Genome-Wide Identification and Characterization of the bZIP Gene Family in Elephant Grass (Cenchrus purpureus) and Its Response to Cold Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cold Stress Treatment
2.2. Identification of bZIP Factors in Cenchrus purpureus
2.3. Chromosomal Localization, Collinearity Analysis, and Phylogenetic Analysis of CpbZIPs
2.4. CpbZIP Analysis of Conserved Domains, Genetic Structure, Conserved Motifs, and Cis-Regulatory Elements
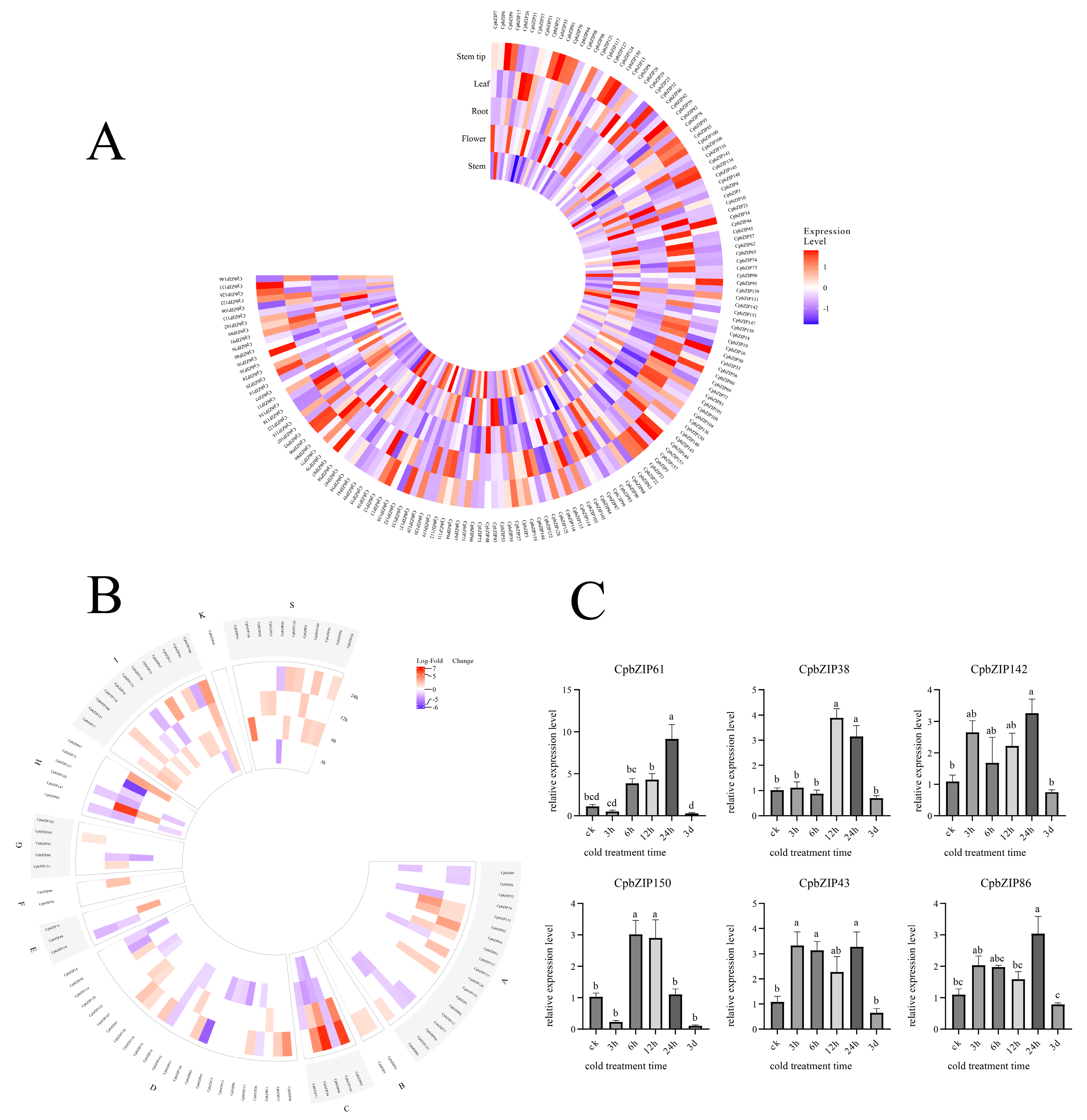
2.5. Analysis of CpbZIPs Expression Patterns in Different Tissues
2.6. Analysis of Cold Stress Transcriptome Expression Data
2.7. Analysis of CpbZIP Expression Patterns Under Low-Temperature Stress
3. Results
3.1. The CpbZIP Family Comprises 158 Members with Diverse Physicochemical Characteristics
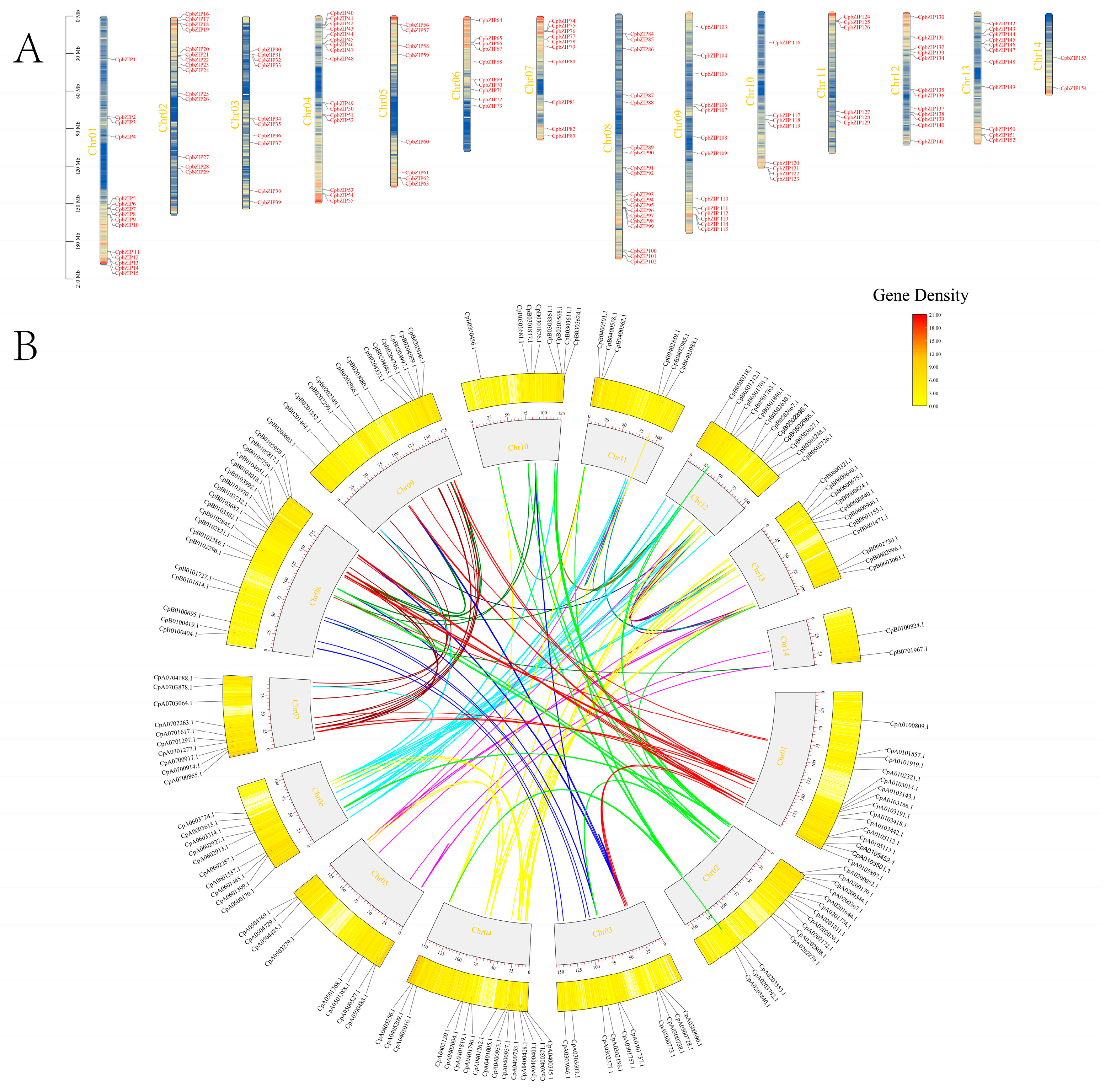
3.2. CpbZIPs Show Uneven Chromosomal Distribution and Expansion Primarily via Segmental Duplication
3.3. Phylogenetic Analysis Reveals 13 CpbZIP Subgroups and Conserved Evolutionary Relationships
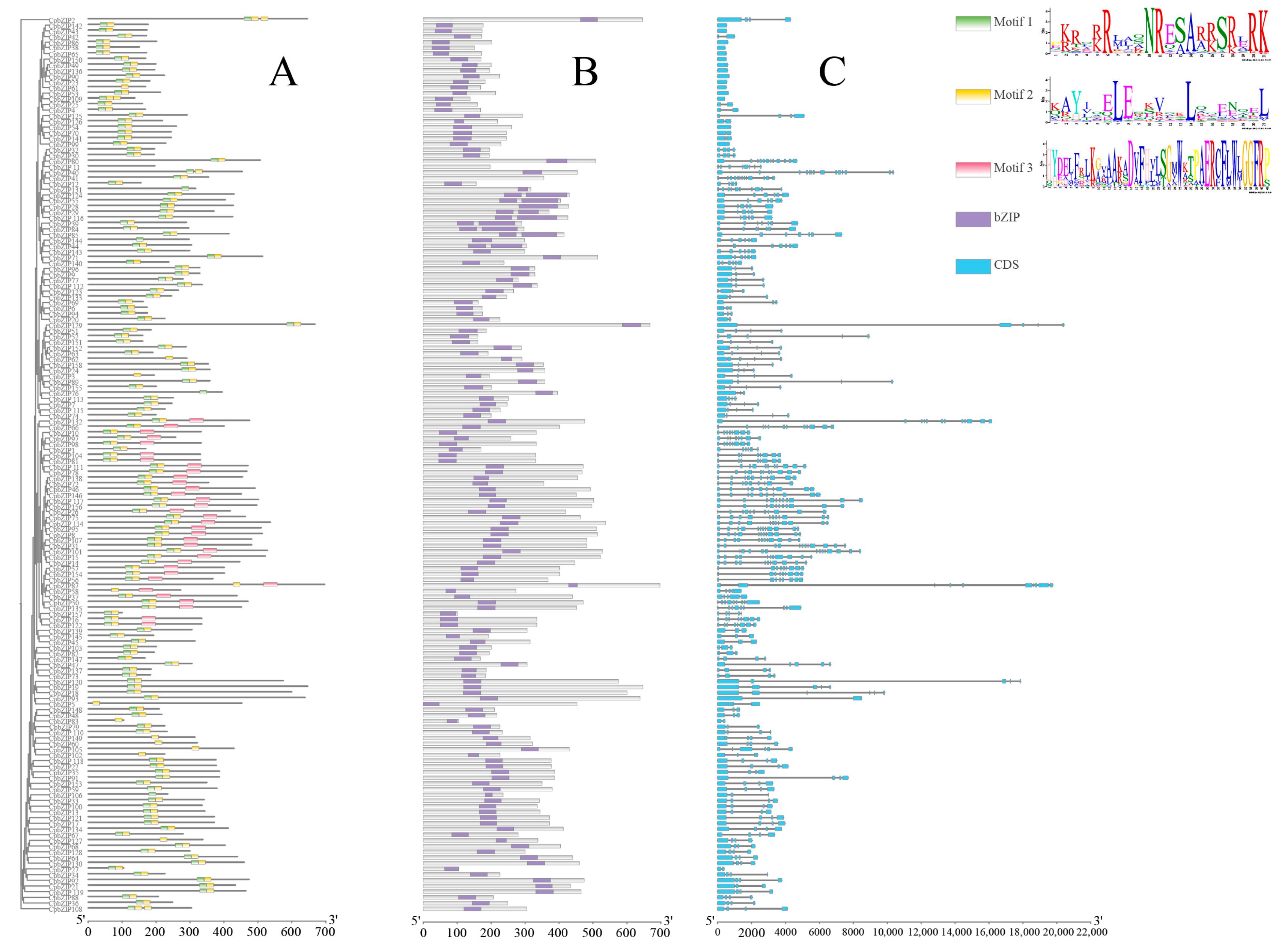
3.4. Conserved Motifs, Domains and Gene Structures Support the CpbZIP Phylogenetic Classification
3.5. Identification and Functional Classification of Cis-Acting Elements in CpbZIP Promoters
3.6. Identification of Key CpbZIP Genes Responsive to Cold Stress in Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bZIP | Basic region-leucine zipper |
| RT-qPCR | Reverse Transcription–Quantitative Polymerase Chain Reaction |
| CK | Control |
| CS | Cold Stress |
| Ka | Non-synonymous substitution rate |
| Ks | Synonymous substitution rate |
| MW | Molecular Weight |
| pI | Theoretical Isoelectric Point |
| kDa | Kilodalton |
| ML | Maximum Likelihood |
| CDS | Coding Sequence |
| ABA | Abscisic Acid |
| ABRE | Abscisic Acid Response Element |
| MeJA | Methyl Jasmonate |
