Species
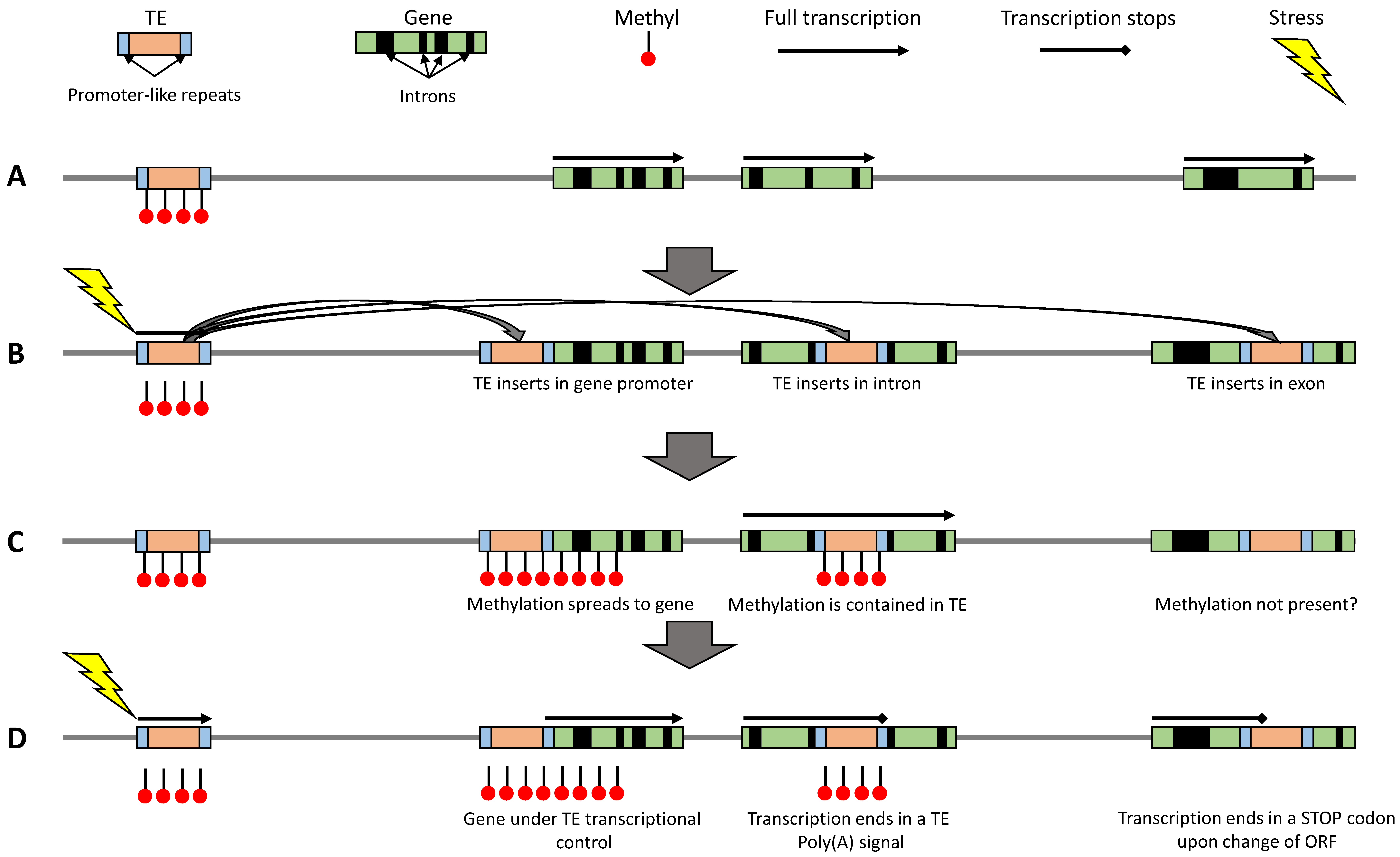
3. Is Extension of TE Methylation into Surrounding Regions detrimental?
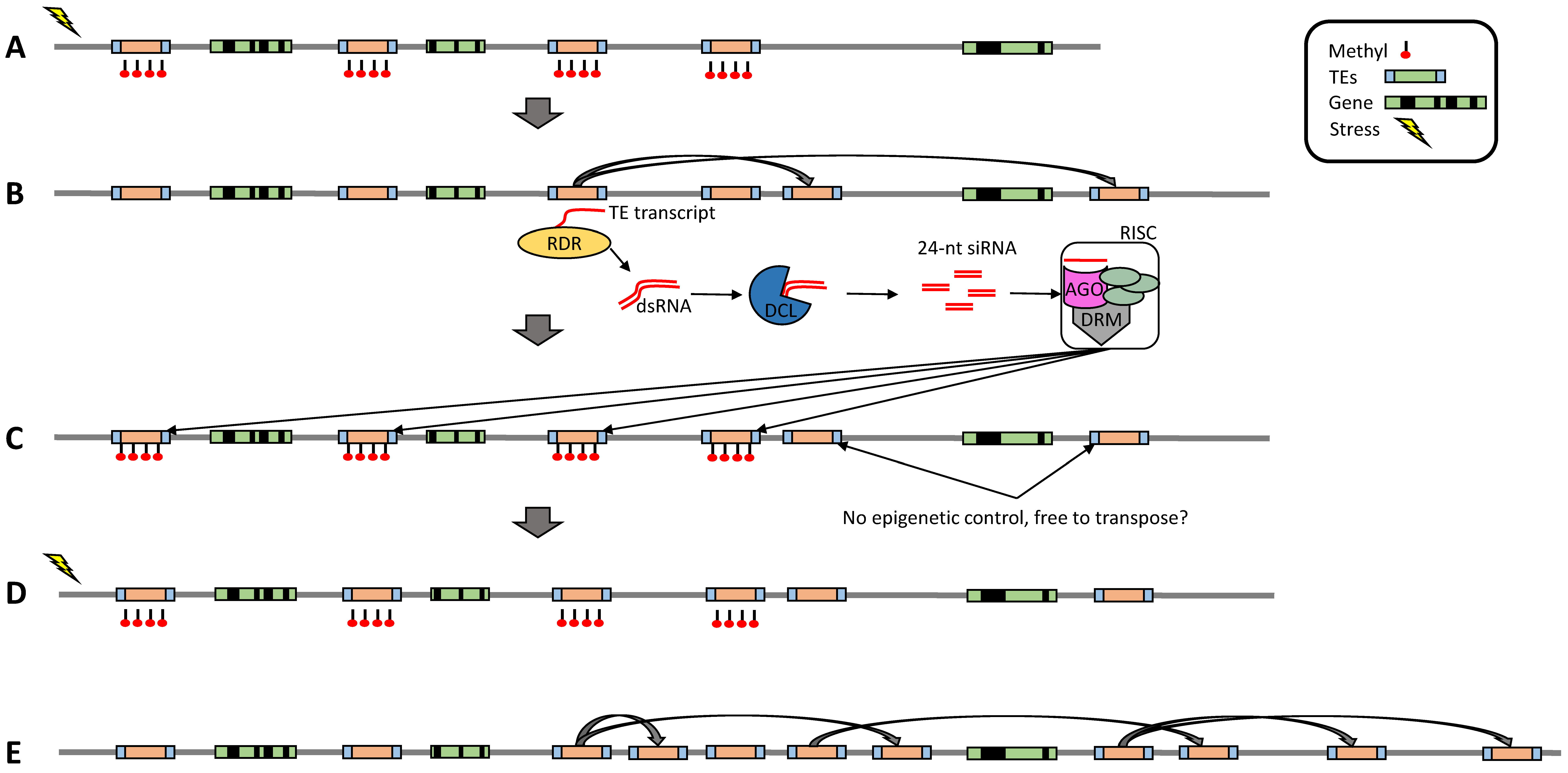
4. Can TEs Escape Epigenetic Control?
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; Scheid, O.M.; Kingston, R.E.; Tamkun, J.W.; Baulcombe, D.C.; Dean, C. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Rando, O.J. Combinatorial complexity in chromatin structure and function: Revisiting the histone code. Curr. Opin. Genet. Dev. 2012, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.; Ho, S.; Murnane, J.P.; Kit, A.; Yeung, Y.; Ng, H.K.; Wing, A.; Lo, I. Report telomeres acquire distinct heterochromatin characteristics during siRNA-induced RNA interference in mouse cells. Curr. Biol. 2008, 18, 183–187. [Google Scholar] [CrossRef]
- Djupedal, I.; Ekwall, K. Epigenetics: Heterochromatin meets RNAi. Cell Res. 2009, 19, 282–295. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Mittelsten Scheid, O. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Ma, Y.; Dai, H.; Li, L.; Liu, Y.; Li, H.; Zhao, G.; Zhang, Z. Characterization of the hormone and stress-induced expression of FaRE1 retrotransposon promoter in strawberry. J. Plant Biol. 2012, 55, 1–7. [Google Scholar] [CrossRef]
- Salazar, M.; González, E.; Casaretto, J.A.; Casacuberta, J.M.; Ruiz-Lara, S. The promoter of the TLC1.1 retrotransposon from Solanum chilense is activated by multiple stress-related signaling molecules. Plant Cell Rep. 2007, 26, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Tanskanen, J.; Immonen, S.; Nevo, E.; Schulman, A.H. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 2000, 97, 6603–6607. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, J.M.; Grandbastien, M.-A. Characterisation of LTR sequences involved in the protoplast specific expression of the tobacco Tnt1 retrotransposon. Nucleic Acids Res. 1993, 21, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Mhiri, C.; Vernhettes, S.; Casacuberta, J.M. The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol. Biol. 1997, 33, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999, 18, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Tapia, G.; Verdugo, I.; Poblete, F.; Gonza, E. Involvement of ethylene in stress-induced expression of the TLC1 1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol. 2005, 138, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Romanish, M.T.; Mager, D.L. Transposable elements: An abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012, 46, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Vitte, C.; Fustier, M.-A.; Alix, K.; Tenaillon, M.I. The bright side of transposons in crop evolution. Brief. Funct. Genom. 2014, 13, 276–295. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.; Fujimoto, R.; Sato, Y.; Nishio, T. A novel wx mutation caused by insertion of a retrotransposon-like sequence in a glutinous cultivar of rice (Oryza sativa). Theor. Appl. Genet. 2007, 115, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, A.; Liu, B.; Kong, F.; Arase, S.; Abe, J. Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J. Mol. Evol. 2009, 69, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.H.; De Melo, D.F.; Gouveia, Z.; Cardoso, H.G.; Peixe, A.; Arnholdt-Schmitt, B. The alternative oxidase family of Vitis vinifera reveals an attractive model to study the importance of genomic design. Physiol. Plant. 2009, 137, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, T.; Eulgem, T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 2013, 110, E3535–E3543. [Google Scholar] [CrossRef] [PubMed]
