A Marigold (Tagetes erecta) MADS-Box Transcription Factor, TeSEP4, Regulates Petal Color by Modulating Chlorophyll and Carotenoid Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction and Gene Expression Quantification
2.3. Vectors Construction and Plant Transformation
2.4. Chlorophyll and Carotenoid Extraction and Analysis
2.5. Transcriptome Sequencing and Bioinformatic Analysis
2.6. cDNA Synthesis and qRT-PCR Validation
2.7. Statistical Analysis
3. Results
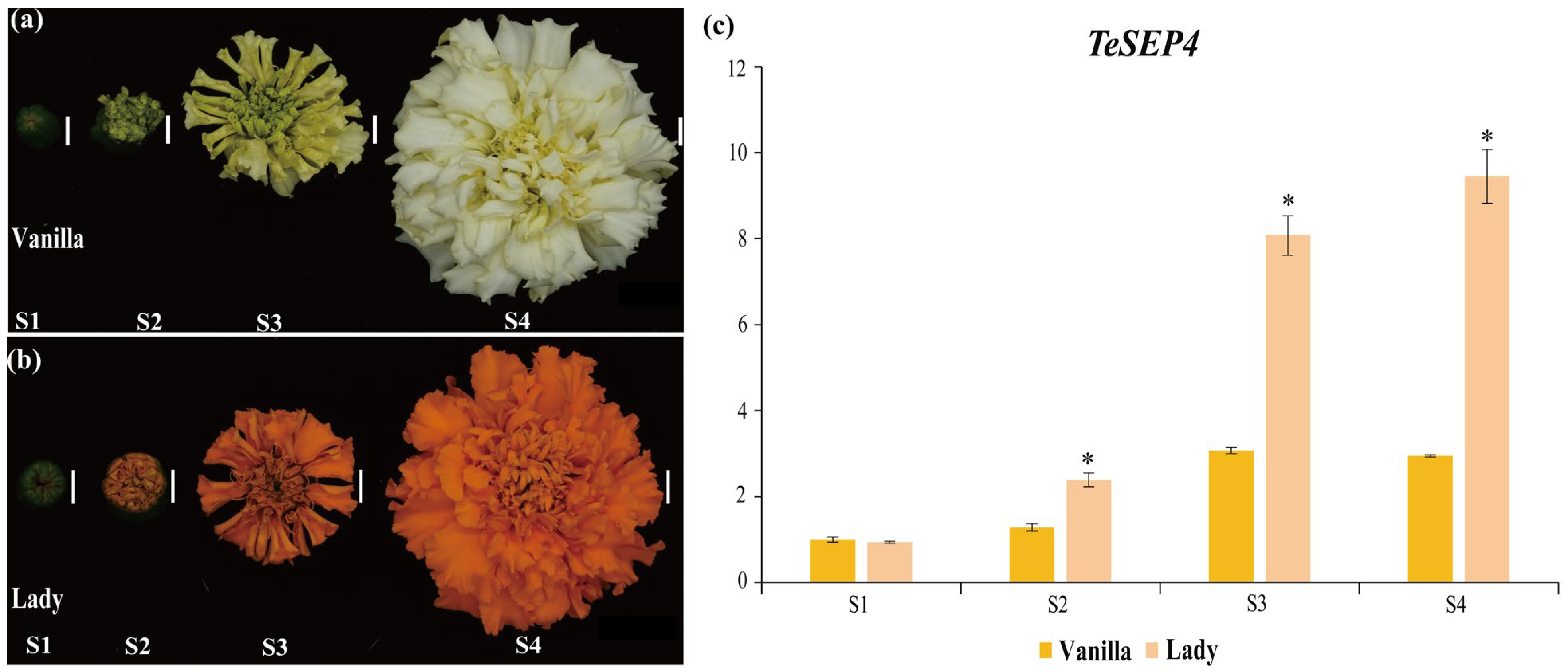
3.1. TeSEP4 Is Highly Expressed in the Orange Petals
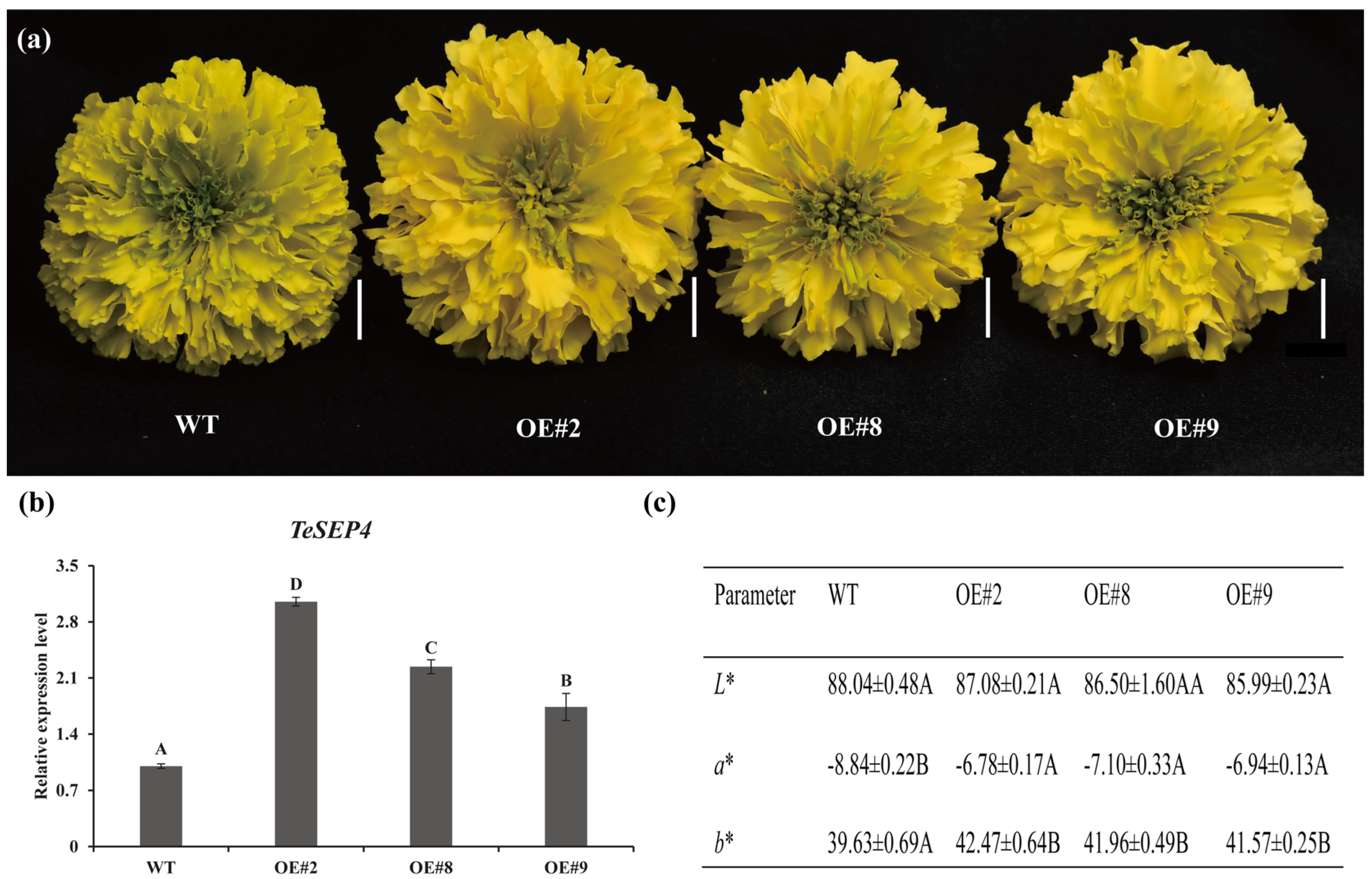
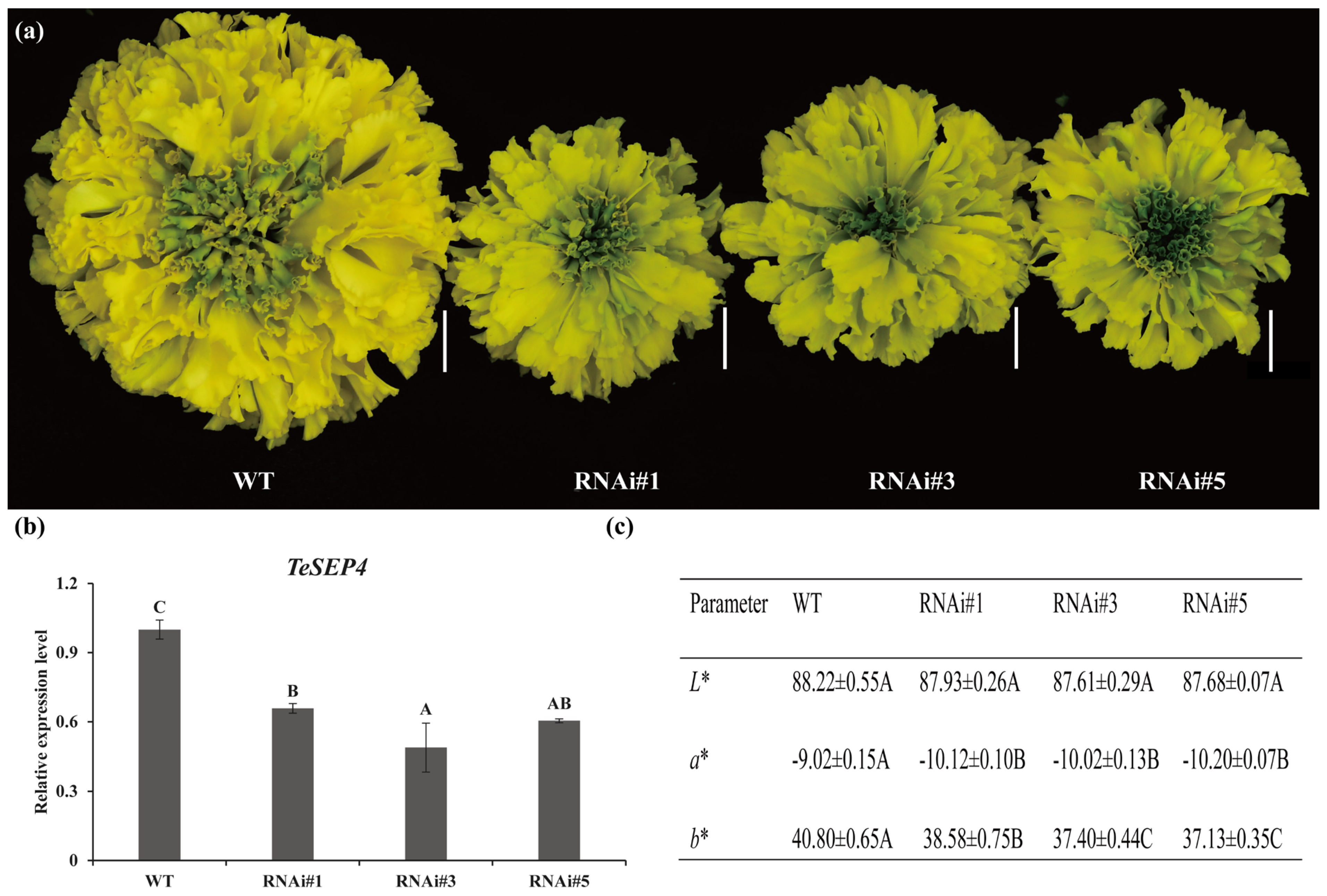
3.2. TeSEP4 Regulates the Marigold Petal Color
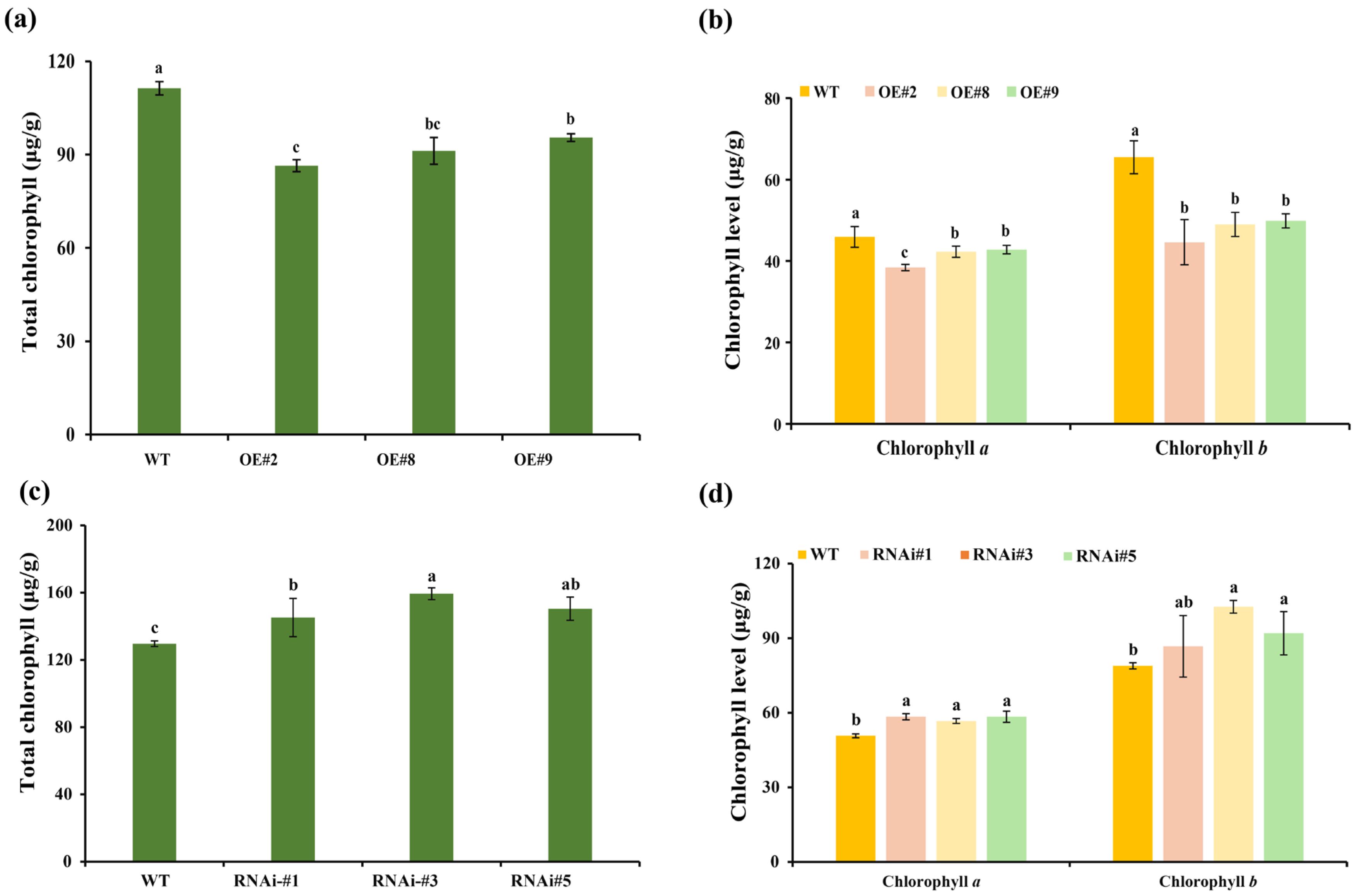
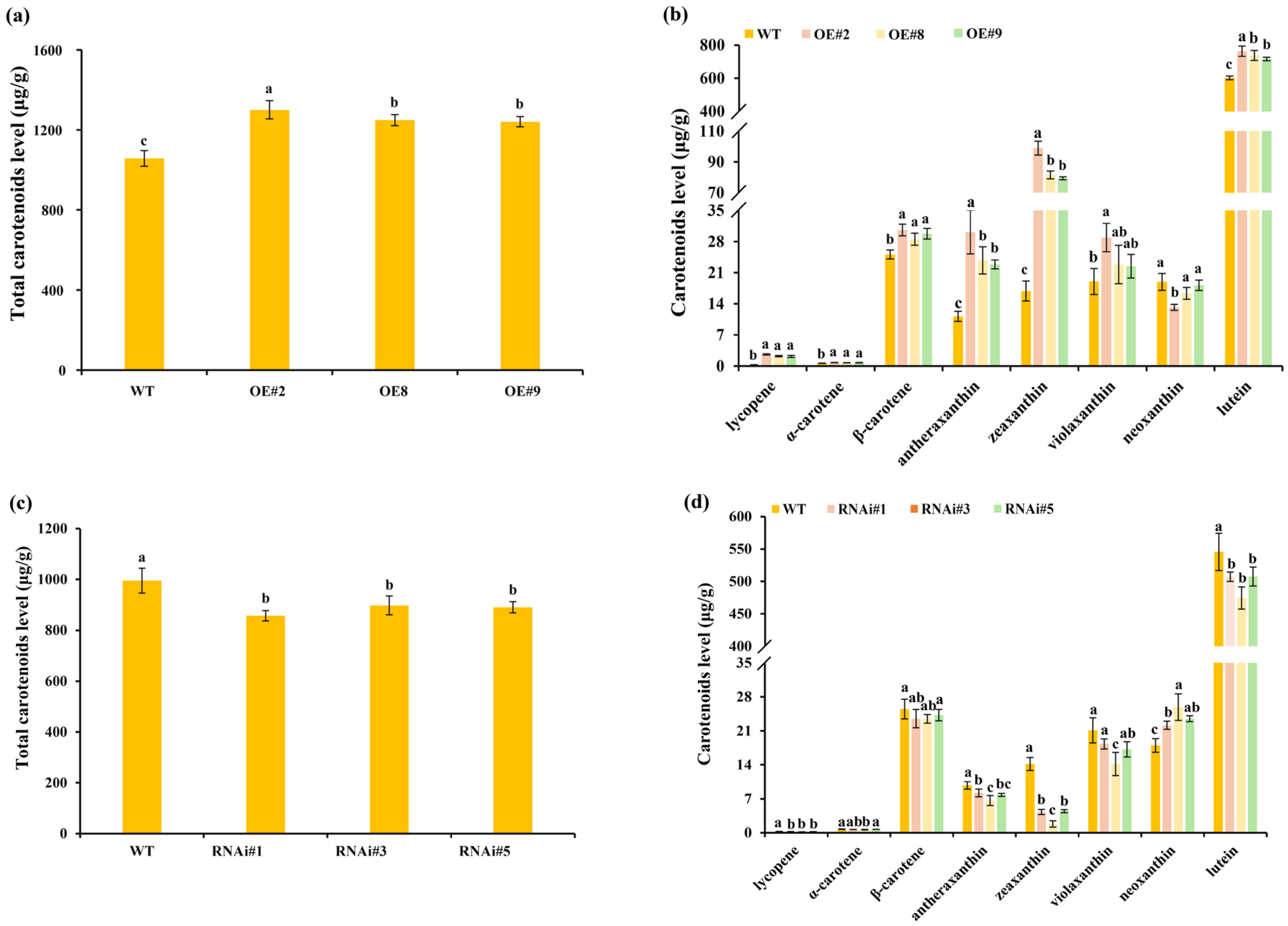
3.3. TeSEP4 Regulates the Chlorophyll and Carotenoid Accumulation in Marigold Petals
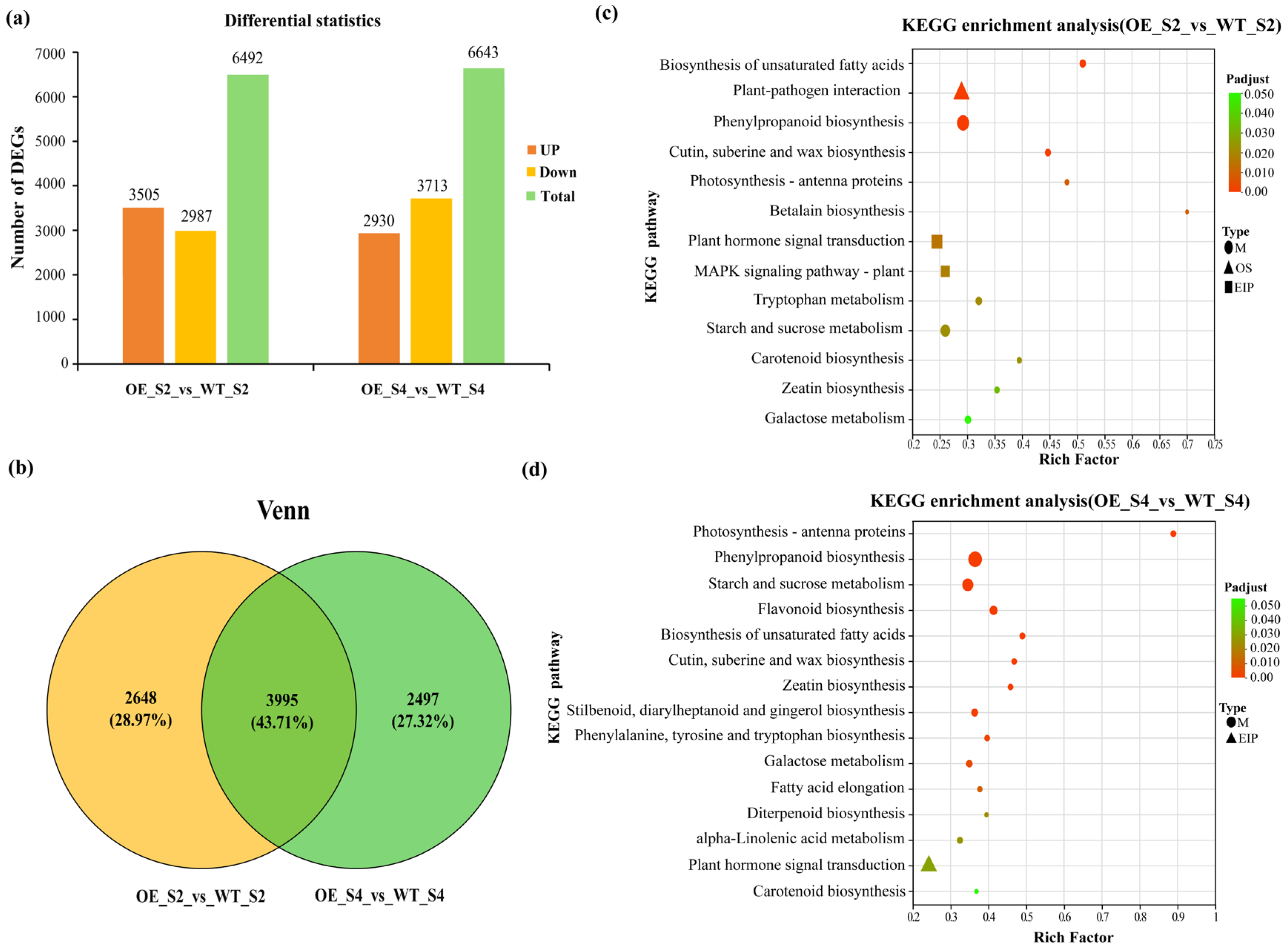
3.4. Transcriptome Sequencing and Functional Enrichment Analysis of DEGs
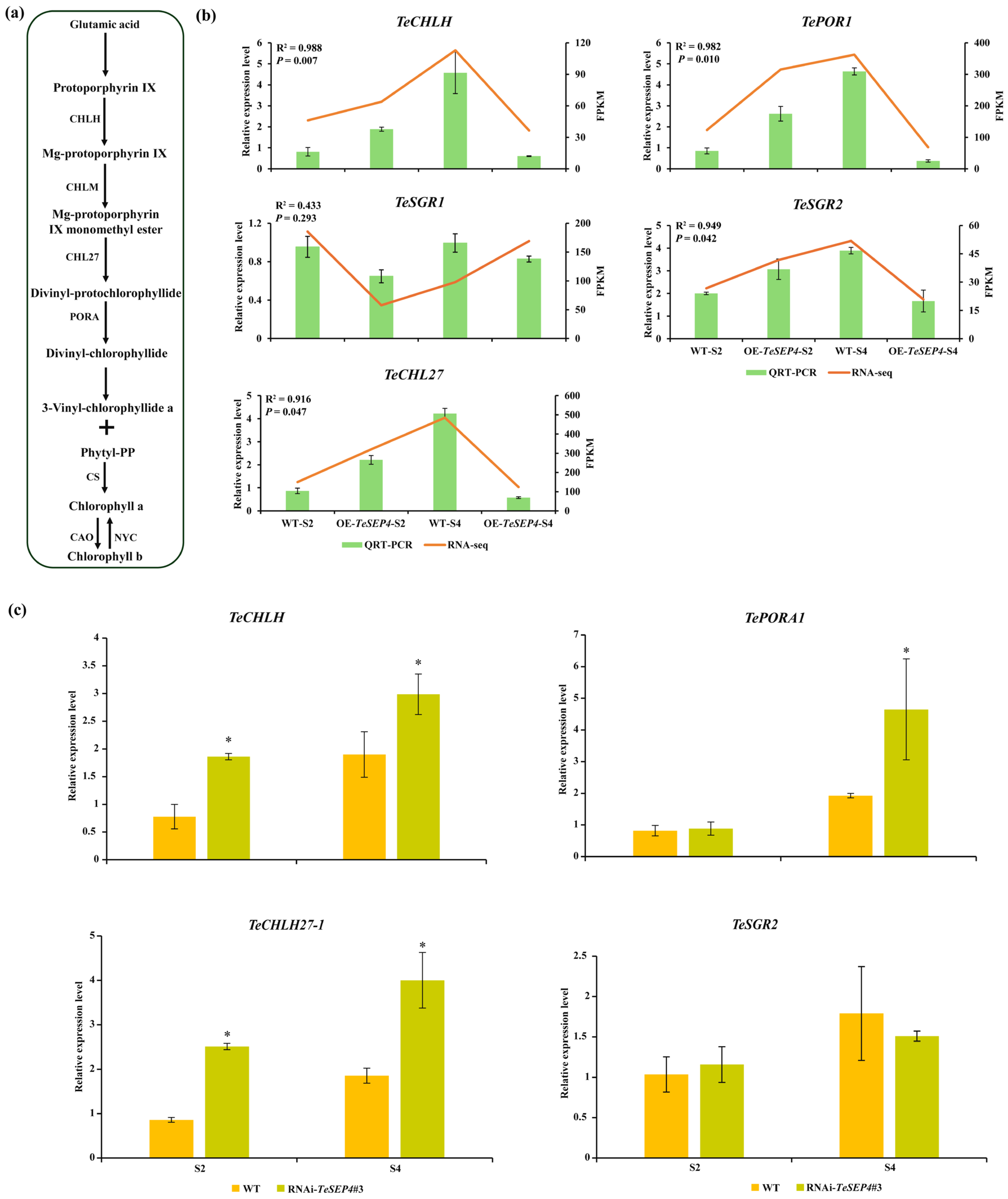
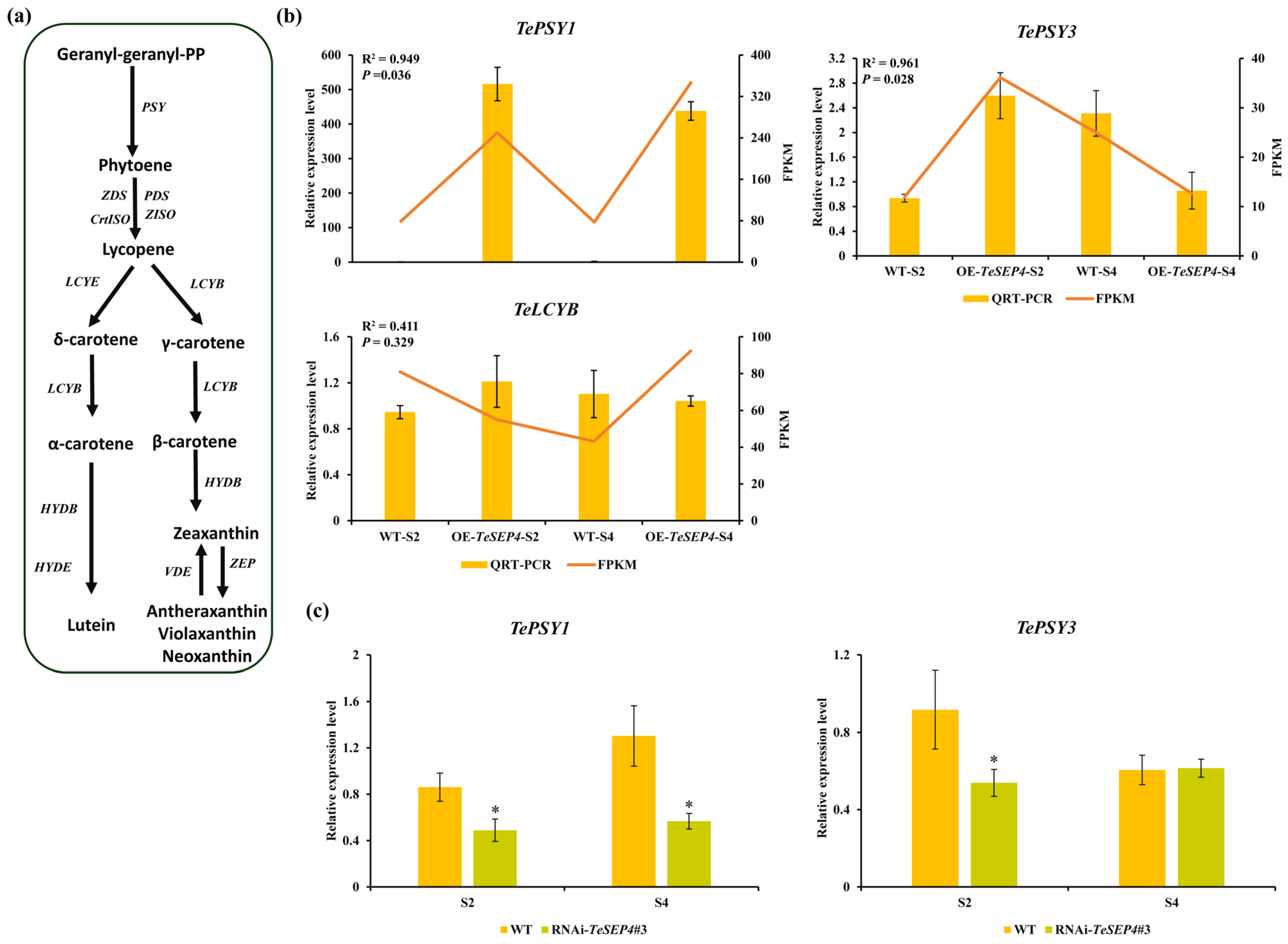
3.5. DEGs Involved in the Carotenoid and Chlorophyll Biosynthesis Pathway and Their Expression Patterns
4. Discussion
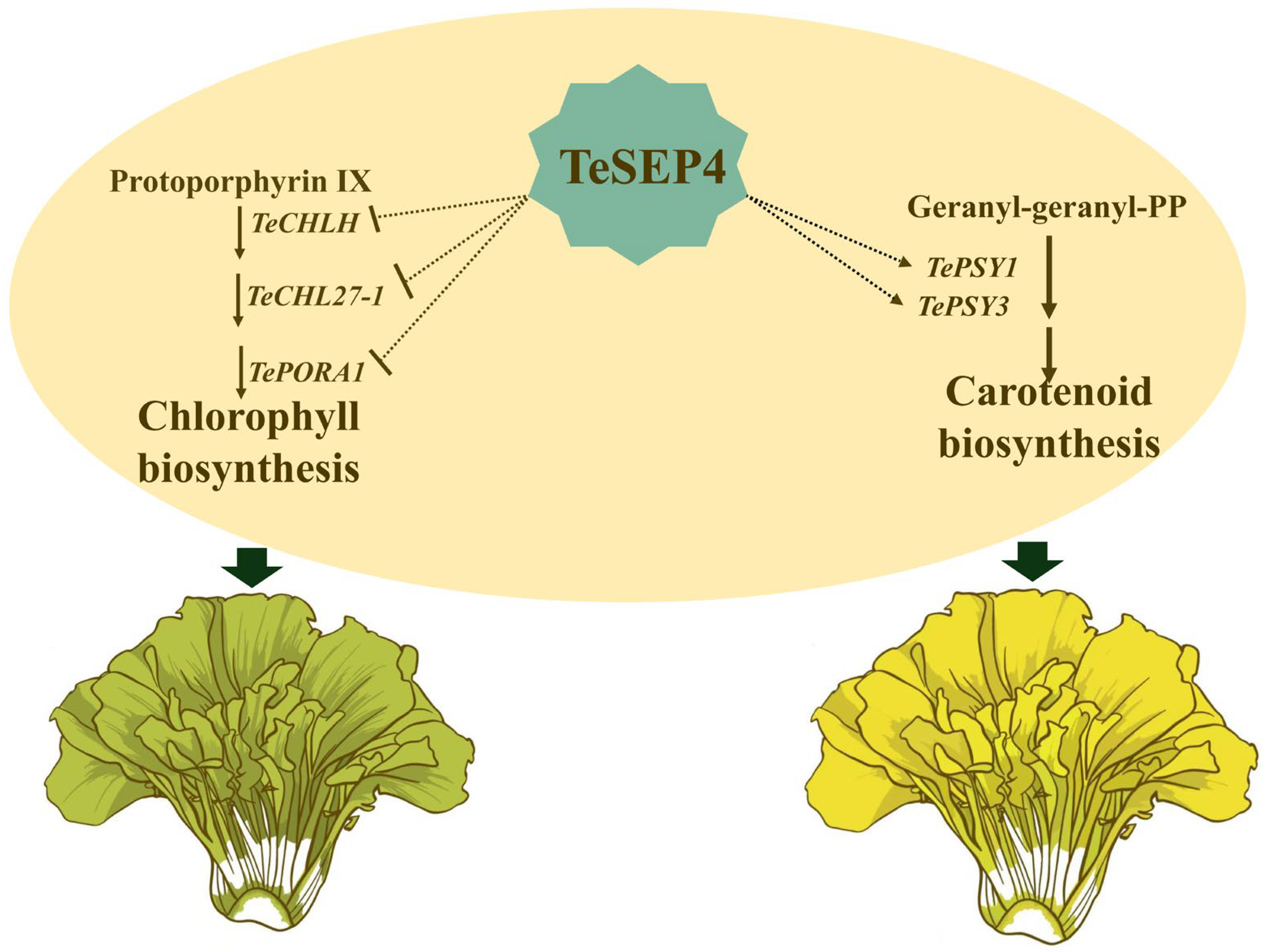
4.1. TeSEP4 Simultaneously Regulates Chlorophyll and Carotenoid Biosynthesis in Marigold Petals
4.2. TeSEP4 Might Be a Key TF Coordinating Chlorophyll and Carotenoid Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Izadpanah, F.; Frede, K.; Soltani, F.; Baldermann, S. Comparison of carotenoid, chlorophyll concentrations and their biosynthetic transcript levels in different coloured cauliflower. Hortic. Plant J. 2024, 10, 743–754. [Google Scholar] [CrossRef]
- Chen, H.; Ji, H.; Zhu, S.; Zhu, K.; Ye, J.; Deng, X. Carotenoid and transcriptome profiles of a novel citrus cultivar ‘Jinlegan’reveal mechanisms of yellowish fruit formation. Hortic. Adv. 2023, 1, 5. [Google Scholar] [CrossRef]
- Hashimoto, H.; Uragami, C.; Cogdell, R.J. Carotenoids and Photosynthesis. Subcell Biochem. 2016, 79, 111–139. [Google Scholar] [CrossRef]
- Green, B.R.; Durnford, D.G. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Biol. 1996, 47, 685–714. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Zheng, X.; Chen, X.; Wang, W.; Liu, A.; Ji, J.; Wang, G.; Guan, C. The potential roles of carotenoids in enhancing phytoremediation of bisphenol A contaminated soil by promoting plant physiology and modulating rhizobacterial community of tobacco. Chemosphere 2023, 316, 137807. [Google Scholar] [CrossRef]
