Abstract
Maize is a globally vital crop for both grain and forage production. Its cultivation and growth are significantly restricted by salt stress. Expansins are non-enzymatic plant cell wall proteins that play pivotal roles in growth, development, and stress responses by mediating cell wall loosening. We identified ZmEXPA6, an α-expansin gene, as exhibiting high expression levels in maize roots under salt stress. In this study, the ZmEXPA6 gene was cloned and functionally characterized. Heterologous overexpression of ZmEXPA6 promoted root elongation and enhanced salt tolerance of Arabidopsis thaliana. Under salt stress, the ZmEXPA6 overexpression lines exhibited elevated levels of anthocyanin (61.70%, 59.70%), proline (16.39%, 15.11%), soluble sugars (11.97%, 8.68%), and soluble proteins (14.83%, 13.74%) compared to the WT. Concurrently, the expression of genes associated with anthocyanin and proline biosynthesis was markedly up-regulated in these overexpression lines. The ZmEXPA6 overexpression lines exhibited elevated activities of SOD (23.81%, 23.51%), CAT (13.86%, 10.93%), and POD (4.27%, 1.39%) compared to the WT, along with significantly reduced accumulation of MDA (23.47%, 24.48%), O2− (21.9%, 19.8%), and H2O2 (27.61%, 18.07%). These results indicate that ZmEXPA6 is involved in the growth and development of Arabidopsis thaliana and improves its salt tolerance through enhanced osmotic adjustment and elevated antioxidant capacity.
1. Introduction
Maize (Zea mays L.) is a globally vital cereal crop that serves critical roles in food security, livestock feed production, and industrial processes. However, soil salinization has become a pressing global issue, currently affecting approximately 20% of arable land worldwide. Projections indicate that over 50% of irrigated soils could be threatened by salinity within the next 25 years [1]. Salinity stress, a major abiotic stress factor, significantly inhibits plant growth and development. Under salt stress, the high concentration of soluble salt in soil causes difficulties in plant roots’ water absorption, resulting in osmotic stress [2]. When salinity exceeds a critical threshold, ion imbalance and reactive oxygen species (ROS) overaccumulation in plant cells induce ionic toxicity or oxidative stress [3]. Together, these effects damage cellular integrity, disrupt metabolic homeostasis, suppress organ growth and seed germination, and may ultimately lead to plant mortality [4].
Expansin is an active protein that relaxes plant cell walls and is considered to be the most important cell wall modifier for cell elongation and growth [5]. It is induced and activated under acidic conditions in a non-enzymatic manner and disrupts hydrogen bonding between hemicellulose and cellulose microfibrils [6,7,8]. This action enhances cell wall extensibility and plasticity, thereby promoting cell expansion and tissue growth [9]. It plays crucial roles in regulating plant growth, development, and stress responses, attracting extensive scientific attention.
Extensive research has demonstrated that expansins participate in multiple facets of plant growth and development, including but not limited to root hair development, fruit ripening and softening, stem and leaf growth, seed germination, yield increase, stomatal movement, and so on [10]. Concurrently, it plays a pivotal role in plants’ defense against various abiotic stresses, particularly drought, salinity, temperature fluctuations, and heavy metal contamination [11]. Previous studies demonstrated that NtEXPA11 overexpression significantly enhanced both primary root elongation and lateral root formation under salt stress conditions [12]. Overexpression of OsEXPA7 promoted an increase in leaf cell size and number, xylem cell elongation, and salt tolerance in overexpression lines [13]. Under drought stress, overexpression of TaEXPA2 in wheat caused up-regulation of ROS-scavenging-enzyme-related genes and improved antioxidant enzyme activity and drought tolerance [14]. Overexpression of AnEXPA1 increased SOD activity and decreased MDA and H2O2 accumulation in transgenic plants under drought stress [15]. TaEXPB7-B overexpression lines showed higher soluble sugar and proline content than the WT under salt stress [16].
The market demand for maize and the harm of salt stress to agricultural production necessitate expanding salt-tolerant maize germplasm resources. The discovery of salt-tolerant genes in maize could provide a theoretical basis for rearing and improving salt-tolerant maize cultivars. In this study, we screened ZmEXPA6, a highly expressed expansin gene from a maize root transcriptome library, under 200 mM NaCl treatment (Figure S1). Salt stress significantly induced ZmEXPA6 expression, indicating its potential involvement in salt stress response regulation. In this study, the ZmEXPA6 gene was cloned successfully, and its function under salt stress was explored through phenotype, physiological index, and expression of salt stress-related genes of ZmEXPA6-overexpression Arabidopsis thaliana lines under salt stress. This study demonstrates that ZmEXPA6 can promote the growth and development of Arabidopsis thaliana and improve salt tolerance. These findings will establish a foundation for the rearing of salt-tolerant cultivars.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Columbia Arabidopsis thaliana col-0 (Columbia-0) was used as the experimental material and as the wild type (WT). The full- and uniform-size Arabidopsis seeds were selected and washed with 75% ethanol 3 times, 4 min each time. Each of the 3 times, they were washed with 95% ethanol, with 30 s of agitation. Finally, they were aseptically sowed on 1/2 MS (half-strength Murashige and Skoog media) solid medium (3% sucrose and 0.9% agar, pH 5.8). Following 48 h of vernalization at 4 °C, the Arabidopsis seedlings were cultured at a temperature of 22 °C/18 °C (day/night), a humidity of 70%, a light intensity of 600 µmol m−2 s−1, and a photoperiod of 16/8 h (day/night). The maize material used in this study was the inbred line B73. After germination, uniform seedlings were transferred to a full-strength Hoagland’s nutrient solution and pre-cultured for 7 days. For the salt stress treatment, NaCl was gradually added to the solution to achieve a final concentration of 200 mM. Root samples were collected at 0, 1, 2, 3, 4, 5, and 6 d after treatment, immediately frozen in liquid nitrogen, and stored at −80 °C for subsequent RNA extraction. The gene expression levels in the roots were analyzed using quantitative real-time polymerase chain reaction (qRT-PCR).
