Genome-Wide Characterization of the Fantastic Four Gene Family Identifies TaFAF-5D.5 Associated with Growth Habit Variation in Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Molecular Characterization of TaFAF Genes
2.2. Phylogenetic and Structural Analyses
2.3. Calculation of Protein Molecular Weight, Isoelectric Point, Ka/Ks Values, and Divergence Time
2.4. cis-Regulatory Element Analysis
2.5. Expression Analysis
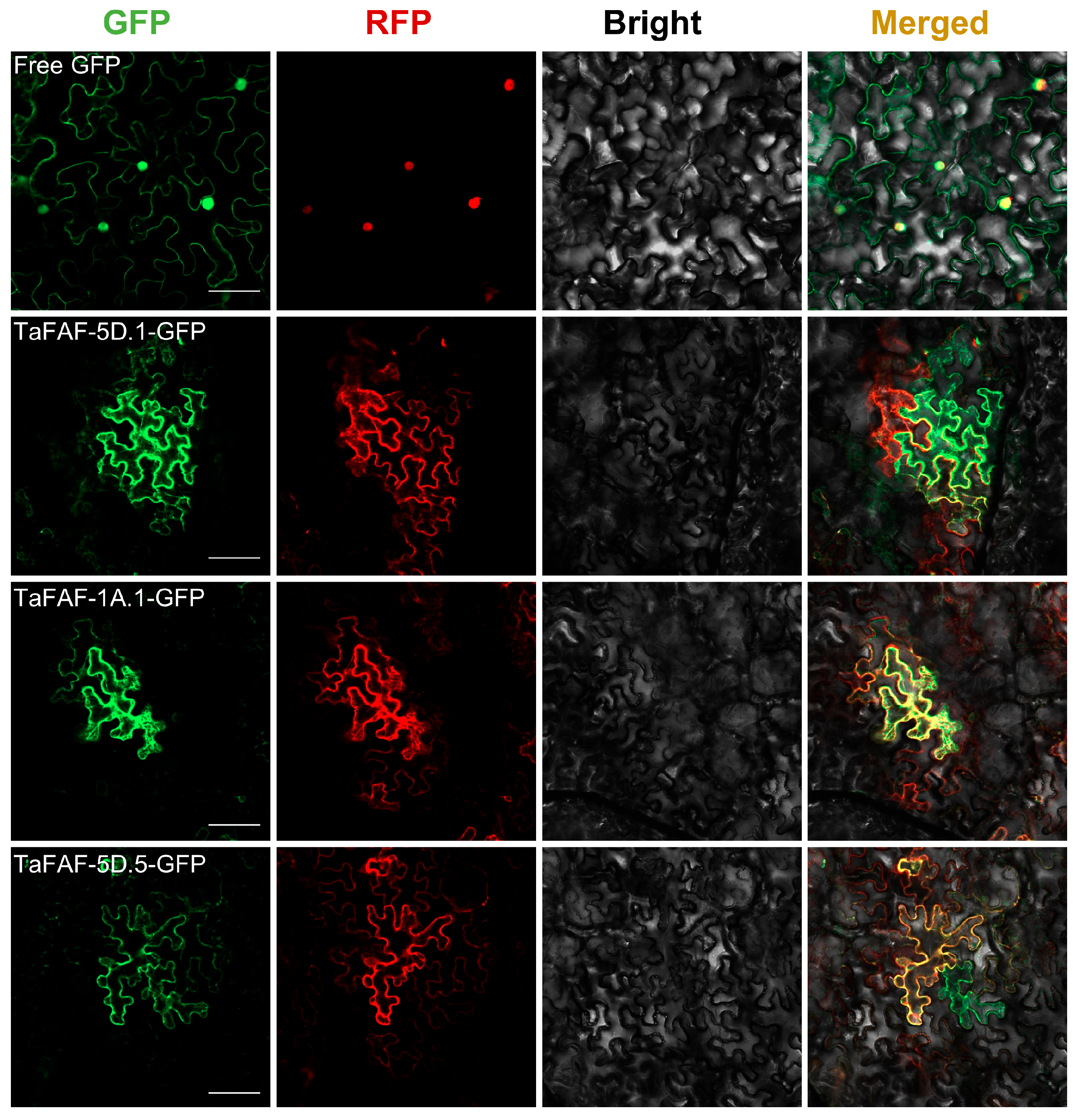
2.6. Subcellular Localization
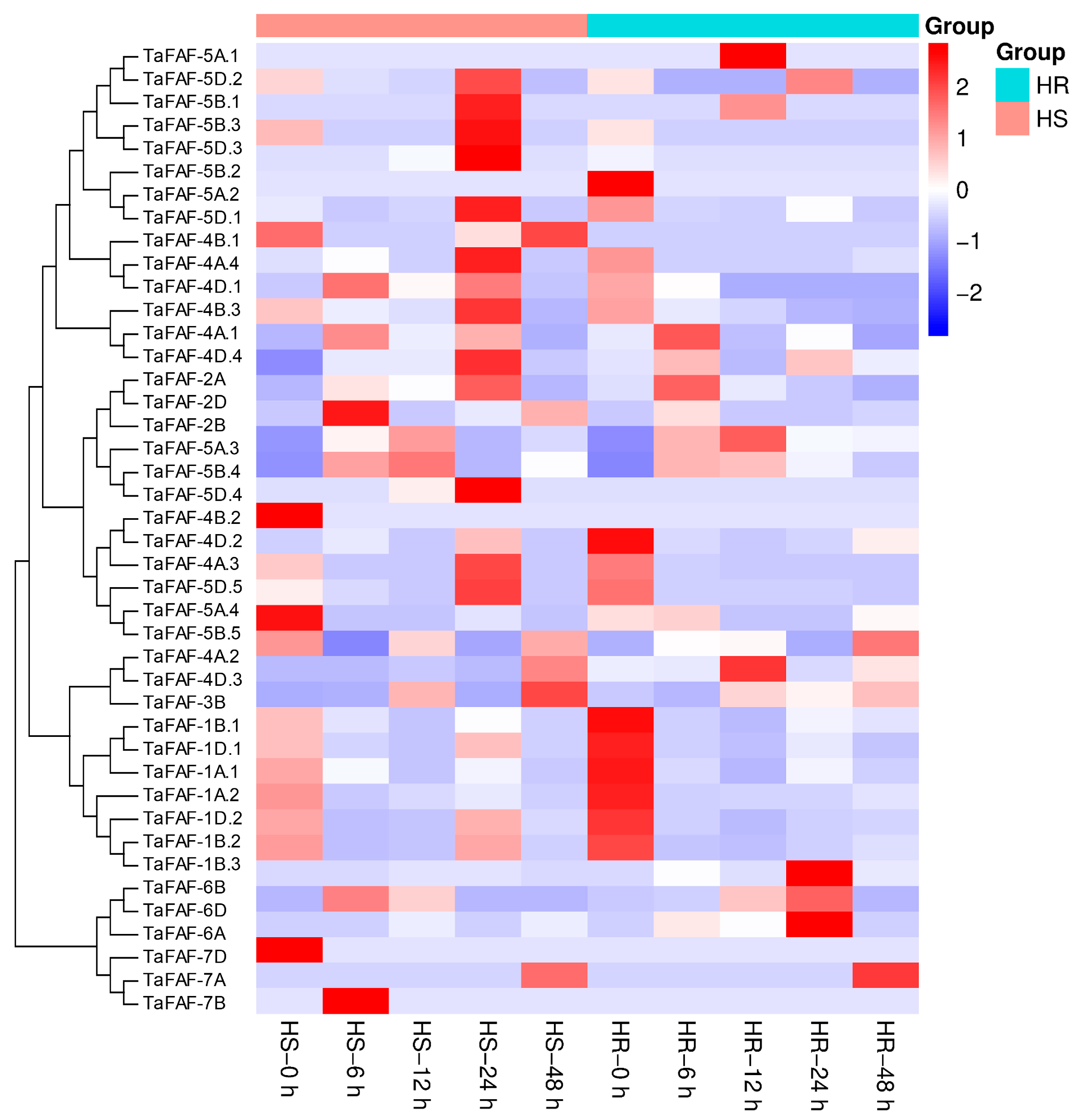
2.7. Analysis of TaFAF Expressions in Response to Heat Stress
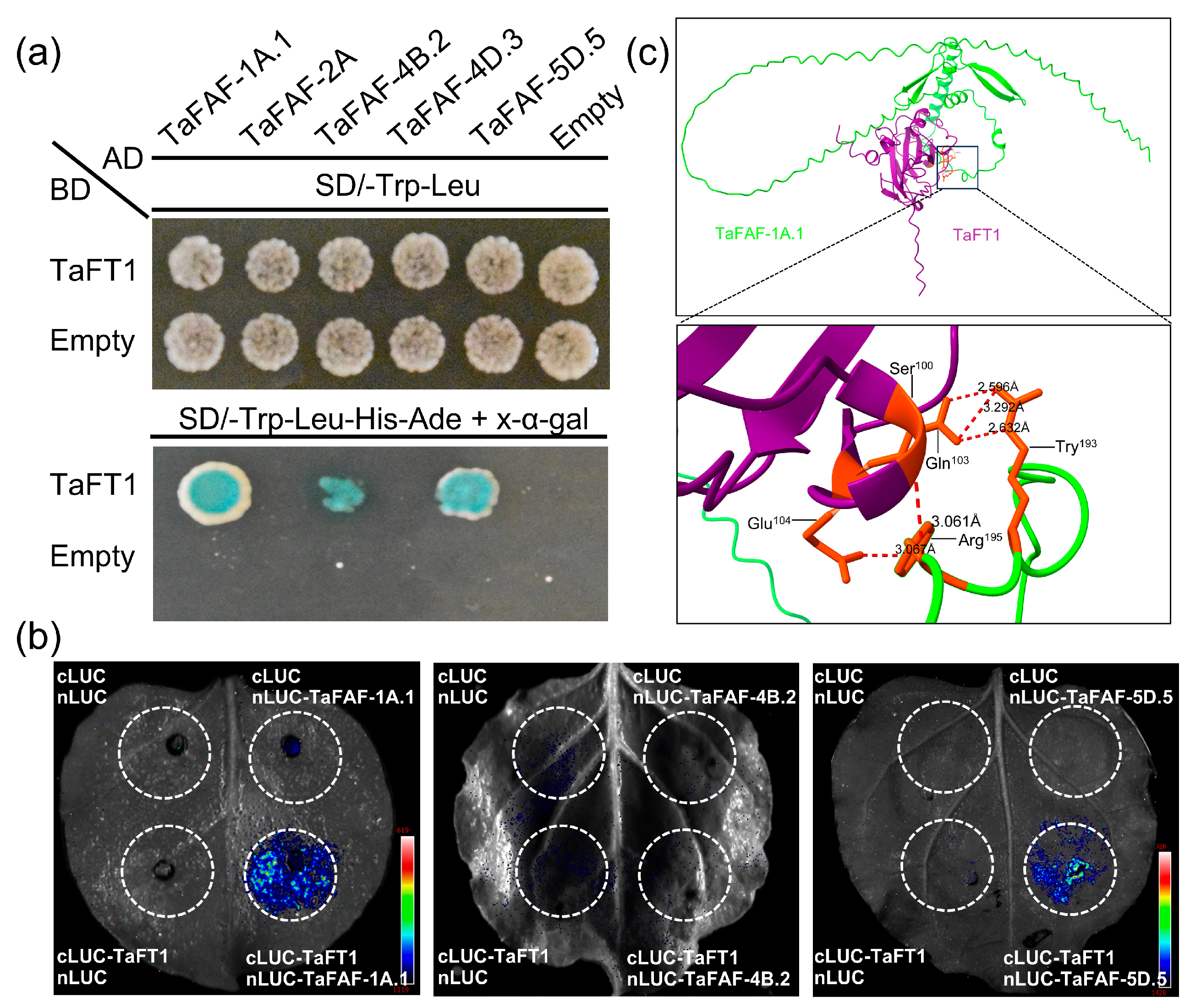
2.8. Yeast Two-Hybrid Assays
2.9. Split-Luciferase Complementation Imaging Assays
2.10. Protein–Protein Interaction Modeling and Structural Analysis
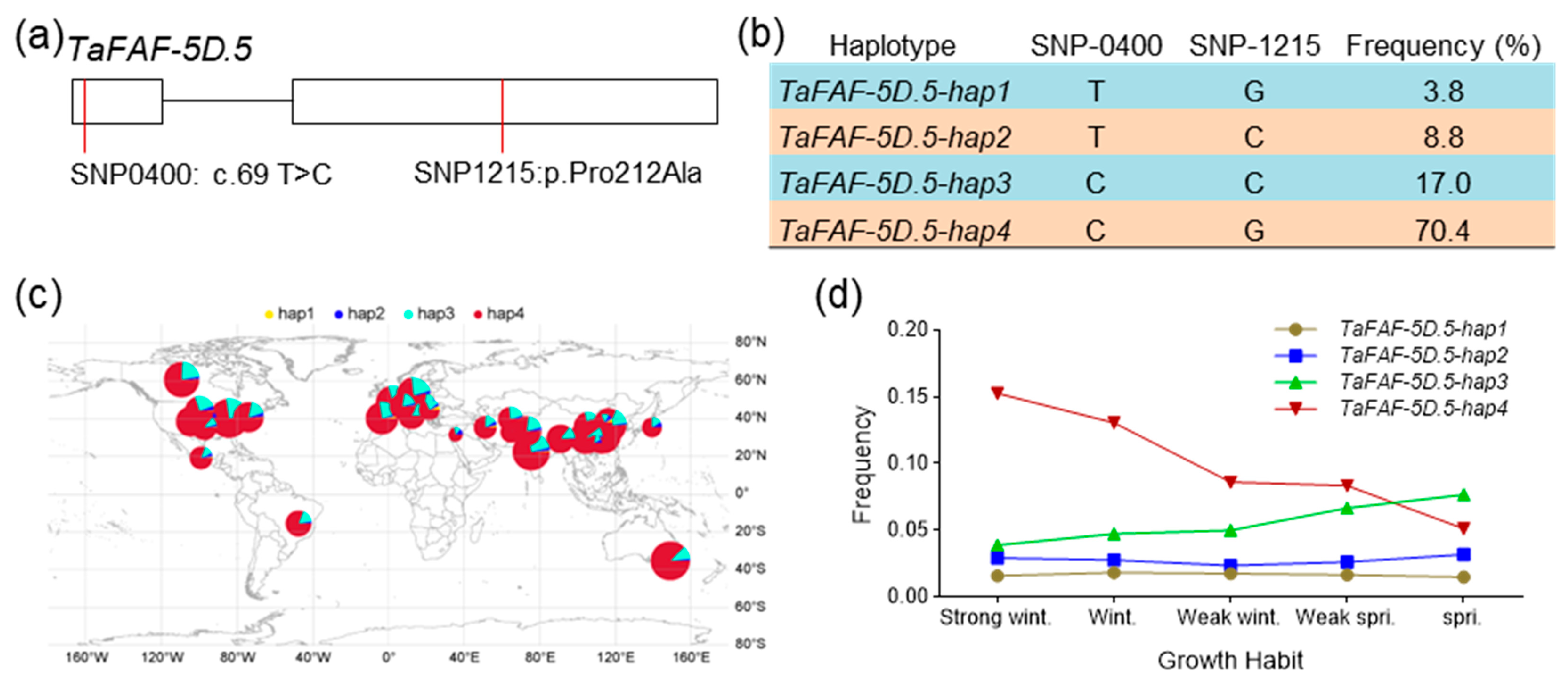
2.11. Haplotype Analysis
2.12. Statistical Analysis
3. Results
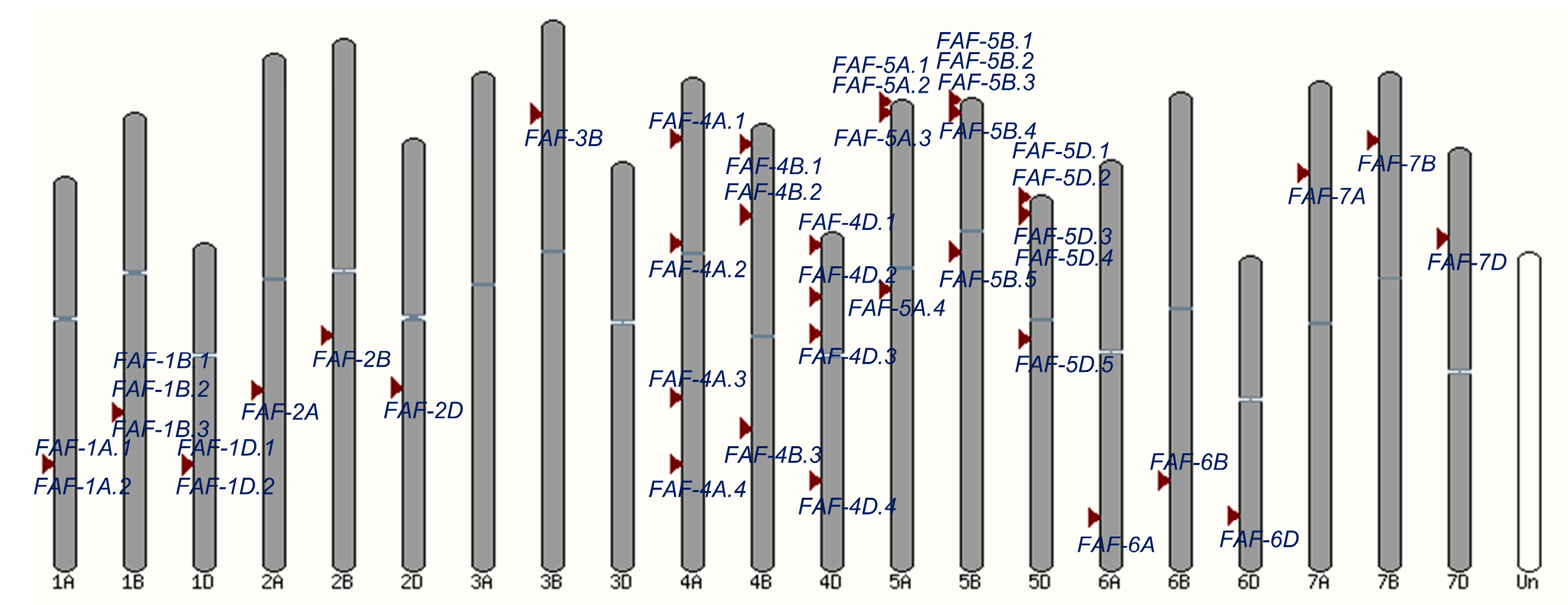
3.1. Characterization of the FAF Genes in Wheat
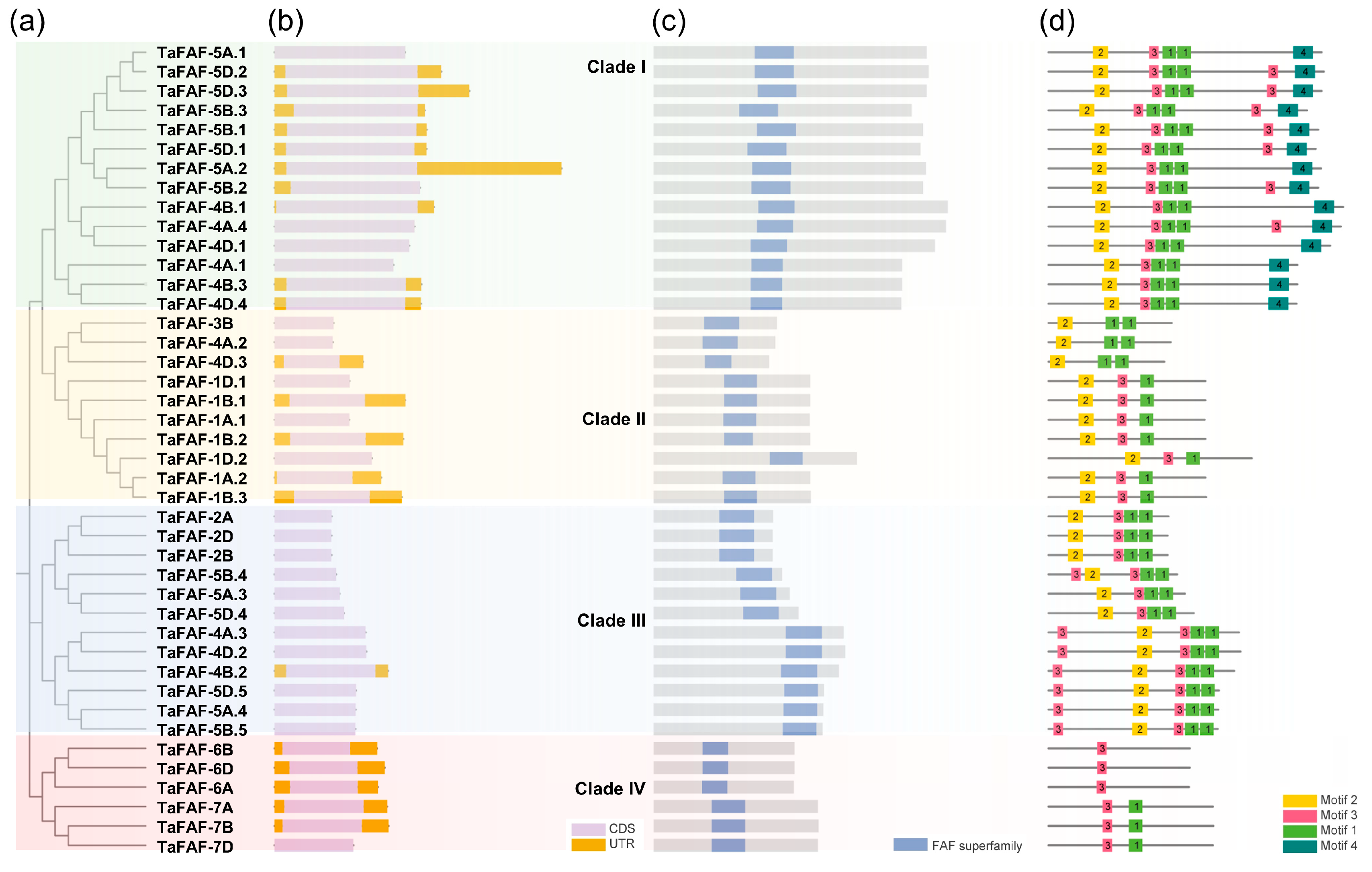
3.2. Phylogenetics and Structure of TaFAF Genes
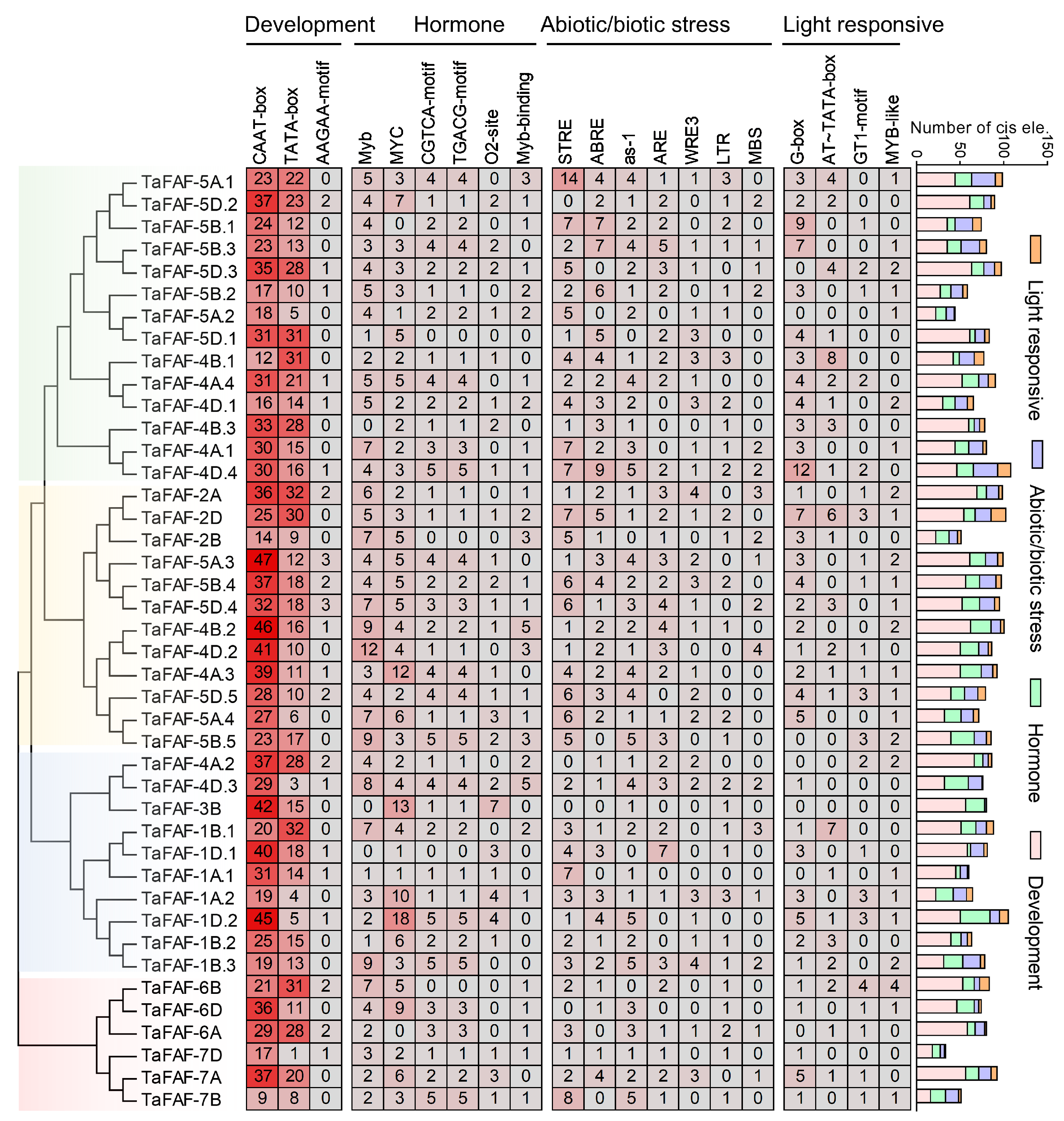
3.3. Identification of the Cis-Regulatory Elements
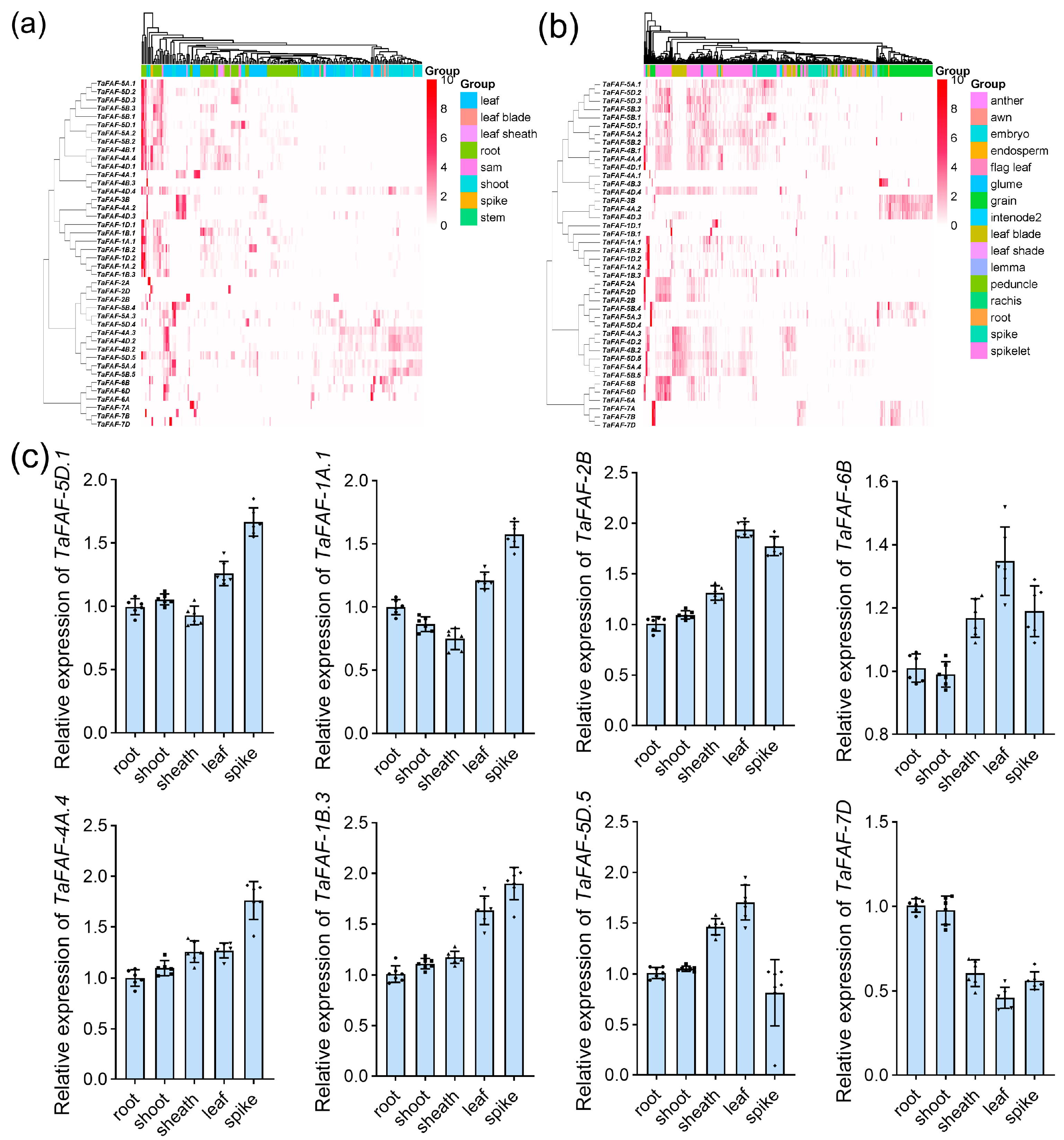
3.4. Expression Patterns and Subcellular Localization of TaFAF Genes
3.5. Expression Characterization of Temperature-Responsive TaFAF Genes