- Sharma, S.; Katoch, V.; Kumar, S.; Chatterjee, S. Functional relationship of vegetable colors and bioactive compounds: Implications in human health. J. Nutr. Biochem. 2021, 92, 108615. [Google Scholar] [CrossRef] [PubMed]
- Tamiaki, H.; Kichishima, S. Chlorophyll pigments and their synthetic analogs. Plant Cell Physiol. 2025, 66, 153–167. [Google Scholar] [CrossRef]
- Martins, T.; Barros, A.N.; Rosa, E.; Antunes, L. Enhancing health benefits through chlorophylls and chlorophyll-rich agro-food: A comprehensive review. Molecules 2023, 28, 5344. [Google Scholar] [CrossRef]
- Ferruzzi, M.G.; Böhm, V.; Courtney, P.D.; Schwartz, S.J. Antioxidant and antimutagenic activity of dietary chlorophyll derivatives determined by radical scavenging and bacterial reverse mutagenesis assays. J. Food Sci. 2002, 67, 2589–2595. [Google Scholar] [CrossRef]
- Cazzonelli, C.I. Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 2011, 38, 833–847. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Van Loo-Bouwman, C.A.; Naber, T.H.; Schaafsma, G. A review of vitamin A equivalency of β-carotene in various food matrices for human consumption. Br. J. Nutr. 2014, 111, 2153–2166. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Lu, Y.; Shen, X.; Li, Y.; Xu, Y.; Chen, Y.; Chen, Y.; Hu, X.; Li, X.; Sun, X.; Gong, J. Regulation of chlorophyll and carotenoid metabolism in citrus fruit. Hortic. Plant J. 2025, 11, 951–962. [Google Scholar] [CrossRef]
- Liang, M.-H.; He, Y.-J.; Liu, D.-M.; Jiang, J.-G. Regulation of carotenoid degradation and production of apocarotenoids in natural and engineered organisms. Crit. Rev. Biotechnol. 2021, 41, 513–534. [Google Scholar] [CrossRef]
- Wang, P.; Grimm, B. Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth. Res. 2015, 126, 189–202. [Google Scholar] [CrossRef]
- Jiao, B.; Meng, Q.; Lv, W. Roles of stay-green (SGR) homologs during chlorophyll degradation in green plants. Bot. Stud. 2020, 61, 25. [Google Scholar] [CrossRef]
- Tanaka, A.; Ito, H. Chlorophyll degradation and its physiological function. Plant Cell Physiol. 2025, 66, 139–152. [Google Scholar] [CrossRef]
- Tian, S.; Yang, Y.; Fang, B.; Uddin, S.; Liu, X. The CrMYB33 transcription factor positively coordinate the regulation of both carotenoid accumulation and chlorophyll degradation in the peel of citrus fruit. Plant Physiol. Biochem. 2024, 209, 108540. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Pang, X.; Liu, W.; Wang, R.; Su, D.; Gao, Y.; Wu, M.; Deng, W.; Liu, Y.; Li, Z. SlZHD17 is involved in the control of chlorophyll and carotenoid metabolism in tomato fruit. Hortic. Res. 2021, 8, 259. [Google Scholar] [CrossRef]