- Varagona, M.J.; Purugganan, M.; Wessler, S.R. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 1992, 4, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, H. Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 2009, 57, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Licciardello, C.; Zhang, Y.; Liu, J.; Mackay, S.; Bailey, P.; Reforgiato-Recupero, G.; Martin, C. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 2012, 24, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L.; Walbot, V. DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 1986, 83, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Martienssen, R.; Barkan, A.; Taylor, W.C.; Freeling, M. Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 1990, 4, 331–343. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Vongs, A.; Kakutani, T.; Martienssen, R.A.; Richards, E.J. Arabidopsis-Thaliana DNA Methylation Mutants. Science 1993, 260, 1926–1928. [Google Scholar] [CrossRef] [PubMed]
- Hirochika, H.; Okamoto, H.; Kakutani, T. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 2000, 12, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Miura, A.; Yonebayashi, S.; Watanabe, K. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 2001, 411, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 2009, 60, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.; Allshire, R.C. RNA silencing and genome regulation. Trends Cell Biol. 2005, 15, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. The stress concept in plants: An introduction. Ann. N. Y. Acad. Sci. 1998, 851, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Sano, H. Inheritance of acquired traits in plants Reinstatement of Lamarck. Plant Signal. Behav. 2010, 5, 346–348. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.P.; Gehring, M. Stable transgenerational epigenetic inheritance requires a DNA methylation-sensing circuit. Nat. Commun. 2017, 8, 2124. [Google Scholar] [CrossRef] [PubMed]
- Civan, P.; Svec, M.; Huptvogel, P. On the coevolution of transposable elements and plant genomes. J. Bot. 2011, 2011, 893546. [Google Scholar] [CrossRef]
- Pereira, V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004, 5, R79. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Du, J. Young but not relatively old retrotransposons are preferentially located in gene-rich euchromatic regions in tomato (Solanum lycopersicum) plants. Plant J. 2014, 80, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- De Souza, F.S.J.; Franchini, L.F.; Rubinstein, M. Exaptation of transposable elements into novel cis-regulatory elements: Is the evidence always strong? Mol. Biol. Evol. 2013, 30, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.D.; Smith, L.M.; Guo, Y.; Ott, F.; Weigel, D.; Gaut, B.S. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 2011, 108, 2322–2327. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Weigel, D.; Smith, L.M. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 2013, 9, e1003255. [Google Scholar] [CrossRef] [PubMed]