3.6. TaFAFs Interact with the Flowering Regulator TaFT1
3.7. TaFAF-5D.5 Haplotypes Are Associated with Wheat Growth Habit
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Pant, K.K.; Naik, J.; Barthakur, S.; Chandra, V. High-temperature stress in wheat (Triticum aestivum L.): Unfolding the impacts, tolerance and methods to mitigate the detrimental effects. Cereal Res. Commun. 2025, 53, 1171–1197. [Google Scholar] [CrossRef]
- Lamba, K.; Kumar, M.; Singh, V.; Chaudhary, L.; Sharma, R.; Yashveer, S.; Dalal, M.S. Heat stress tolerance indices for identification of the heat tolerant wheat genotypes. Sci. Rep. 2023, 13, 10842. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, H.; Wei, H.; Li, S.; Zhang, N.; Si, H. Roles of TCP transcription factors in plant growth and development. Physiol. Plant 2025, 177, e70357. [Google Scholar] [CrossRef]
- Wahl, V.; Brand, L.H.; Guo, Y.-L.; Schmid, M. The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ai, G.; Ji, K.; Huang, R.; Chen, C.; Yang, Z.; Wang, J.; Cui, L.; Li, G.; Tahira, M.; et al. EARLY FLOWERING is a dominant gain-of-function allele of FANTASTIC FOUR 1/2c that promotes early flowering in tomato. Plant Biotechnol. J. 2023, 22, 698–711. [Google Scholar] [CrossRef]