- Chen, H.; Ji, H.; Huang, W.; Zhang, Z.; Zhu, K.; Zhu, S.; Chai, L.; Ye, J.; Deng, X. Transcription factor CrWRKY42 coregulates chlorophyll degradation and carotenoid biosynthesis in citrus. Plant Physiol. 2024, 195, 728–744. [Google Scholar] [CrossRef]
- Wang, J.; Shan, Q.; Yuan, Q.; Pan, L.; Wang, M.; Zhao, P.; Yu, F.; Dai, L.; Xie, L.; Wang, Z.; et al. The transcription factor CaBBX10 promotes chlorophyll and carotenoid pigment accumulation in Capsicum annuum fruit. Plant Physiol. 2025, 197, kiae592. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, W.; Lin, P.; Wang, Z.; Chen, Y.; Yang, X.; Zhao, W.; Zhang, Y.; Wang, D.; Que, Y. Evolution and function of MADS-box transcription factors in plants. Int. J. Mol. Sci. 2024, 25, 13278. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.; Ma, H.; Nei, M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, Y.; Yang, X.; Pan, Q.; Ma, G.; Bao, M.; Zheng, B.; Duanmu, D.; Lin, R.; Larkin, R.; et al. Overexpression of particular MADS-box transcription factors in heat-stressed plants induces chloroplast biogenesis in petals. Plant Cell Environ. 2019, 42, 1545–1560. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.-Z.; Zheng, T.; Liu, G.; Wang, W.; Zang, L.; Liu, H.; Yang, C. Overexpression of a MADS-Box gene from birch (Betula platyphylla) promotes flowering and enhances chloroplast development in transgenic tobacco. PLoS ONE 2013, 8, e63398. [Google Scholar] [CrossRef]
- Cong, D.; Zhao, X.; Ni, C.; Li, M.; Han, L.; Cheng, J.; Liu, H.; Liu, H.; Yao, D.; Liu, S. The SEPALLATA-like gene HrSEP1 in Hippophae rhamnoides regulates flower development by interacting with other MADS-box subfamily genes. Front. Plant Sci. 2025, 15, 1503346. [Google Scholar] [CrossRef]
- Ireland, H.S.; Yao, J.L.; Tomes, S.; Sutherland, P.W.; Nieuwenhuizen, N.; Gunaseelan, K.; Winz, R.A.; David, K.M.; Schaffer, R.J. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 2013, 73, 1044–1056. [Google Scholar] [CrossRef]
- Puranik, S.; Acajjaoui, S.; Conn, S.; Costa, L.; Conn, V.; Vial, A.; Marcellin, R.; Melzer, R.; Brown, E.; Hart, D. Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 2014, 26, 3603–3615. [Google Scholar] [CrossRef]