| MBS | MYB Binding Site |
| ARE | Anaerobic-Related Element |
| Log2FC | Log2-Fold Change |
| TPM | Transcripts Per Million |
| EF1α | Elongator Factor-1-Alpha |
| RNAi | RNA Interference |
| SD | Standard Deviation |
| ANOVA | Analysis of Variance |
References
- Liang, X.; Erickson, J.E.; Sollenberger, L.E.; Rowland, D.L.; Silveira, M.L.; Vermerris, W. Growth and transpiration responses of elephantgrass and energycane to soil drying. Crop Sci. 2018, 58, 354–363. [Google Scholar] [CrossRef]
- Anderson, W.F.; Dien, B.S.; Brandon, S.K.; Peterson, J.D. Assessment of Bermudagrass and Bunch Grasses as Feedstock for Conversion to Ethanol. In Proceedings of the Biotechnology for Fuels and Chemicals, Denver, CO, USA, 29 April–2 May 2007; Humana Press: Totowa, NJ, USA, 2008; pp. 13–21. [Google Scholar]
- Kebede, G.; Feyissa, F.; Assefa, G.; Alemayehu, M.; Mengistu, A.; Kehaliew, A.; Melese, K.; Mengistu, S.; Tadesse, E.; Wolde, S.; et al. Agronomic performance, dry matter yield stability and herbage quality of Napier grass (Pennisetum purpureum (L.) Schumach) accessions in different agro-ecological zones of Ethiopia. J. Agric. Crop Res. 2017, 5, 49–65. [Google Scholar]
- Muktar, M.S.; Teshome, A.; Hanson, J.; Negawo, A.T.; Habte, E.; Domelevo Entfellner, J.-B.; Lee, K.-W.; Jones, C.S. Genotyping by sequencing provides new insights into the diversity of Napier grass (Cenchrus purpureus) and reveals variation in genome-wide LD patterns between collections. Sci. Rep. 2019, 9, 6936. [Google Scholar] [CrossRef]
- Adhikari, L.; Baral, R.; Paudel, D.; Min, D.; Makaju, S.O.; Poudel, H.P.; Acharya, J.P.; Missaoui, A.M. Cold stress in plants: Strategies to improve cold tolerance in forage species. Plant Stress 2022, 4, 100081. [Google Scholar] [CrossRef]