- Shang, L.; Tao, J.; Song, J.; Wang, Y.; Zhang, X.; Ge, P.; Li, F.; Dong, H.; Gai, W.; Grierson, D.; et al. CRISPR/Cas9-mediated mutations of FANTASTIC FOUR gene family for creating early flowering mutants in tomato. Plant Biotechnol. J. 2023, 22, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.; Baek, W.; Lim, C.W.; Lee, S.C. A pepper RING-finger E3 ligase, CaFIRF1, negatively regulates the high-salt stress response by modulating the stability of CaFAF1. Plant Cell Environ. 2024, 47, 1319–1333. [Google Scholar] [CrossRef]
- de Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.V.; Bodanese Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Kerbler, S.M.; Wigge, P.A. Wigge, Temperature sensing in plants. Annu. Rev. Plant Biol. 2023, 74, 341–366. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.-R.; Gao, J.; Lin, H.-X.; Lin, Y. The molecular basis of heat stress responses in plants. Mol. Plant 2023, 16, 1612–1634. [Google Scholar] [CrossRef]
- Farhad, M.; Kumar, U.; Tomar, V.; Bhati, P.K.; Krishnan, J.N.; Barek, V.; Brestic, M.; Hossain, A. Heat stress in wheat: A global challenge to feed billions in the current era of the changing climate. Front. Sustain. Food Syst. 2023, 7, 1203721. [Google Scholar] [CrossRef]
- Chaudhary, R.; Baranwal, V.K.; Kumar, R.; Sircar, D.; Chauhan, H. Genome-wide identification and expression analysis of Hsp70, Hsp90, and Hsp100 heat shock protein genes in barley under stress conditions and reproductive development. Funct. Integr. Genom. 2019, 19, 1007–1022. [Google Scholar] [CrossRef]