- Morel, P.; Chambrier, P.; Boltz, V.; Chamot, S.; Rozier, F.; Rodrigues Bento, S.; Trehin, C.; Monniaux, M.; Zethof, J.; Vandenbussche, M. Divergent functional diversification patterns in the SEP/AGL6/AP1 MADS-box transcription factor superclade. Plant Cell 2019, 31, 3033–3056. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.J.; Chen, Y.Y.; Du, J.S.; Chen, Y.Y.; Chung, M.C.; Tsai, W.C.; Wang, C.N.; Chen, H.H. Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 2014, 202, 1024–1042. [Google Scholar] [CrossRef]
- Matsubara, K.; Shimamura, K.; Kodama, H.; Kokubun, H.; Watanabe, H.; Basualdo, I.L.; Ando, T. Green corolla segments in a wild Petunia species caused by a mutation in FBP2, a SEPALLATA-like MADS box gene. Planta 2008, 228, 401–409. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Juntheikki, I.; Mouhu, K.; Broholm, S.K.; Rijpkema, A.S.; Kins, L.; Lan, T.; Albert, V.A.; Teeri, T.H. Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 2017, 216, 939–954. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Chen, J.; Ren, X.; Yuan, J.; Han, X.; Mao, L.; Ying, T.; Luo, Z. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci. Hortic. 2018, 227, 124–131. [Google Scholar] [CrossRef]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, Y.; Jia, H.; Wang, K.; Chen, S.; Ma, T.; Deng, Y.; Lang, Z.; Niu, Q. Tomato MADS-RIN regulates GAME5 expression to promote non-bitter glycoalkaloid biosynthesis in fruit. Plant J. 2024, 120, 2500–2514. [Google Scholar] [CrossRef]
- Yuan, S.; Piao, C.-L.; Zhang, X.; Cui, M.-L. Overexpression of SEPALLATA3-like Gene SnMADS37 Generates Green Petal-Tip Flowers in Solanum nigrum. Plants 2025, 14, 1891. [Google Scholar] [CrossRef]
- Chauhan, A.S.; Chen, C.-W.; Singhania, R.R.; Tiwari, M.; Sartale, R.G.; Dong, C.-D.; Patel, A.K. Valorizations of marigold waste for high-value products and their industrial importance: A comprehensive review. Resources 2022, 11, 91. [Google Scholar] [CrossRef]
- Lopresti, A.L.; Smith, S.J.; Drummond, P.D. The Effects of Lutein and Zeaxanthin Supplementation on Cognitive Function in Adults with Self-Reported Mild Cognitive Complaints: A Randomized, Double-Blind, Placebo-Controlled Study. Front. Nutr. 2022, 9, 843512. [Google Scholar] [CrossRef]