- Al-Shoaibi, A. Growth response of elephant grass (Pennisetum purpureum) and Zea mays to chilling temperature. Biosci. Biotechnol. Res. Asia 2007, 4, 389–394. [Google Scholar]
- Kour, S.; Kapil, P.; Sood, Y. Harnessing the Potential of Napier Grass (Pennisetum purpureum) for Sustainable Biofuel Production. J. Sci. Res. Rep. 2025, 31, 734–745. [Google Scholar] [CrossRef]
- Nimmanterdwong, P.; Chalermsinsuwan, B.; Østergård, H.; Piumsomboon, P. Environmental performance assessment of Napier grass for bioenergy production. J. Clean. Prod. 2017, 165, 645–655. [Google Scholar] [CrossRef]
- Rahul Kapoor, R.K. Genetic variability and association studies in Napier grass (Pennisetum purpureum Schumach.) for green fodder yield and quality traits. Electron. J. Plant Breed. 2017, 8, 885–891. [Google Scholar] [CrossRef]
- Osman, N.; Roslan, A.; Ibrahim, M.; Hassan, M. Potential use of Pennisetum purpureum for phytoremediation and bioenergy production: A mini review. Asia Pac. J. Mol. Biol. Biotechnol. 2020, 28, 14–26. [Google Scholar] [CrossRef]
- Kong, S.; Park, S.Y.; Lee, Y.H. Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 1425–1443. [Google Scholar] [CrossRef]
- Ptashne, M.; Gann, A. Transcriptional activation by recruitment. Nature 1997, 386, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Hurst, H.C. Transcription factors 1: bZIP proteins. Protein Profile 1995, 2, 101–168. [Google Scholar] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.y.; Cho, D.I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Qian, W.; Yao, L.; Hao, X.; Li, N.; Yang, Y.; Wang, X. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis. Ann. Bot. 2017, 119, 1195–1209. [Google Scholar] [CrossRef]
- Liang, Y.; Xia, J.; Jiang, Y.; Bao, Y.; Chen, H.; Wang, D.; Zhang, D.; Yu, J.; Cang, J. Genome-Wide Identification and Analysis of bZIP Gene Family and Resistance of TaABI5 (TabZIP96) under Freezing Stress in Wheat (Triticum aestivum). Int. J. Mol. Sci. 2022, 23, 2351. [Google Scholar] [CrossRef]