- Zang, X.; Geng, X.; Wang, F.; Liu, Z.; Zhang, L.; Zhao, Y.; Tian, X.; Ni, Z.; Yao, Y.; Xin, M.; et al. Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 2017, 17, 14. [Google Scholar] [CrossRef]
- Wang, X.; Xu, C.; Cai, X.; Wang, Q.; Dai, S. Heat-responsive photosynthetic and signaling pathways in plants: Insight from proteomics. Int. J. Mol. Sci 2017, 18, 2191. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Bae, Y.; Lee, S.C. Differential role of Capsicum annuum FANTASTIC FOUR-like gene CaFAF1 on drought and salt stress responses. Environ. Exp. Bot. 2022, 199, 104887. [Google Scholar] [CrossRef]
- Pont, C.; Leroy, T.; Seidel, M.A.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goué, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zheng, X.; Yang, D.; Li, L.; Yin, H. Genome-wide identification of the nuclear redox protein gene family revealed its potential role in drought stress tolerance in rice. Front. Plant Sci. 2025, 16, 1562718. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, H.; Guo, H.; Zhao, L.; Xie, Y.; Gu, J.; Zhao, S.; Ding, Y.; Li, H.; Zhou, C.; et al. Genome-wide characterization of two homeobox families identifies key genes associated with grain-related traits in wheat. Plant Sci. 2023, 336, 111862. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, H.; Guo, H.; Xie, Y.; Zhao, L.; Gu, J.; Li, H.; Zhao, S.; Ding, Y.; Zhou, C.; et al. A gain-of-function mutation at the C-terminus of FT-D1 promotes heading by interacting with 14-3-3A and FDL6 in wheat. Plant Biotechnol. J. 2024, 23, 20–35. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, H.; Guo, H.; Zhou, C.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. Identification of the vernalization gene VRN-B1 responsible for heading date variation by QTL mapping using a RIL population in wheat. BMC Plant Biol. 2020, 20, 331. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, Z.; Li, X.; Ni, Z.; Hu, Z.; Xin, M.; Peng, H.; Yao, Y.; Sun, Q.; Guo, W. SnpHub: An easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat. Gigascience 2020, 9, giaa060. [Google Scholar] [CrossRef]
- Jiang, Q.; Hou, J.; Hao, C.; Wang, L.; Ge, H.; Dong, Y.; Zhang, X. The wheat (T. aestivum) sucrose synthase 2 gene (TaSus2) active in endosperm development is associated with yield traits. Funct. Integr. Genom. 2010, 11, 49–61. [Google Scholar] [CrossRef]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Xie, Y.; Zeng, W.; Wang, C.; Xu, D.; Guo, H.; Xiong, H.; Fang, H.; Zhao, L.; Gu, J.; Zhao, S.; et al. Fine mapping of qd1, a dominant gene that regulates stem elongation in bread wheat. Front. Genet. 2021, 12, 793572. [Google Scholar] [CrossRef]