- Johra, F.T.; Bepari, A.K.; Bristy, A.T.; Reza, H.M. A Mechanistic Review of β-Carotene, Lutein, and Zeaxanthin in Eye Health and Disease. Antioxidants 2020, 9, 1046. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, K.; Huang, C.; Yue, Y.; He, Y. Identification of a carotenoid cleavage dioxygenase gene TeCCD4a regulating flower color and carotenoid content of marigold. Gene 2025, 969, 149760. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Martínez, A.A.; García-Saucedo, P.A.; Carabez-Trejo, A.; Cruz-Hernández, A.; Paredes-López, O. Carotenogenic gene expression and ultrastructural changes during development in marigold. J. Plant Physiol. 2005, 162, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Wang, R.; Zhang, E.; Shang, Y.; Feng, G.; Wang, W.; Ma, Y.; Bai, W.; Zhang, W.; Xu, Z. Carotenoid biosynthesis profiling unveils the variance of flower coloration in Tagetes erecta and enhances fruit pigmentation in tomato. Plant Sci. 2024, 347, 112207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Y.; Wang, W.; Cao, Z.; Fu, Q.; Bao, M.; He, Y. Functional analysis of the marigold (Tagetes erecta) lycopene ε-cyclase (TeLCYe) promoter in transgenic tobacco. Mol. Biotechnol. 2019, 61, 703–713. [Google Scholar] [CrossRef]
- Zhang, C.; Wei, L.; Yu, X.; Li, H.; Wang, W.; Wu, S.; Duan, F.; Bao, M.; Chan, Z.; He, Y. Functional conservation and divergence of SEPALLATA-like genes in the development of two-type florets in marigold. Plant Sci. 2021, 309, 110938. [Google Scholar] [CrossRef]
- Li, L.; Wang, X.; Zhang, X.; Guo, M.; Liu, T. Unraveling the target genes of RIN transcription factor during tomato fruit ripening and softening. J. Sci. Food Agric. 2017, 97, 991–1000. [Google Scholar] [CrossRef]
- Mitoma, M.; Kanno, A. The Greenish Flower Phenotype of Habenaria radiata (Orchidaceae) Is Caused by a Mutation in the SEPALLATA-Like MADS-Box Gene HrSEP-1. Front. Plant Sci. 2018, 9, 831. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, C.; Zhu, K.; Li, H.; Xu, S.; Tao, Z.; He, Y. Functional Conservation and Redundancy of Duplicated AGAMOUS Homologs in Regulating Floral Organ Development of Tagetes erecta. Agronomy 2025, 15, 2379. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Y.; Liu, Y.; Yi, Q.; Chen, W.; Zhu, Y.; Duan, F.; Zhang, L.; He, Y. Establishment of Agrobacterium tumefaciens-mediated Genetic Transformation System of Marigold (Tagetes erecta). Chin. Bull. Bot. 2023, 58, 760–769. (In Chinese) [Google Scholar] [CrossRef]
- Tsaniklidis, G.; Makraki, T.; Papadimitriou, D.; Nikoloudakis, N.; Taheri-Garavand, A.; Fanourakis, D. Non-destructive estimation of area and greenness in leaf and seedling scales: A case study in cucumber. Agronomy 2025, 15, 2294. [Google Scholar] [CrossRef]