- Galindo-González, L.; Mhiri, C.; Deyholos, M.K.; Grandbastien, M.-A. LTR-retrotransposons in plants: Engines of evolution. Gene 2017, 626, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Sarazin, A.; Bowler, C.; Colot, V.; Quesneville, H. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acid Res. 2011, 39, 6919–6931. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Lepère, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Slotkin, R.K. Developmental relaxation of transposable element silencing in plants: Functional or byproduct? Curr. Opin. Plant Biol. 2012, 15, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Sarilar, V.; Palacios, P.M.; Rousselet, A.; Ridel, C.; Falque, M.; Eber, F.; Chèvre, A.M.; Joets, J.; Brabant, P.; Alix, K. Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids. New Phytol. 2013, 198, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Salmon, A.; Zerjal, T.; Tenaillon, M.; Grandbastien, M.-A.; Ainouche, M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009, 184, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, W.; Ohama, N.; Tanabe, N.; Masuta, Y.; Masuda, S.; Mitani, N.; Yamaguchi-Shinozaki, K.; Ma, J.F.; Kato, A.; Ito, H. A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front. Plant Sci. 2015, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 2017, 18, 134. [Google Scholar] [CrossRef] [PubMed]
- Pietzenuk, B.; Markus, C.; Gaubert, H.; Bagwan, N.; Merotto, A.; Bucher, E. Recurrent evolution of heat-responsiveness in Brassicaceae COPIA elements. Genome Biol. 2016, 17, 209. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Hoen, D.R.; Bureau, T.E. Discovery of novel genes derived from transposable elements using integrative genomic analysis. Mol. Biol. Evol. 2015, 32, 1487–1506. [Google Scholar] [CrossRef] [PubMed]
- Le, T.-N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.; Zhu, Q.-H.; Taylor, J.M.; Kazan, K.; Llewellyn, D.J.; Zhang, R.; et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Seidl, M.F.; Thomma, B.P.H.J. Transposable elements direct the doevolution between plants and microbes. Trends Genet. 2017, 33, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.J.; Fischer, R.L. Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant Biol. 2011, 14, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Vaughn, M.; Tanurdžic, M.; Borges, F.; Becker, J.D.; Feijó, A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Bubb, K.L.; Henikoff, S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 2009, 324, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, Y.; Saze, H.; Kinoshita, T.; Miura, A.; Soppe, W.J.J.; Koornneef, M. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2006, 49, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Castelletti, S.; Tuberosa, R.; Pindo, M.; Salvi, S. A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL vgt1. G3 2014, 4, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005, 41, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Kenan-Eichler, M.; Leshkowitz, D.; Tal, L.; Noor, E.; Melamed-bessudo, C.; Feldman, M.; Levy, A.A. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 2011, 188, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Kraitshtein, Z.; Yaakov, B.; Khasdan, V.; Kashkush, K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 2010, 186, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Xia, E.; Yao, Q.; Liu, X.; Gao, L. Autotetraploid rice methylome analysis reveals methylation variation of transposable elements and their effects on gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, E7022–E7029. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Grandbastien, M.-A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010, 186, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef] [PubMed]
- Waugh, R.; McLean, K.; Flavell, A.J.; Pearce, S.R.; Kumar, A.; Thomas, B.B.T.; Powell, W. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol. Gen. Genet. 1997, 253, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.T.; Pattyn, P.; Bakker, E.G.; Cao, J.; Cheng, J.-F.; Clark, R.M.; Fahlgren, N.; Fawcett, J.A.; Grimwood, J.; Gundlach, H.; et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011, 43, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Kawanabe, T.; Fujimoto, R.; Sasaki, T.; Taylor, J.M.; Dennis, E.S. A comparison of transcriptome and epigenetic status between closely related species in the genus Arabidopsis. Gene 2012, 506, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Hufford, M.B.; Gaut, B.S.; Ross-ibarra, J. Genome size and transposable element content as determined by high-throughput sequencing in Maize and Zea luxurians. Genome Biol. Evol. 2011, 3, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Stetter, M.G.; Stitzer, M.C. Adaptation in plant genomes: Bigger is different. Am. J. Bot. 2018, 105, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.-V.; Black, M.; Vaughn, M.W.; Dedhia, N.; Mccombie, W.R.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Ellis, N.A.; Makarevitch, I.; Yeh, C.; Gent, J.I.; Guo, L.; Mcginnis, K.M.; Zhang, X.; Schnable, P.S.; Vaughn, M.W.; et al. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 2012, 8, e1003127. [Google Scholar] [CrossRef] [PubMed]
- West, P.T.; Li, Q.; Ji, L.; Eichten, S.R.; Song, J.; Vaughn, M.W.; Schmitz, R.J.; Springer, N.M. Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D.; Slotkin, R.K. Strategies for silencing and escape: The ancient struggle between transposable elements and their hosts. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2011; Volume 292, pp. 119–152. ISBN 9780123860330. [Google Scholar]
- Jiang, N.; Bao, Z.; Zhang, X.; Eddy, S.R.; Wessler, S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 2004, 431, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Guermonprez, H.; Henaff, E.; Cifuentes, M.; Casacuberta, J.M. Chapter 7—MITEs, Miniature elements with a major role in plant genome evolution. In Plant Transposable Elements; Grandbastien, M.A., Casacubertal, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 113–124. ISBN 9783642318429. [Google Scholar]