- Li, M.; Hwarari, D.; Li, Y.; Ahmad, B.; Min, T.; Zhang, W.; Wang, J.; Yang, L. The bZIP transcription factors in Liriodendron chinense: Genome-wide recognition, characteristics and cold stress response. Front. Plant Sci. 2022, 13, 1035627. [Google Scholar] [CrossRef]
- Liu, C.; Schläppi, M.R.; Mao, B.; Wang, W.; Wang, A.; Chu, C. The bZIP 73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019, 17, 1834–1849. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.; Liu, L.; Jia, B.; Ye, Z.; Tang, X.; Heng, W.; Liu, L. Functional analysis of PbbZIP11 transcription factor in response to cold stress in Arabidopsis and pear. Plants 2024, 13, 24. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Feng, X.; Li, Y.; Guzmán, C.; Lin, N.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; et al. Genome-wide identification of bZIP transcription factor genes related to starch synthesis in barley (Hordeum vulgare L.). Genome 2021, 64, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sun, M.; Luo, D.; Yang, D.; Zhang, Y.; Jia, J.; Ding, Q.; Najeeb, A.; Wang, X.; Ji, Y.; et al. Transcriptome profiling the response of elephant grass (Cenchrus purpureus) root to cold stress. Grass Res. 2025, 5, e014. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Paterson, A.H. Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 2012, 1256, 1–14. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A pivotal regulator of light-dependent development in higher plants. Front. Plant Sci. 2022, 12, 800989. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251. [Google Scholar]
- Khan, M.; Gemenet, D.C.; Villordon, A. Root system architecture and abiotic stress tolerance: Current knowledge in root and tuber crops. Front. Plant Sci. 2016, 7, 1584. [Google Scholar] [CrossRef]