- Xiong, B.; Li, L.; Li, Q.; Mao, H.; Wang, L.; Bie, Y.; Zeng, X.; Liao, L.; Wang, X.; Deng, H. Identification of photosynthesis characteristics and chlorophyll metabolism in leaves of citrus cultivar (Harumi) with varying degrees of chlorosis. Int. J. Mol. Sci. 2023, 24, 8394. [Google Scholar] [CrossRef]
- Zhang, C.; Zhu, K.; Huang, C.; Ke, L.; Wen, Y.; Li, H.; Yang, C.; Tao, Z.; He, Y. Transcription Factor TeMADS6 Coregulates Carotenoid Biosynthesis and Chlorophyll Degradation Resulting in Yellow-Green Petal Color of Marigold (Tagetes erecta). Plants 2025, 14, 3763. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Dong, B.; Fu, J.; Hu, S.; Zhao, H. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans. Front. Plant Sci. 2018, 9, 1499. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413. [Google Scholar] [CrossRef]
- Song, Y.; Cheng, Q.; Li, X.; Ma, S.; Shen, H.; Sun, L. The MADS-Box Transcription Factor CaRIN Positively Regulates Chlorophyll Degradation During Pepper (Capsicum annuum L.) Fruit Ripening by Repressing the Expression of CaLhcb-P4. Plants 2025, 14, 445. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 2021, 72, 3028–3043. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Blanc-Mathieu, R.; Janeau, A.; Paul, M.; Lucas, J.; Xu, X.; Ye, H.; Lai, X.; Le Hir, S.; Guillotin, A. SEPALLATA-driven MADS transcription factor tetramerization is required for inner whorl floral organ development. Plant Cell 2024, 36, 3435–3450. [Google Scholar] [CrossRef]
- Lv, G.; Zang, Y.; Tao, Z.; Liu, C.; Zhang, H.; Bao, M.; He, Y. The Quantitative Classification of Flower Color Phenotype and Genetic Study for Population Flower Color in Marigold. Hortic. Plant J. 2025, 52, 646–654. (In Chinese) [Google Scholar] [CrossRef]
- Zhu, K.; Chen, H.; Mei, X.; Lu, S.; Xie, H.; Liu, J.; Chai, L.; Xu, Q.; Wurtzel, E.T.; Ye, J.; et al. Transcription factor CsMADS3 coordinately regulates chlorophyll and carotenoid pools in Citrus hesperidium. Plant Physiol. 2023, 193, 519–536. [Google Scholar] [CrossRef]
- Luo, J.; Wang, H.; Chen, S.; Ren, S.; Fu, H.; Li, R.; Wang, C. CmNAC73 mediates the formation of green color in chrysanthemum flowers by directly activating the expression of chlorophyll biosynthesis genes HEMA1 and CRD1. Genes 2021, 12, 704. [Google Scholar] [CrossRef]
- Zhang, J.; Sui, C.; Liu, H.; Chen, J.; Han, Z.; Yan, Q.; Liu, S.; Liu, H. Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and tobacco (Nicotiana tabacum L.). BMC Plant Biol. 2021, 21, 45. [Google Scholar] [CrossRef]