- Liu, J.; Yao, Y.; Xin, M.; Peng, H.; Ni, Z.; Sun, Q. Shaping polyploid wheat for success: Origins, domestication, and the genetic improvement of agronomic traits. J. Integr. Plant Biol. 2022, 64, 536–563. [Google Scholar] [CrossRef]
- Gao, J.; Ma, G.; Chen, J.; Gichovi, B.; Cao, L.; Liu, Z.; Chen, L. The B3 gene family in Medicago truncatula: Genome-wide identification and the response to salt stres. Plant Physiol. Biochem. 2024, 206, 108260. [Google Scholar] [CrossRef]
- Li, S.; Yao, Y.; Ye, W.; Wang, S.; Zhang, C.; Liu, S.; Sun, F.; Xi, Y. Genome-wide identification of wheat KNOX gene family and functional characterization of TaKNOX14-D in plants. Int. J. Mol. Sci. 2022, 23, 15918. [Google Scholar] [CrossRef] [PubMed]
- Sang, N.; Liu, H.; Ma, B.; Huang, X.; Zhuo, L.; Sun, Y. Roles of the 14-3-3 gene family in cotton flowering. BMC Plant Biol. 2021, 21, 1–17. [Google Scholar] [CrossRef]
- Song, X.; Li, N.; Guo, Y.Y.; Bai, Y.; Wu, T.; Yu, T.; Feng, S.Y.; Zhang, Y.; Wang, Z.Y.; Liu, Z.; et al. Comprehensive identification and characterization of simple sequence repeats based on the whole-genome sequences of 14 forest and fruit trees. Forestry Res. 2021, 1, 7. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, T.; Liu, L.T. The rice VCS1 is identified as a molecula tool to mark and visualize the vegetative cell of pollen. Plant Signal. Behav. 2021, 16, e1924502. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Wang, Y.; Si, X.; Pan, Y.; Guo, M.; Wu, M.; Li, Y.; Liu, H.; Zhang, X.; et al. TaFT-D1 positively regulates grain weight by acting as a coactivator of TaFDL2 in wheat. Plant Biotechnol. J. 2025, 23, 2207–2223. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.Y.; Dong, L.L.; Wu, J.; Liu, Y.L.; Yan, H.W. Comprehensive evolutionary analysis of the MAPK gene family from non-seed plants to angiosperm and interaction of PagMAPK3-1 with PagVQ13. Biochem. Biophys. Res. Commun. 2025, 781, 152554. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Shen, C.C.; Li, W.J.; Peng, L.; Hu, M.Y.; Zhang, Y.J.; Zhao, X.Q.; Teng, W.; Tong, Y.P.; He, X. TaLBD41 interacts with TaNAC2 to regulate nitrogen uptake and metabolism in response to nitrate availability. New Phytol. 2024, 242, 641–657. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, S.; Song, W.W.; Abdul, M.; Khan, A.; Sun, S.; Zhang, C.S.; Wu, T.T.; Wu, C.X.; Han, T.F. Natural variations of FT family genes in soybean varieties covering a wide range of maturity groups. BMC Genom. 2019, 20, 230. [Google Scholar] [CrossRef]
- Luo, X.; Liu, B.; Xie, L.; Wang, K.; Xu, D.; Tian, X.; Xie, L.; Li, L.; Ye, X.; He, Z.; et al. The TaSOC1-TaVRN1 module integrates photoperiod and vernalization signals to regulate wheat flowering. Plant Biotechnol. J. 2023, 22, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, Y.; Wang, K.; Luo, X.; Xu, D.; Tian, X.; Li, L.; Ye, X.; Xia, X.; Li, W.; et al. TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytol. 2021, 231, 834–848. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Jiang, J.; Hou, Z.; Wang, S.; Zhang, Y.; Li, Y.; Fang, Z. Genome-Wide Characterization of the Fantastic Four Gene Family Identifies TaFAF-5D.5 Associated with Growth Habit Variation in Wheat. Agronomy 2026, 16, 221. https://doi.org/10.3390/agronomy16020221
Jiang J, Hou Z, Wang S, Zhang Y, Li Y, Fang Z. Genome-Wide Characterization of the Fantastic Four Gene Family Identifies TaFAF-5D.5 Associated with Growth Habit Variation in Wheat. Agronomy. 2026; 16(2):221. https://doi.org/10.3390/agronomy16020221
Chicago/Turabian StyleJiang, Junlong, Zehao Hou, Shuping Wang, Yingxin Zhang, Yuting Li, and Zhengwu Fang. 2026. "Genome-Wide Characterization of the Fantastic Four Gene Family Identifies TaFAF-5D.5 Associated with Growth Habit Variation in Wheat" Agronomy 16, no. 2: 221. https://doi.org/10.3390/agronomy16020221
APA StyleJiang, J., Hou, Z., Wang, S., Zhang, Y., Li, Y., & Fang, Z. (2026). Genome-Wide Characterization of the Fantastic Four Gene Family Identifies TaFAF-5D.5 Associated with Growth Habit Variation in Wheat. Agronomy, 16(2), 221. https://doi.org/10.3390/agronomy16020221