- Wang, J.; Yu, Y.; Tao, F.; Zhang, J.; Copetti, D.; Kudrna, D.; Talag, J.; Lee, S.; Wing, R.A.; Fan, C. DNA methylation changes facilitated evolution of genes derived from Mutator-like transposable elements. Genome Biol. 2016, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.T.; Cardin, S.E.; Borchert, G.M. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob. Genet. Elem. 2014, 4, e29255. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Xia, J.; Jin, Y. Domestication of transposable elements into MicroRNA genes in plants. PLoS ONE 2011, 6, e19212. [Google Scholar] [CrossRef] [PubMed]
- Piriyapongsa, J.; Jordan, I.K. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 2008, 14, 814–821. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Slotkin, R.K. Transposable element small RNAs as regulators of gene expression. Trends Genet. 2012, 28, 616–623. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Nuthikattu, S.; Slotkin, R.K.; McCue, A.D.; Nuthikattu, S.; Slotkin, R.K. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 2013, 10, 1379–1395. [Google Scholar] [CrossRef] [PubMed]
- Juretic, N.; Hoen, D.R.; Huynh, M.L.; Harrison, P.M.; Bureau, T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005, 15, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Nosaka, M.; Itoh, J.; Nagato, Y.; Ono, A.; Ishiwata, A.; Sato, Y. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 2012, 8, e1002953. [Google Scholar] [CrossRef] [PubMed]
- McCue, A.D.; Nuthikattu, S.; Reeder, S.H.; Slotkin, R.K. Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Kawabe, A.; Etcheverry, M.; Ito, T.; Toyoda, A.; Fujiyama, A.; Colot, V.; Tarutani, Y.; Kakutani, T. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J. 2013, 32, 2407–2417. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, A.; Saito, R.; Takashima, K.; Sasaki, T.; Fu, Y.; Kawabe, A.; Toyoda, A.; Fujiyama, A.; Tarutani, Y.; Kakutani, T.; et al. Evolution of sequence-specific anti-silencing systems in Arabidopsis. Nat. Commun. 2017, 8, 2161. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galindo-González, L.; Sarmiento, F.; Quimbaya, M.A. Shaping Plant Adaptability, Genome Structure and Gene Expression through Transposable Element Epigenetic Control: Focus on Methylation. Agronomy 2018, 8, 180. https://doi.org/10.3390/agronomy8090180
Galindo-González L, Sarmiento F, Quimbaya MA. Shaping Plant Adaptability, Genome Structure and Gene Expression through Transposable Element Epigenetic Control: Focus on Methylation. Agronomy. 2018; 8(9):180. https://doi.org/10.3390/agronomy8090180
Chicago/Turabian StyleGalindo-González, Leonardo, Felipe Sarmiento, and Mauricio A. Quimbaya. 2018. "Shaping Plant Adaptability, Genome Structure and Gene Expression through Transposable Element Epigenetic Control: Focus on Methylation" Agronomy 8, no. 9: 180. https://doi.org/10.3390/agronomy8090180
APA StyleGalindo-González, L., Sarmiento, F., & Quimbaya, M. A. (2018). Shaping Plant Adaptability, Genome Structure and Gene Expression through Transposable Element Epigenetic Control: Focus on Methylation. Agronomy, 8(9), 180. https://doi.org/10.3390/agronomy8090180