- Hiriart, J.B.; Lehto, K.; Tyystjrvi, E.; Junttila, T.; Aro, E.M. Suppression of a key gene involved in chlorophyll biosynthesis by means of virus-inducing gene silencing. Plant Mol. Biol. 2002, 50, 213–224. [Google Scholar] [CrossRef]
- Xu, D.; Lin, L.; Liu, X.; Wangzha, M.; Pang, X.; Feng, L.; Wan, B.; Wu, G.Z.; Yu, J.; Rochaix, J.D. Characterization of a tomato chlh mis-sense mutant reveals a new function of ChlH in fruit ripening. Plant Biotechnol. J. 2025, 23, 911–926. [Google Scholar] [CrossRef]
- Hu, L.; Zhu, Y.; Yu, L.; Lu, L.; Ma, Y.; Zheng, R.; Zhang, J.; Pan, L.; Chen, J.; Hao, Z. PORA1/2-dependent chlorophyll biosynthesis coordinates with carotenoid accumulation to drive petal color patterning in Liriodendron. For. Res. 2025, 5, e013. [Google Scholar] [CrossRef]
- Tottey, S.; Block, M.A.; Allen, M.; Westergren, T.; Albrieux, C.; Scheller, H.M.S.; Jensen, P.E. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 2003, 100, 16119–16124. [Google Scholar] [CrossRef]
- Bang, W.Y.; Jeong, I.S.; Kim, D.W.; Im, C.H.; Ji, C.; Hwang, S.M.; Kim, S.W.; Son, Y.S.; Jeong, J.; Shiina, T. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol. 2008, 49, 1350–1363. [Google Scholar] [CrossRef]
- Fang, Y.; Yushuo, G.; Xiaoqin, P.; Xin, X.; Ning, Z.; Helen, C.; Guojian, H.; Mengbo, W.; Yujin, Y.; Honghai, L. BEL1-LIKE HOMEODOMAIN4 regulates chlorophyll accumulation, chloroplast development, and cell wall metabolism in tomato fruit. J. Exp. Bot. 2020, 71, 5549–5561. [Google Scholar] [CrossRef]
- Lim, C.; Kim, Y.; Shim, Y.; Cho, S.H.; Yang, T.J.; Song, Y.H.; Kang, K.; Paek, N.C. Rice OsGATA16 is a positive regulator for chlorophyll biosynthesis and chloroplast development. Plant J. 2024, 117, 599–615. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, D.; Wang, K.; Geng, X.; Guo, H.; Li, K.; Chen, B.; Guo, J.; Chu, Z.; Li, H. PSY4-mediated carotenoid biosynthesis confers yellow anther and enhances heat tolerance in cotton. Crop J. 2025, in press. [Google Scholar] [CrossRef]
- Cao, X.; Du, R.; Xu, Y.; Wu, Y.; Ye, K.; Ma, J.; Lyu, Y.; Sun, T.; Zhu, X.; Liu, Z. Phytoene synthases 1 modulates tomato fruit quality through influencing the metabolic flux between carotenoid and flavonoid pathways. Hortic. Plant J. 2024, 10, 1383–1397. [Google Scholar] [CrossRef]