- Meng, X.; Li, L.; De Clercq, I.; Narsai, R.; Xu, Y.; Hartmann, A.; Claros, D.L.; Custovic, E.; Lewsey, M.G.; Whelan, J.; et al. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019, 180, 634–653. [Google Scholar] [CrossRef]
- Jin, Y.; Yan, H.; Zhu, X.; Yang, Y.; Jia, J.; Sun, M.; Najeeb, A.; Luo, J.; Wang, X.; He, M.; et al. Single-cell transcriptomes reveal spatiotemporal heat stress response in pearl millet leaves. New Phytol. 2025, 247, 637–650. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Yang, R.; Fan, H.; Yang, D.; Mao, C.; Li, K.; Liu, Y.; Zhou, Z.; Zhong, Y.; Peng, S.; Jin, Y.; et al. Genome-Wide Identification and Characterization of the bZIP Gene Family in Elephant Grass (Cenchrus purpureus) and Its Response to Cold Stress. Agronomy 2026, 16, 43. https://doi.org/10.3390/agronomy16010043
Yang R, Fan H, Yang D, Mao C, Li K, Liu Y, Zhou Z, Zhong Y, Peng S, Jin Y, et al. Genome-Wide Identification and Characterization of the bZIP Gene Family in Elephant Grass (Cenchrus purpureus) and Its Response to Cold Stress. Agronomy. 2026; 16(1):43. https://doi.org/10.3390/agronomy16010043
Chicago/Turabian StyleYang, Ruiming, Hengrui Fan, Dan Yang, Chunli Mao, Kewei Li, Yuhan Liu, Zhiyao Zhou, Yun Zhong, Shiyi Peng, Yarong Jin, and et al. 2026. "Genome-Wide Identification and Characterization of the bZIP Gene Family in Elephant Grass (Cenchrus purpureus) and Its Response to Cold Stress" Agronomy 16, no. 1: 43. https://doi.org/10.3390/agronomy16010043
APA StyleYang, R., Fan, H., Yang, D., Mao, C., Li, K., Liu, Y., Zhou, Z., Zhong, Y., Peng, S., Jin, Y., He, J., Huang, L., & Yan, H. (2026). Genome-Wide Identification and Characterization of the bZIP Gene Family in Elephant Grass (Cenchrus purpureus) and Its Response to Cold Stress. Agronomy, 16(1), 43. https://doi.org/10.3390/agronomy16010043