- Li, J.-W.; Zhou, P.; Deng, Y.-J.; Hu, Z.-H.; Li, X.-H.; Chen, X.; Xiong, A.-S.; Zhuang, J. Overexpressing CsPSY1 gene of tea plant, encoding a phytoene synthase, improves α-carotene and β-carotene contents in carrot. Mol. Biotechnol. 2024, 66, 3311–3322. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Driedonks, N.; Lewis, D.; Shumskaya, M.; Chen, X.; Wurtzel, E.T.; Espley, R.V.; Allan, A.C. The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol. 2015, 15, 185. [Google Scholar] [CrossRef]
- Yu, Y.; Cao, Y.; Chen, Y.; Hussain, H.; Lu, X.; Zhu, K.; Xu, Y.; Feng, L.; Wei, G. Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers. Horticulturae 2025, 11, 1137. [Google Scholar] [CrossRef]
- Lisboa, M.P.; Canal, D.; Filgueiras, J.P.C.; Turchetto-Zolet, A.C. Molecular evolution and diversification of phytoene synthase (PSY) gene family. Genet. Mol. Biol. 2022, 45, e20210411. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, L.; Dong, C.; Guo, J.; Jin, L.; Wei, P.; Li, F.; Zhang, X.; Wang, R. Characterization and functional analysis of phytoene synthase gene family in tobacco. BMC Plant Biol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Fang, X.; Gao, P.; Luan, F.; Liu, S. Identification and characterization roles of phytoene synthase (PSY) genes in watermelon development. Genes 2022, 13, 1189. [Google Scholar] [CrossRef]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef]
- Villa-Rivera, M.G.; Castañeda-Marín, A.; Martínez, O.; Ochoa-Alejo, N. The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits. Plants 2025, 14, 2099. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhang, C.; Huang, C.; Zhu, K.; Ke, L.; Li, H.; He, Y. A Marigold (Tagetes erecta) MADS-Box Transcription Factor, TeSEP4, Regulates Petal Color by Modulating Chlorophyll and Carotenoid Biosynthesis. Agronomy 2026, 16, 88. https://doi.org/10.3390/agronomy16010088
Zhang C, Huang C, Zhu K, Ke L, Li H, He Y. A Marigold (Tagetes erecta) MADS-Box Transcription Factor, TeSEP4, Regulates Petal Color by Modulating Chlorophyll and Carotenoid Biosynthesis. Agronomy. 2026; 16(1):88. https://doi.org/10.3390/agronomy16010088
Chicago/Turabian StyleZhang, Chunling, Chujun Huang, Ke Zhu, Luan Ke, Hang Li, and Yanhong He. 2026. "A Marigold (Tagetes erecta) MADS-Box Transcription Factor, TeSEP4, Regulates Petal Color by Modulating Chlorophyll and Carotenoid Biosynthesis" Agronomy 16, no. 1: 88. https://doi.org/10.3390/agronomy16010088
APA StyleZhang, C., Huang, C., Zhu, K., Ke, L., Li, H., & He, Y. (2026). A Marigold (Tagetes erecta) MADS-Box Transcription Factor, TeSEP4, Regulates Petal Color by Modulating Chlorophyll and Carotenoid Biosynthesis. Agronomy, 16(1), 88. https://doi.org/10.3390/agronomy16010088