In order to study root phenotypes under salt stress, the three-day-old Arabidopsis seedlings growing on 1/2 MS solid medium were transferred onto 1/2 MS solid medium containing different concentrations of NaCl (0, 50, 100, and 150 mM) for 10 days. The root length was measured, and tissue samples were collected for the quantification of associated gene expression levels. In order to study the difference between the growth phenotype in the WT and in overexpression lines under salt stress, the seven-day-old Arabidopsis seedlings germinated on 1/2 MS medium were transplanted into 10 cm × 10 cm pots (four plants per pot) containing matrix (soil: vermiculite: perlite, 3:1:1) and irrigated with NaCl solution every 3 days. Twenty-day-old seedlings were treated with different concentrations of NaCl (0, 50, 100, 150 mM) for 15 days. Each treatment group had five pots. Each pot was irrigated with 100 mL treatment solution every 3 days. Subsequently, images of seedlings were captured. The fresh weight and dry weight of seedlings were measured, and samples were taken for physiological index determination.
2.2. Bioinformatics Analysis
Protparam (https://web.expasy.org/translate/, (accessed on 2 December 2024)) and Prabi (https://npsa-prabi.ibcp.fr/cgi-bin/secpred_hnn.pl, (accessed on 2 December 2024)) were used to predict the primary and secondary structures of ZmEXPA6; SMART (https://swissmodel.expasy.org/, (accessed on 2 December 2024)) was used to predict the conserved domain of the protein; TMHMM (http://www.cbs.dtu.dk/services/TMHMM/, (accessed on 2 December 2024)) was used to predict transmembrane domains; and ProtScale (https://web.expasy.org/protscale/, (accessed on 2 December 2024)) was used to predict protein hydrophilicity and hydrophobicity. The BLASTX tool was used on NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 2 December 2024) to obtain sequences with high homology to ZmEXPA6, and the amino acid sequence of ZmEXPA6 was analyzed for homology using DNAMAN7.0 software. At the same time, we used MEGA11.0 software to create a phylogenetic tree of ZmEXPA6 with the neighbor-joining method.
2.3. Gene Cloning
RNA extraction from maize B73 tissues was performed using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China), with subsequent cDNA synthesis conducted using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The open reading frame (ORF) sequence of ZmEXPA6 was obtained from MaizeGDB and used to design gene-specific primers (Table S1) with Primer Premier 5.0 software. The PCR amplification of ZmEXPA6 was performed using 2 × Pro Taq Master Mix DNA polymerase (Vazyme, Nanjing, China) with cDNA as the template and ZmEXPA6-S/A as the specific primers.
2.4. Vector Construction and Arabidopsis Transformation
The ZmEXPA6 sequence was ligated into the pCAMBIA3301 vector (CaMV 35S promoter) using the In-Fusion HD Cloning Kit (TaKaRa, Beijing, China), and the recombinant plasmid was subsequently transformed into Agrobacterium (GV3101). The primers ZmEXPA6-F/R were used for colony PCR verification. The Agrobacterium-mediated floral dip method [17] was used to perform transformation. The homozygous Arabidopsis thaliana lines were retained for qRT-PCR after three successive generations of screening using herbicides.
2.5. Identification of ZmEXPA6 Overexpression Lines
In order to screen and identify heterologous ZmEXPA6 overexpression lines, we used genomic DNA to identify positive transgenic plants. The sequence of primers (ZmEXPA6-F/R) are listed in Table S1. Then, qRT-PCR was carried out to determine the relative expression level of ZmEXPA6 in heterologous expression lines (ZmEXPA6-qF/qR as primers, Table S1). The Arabidopsis Actin gene (primers AtActin-F/R) was used as an internal reference, and each line was repeated three times. Based on the final results, the line with the highest expression level (OE1) and the line with the second-highest expression level (OE2) were selected for subsequent experiments.
2.6. Determination of Physiological Indices
Seedlings of WT and overexpression lines treated with varying NaCl concentrations (0, 50, 100, or 150 mM) for 15 days were used to determine physiological indices. Fresh leaves were frozen in liquid nitrogen and stored at −80 °C for determination of physiological indices of salt tolerance. The contents of proline, soluble protein, and soluble sugars were determined using the acid ninhydrin method, Coomassie Brilliant Blue assay, and the 3,5-dinitrosalicylic acid (DNS) method, respectively. The specific operation steps were carried out according to the kit instructions of Solarbio (Beijing, China). Samples were ground into a fine powder using liquid nitrogen. Subsequently, 0.1 g of the powdered tissue was homogenized in 0.5 mL of phosphate-buffered saline (PBS) on ice. The homogenate was centrifuged at 8000× g and 4 °C for 10 min, and the resulting supernatant was collected for determination of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activities, according to the method described by Xu et al. [18].
The contents of superoxide anion (O2−) and hydrogen peroxide (H2O2) were determined by hydroxylamine oxidation assay and titanium sulfate spectrophotometry, respectively [19]. Leaves (0.1 g) frozen rapidly with liquid nitrogen were briefly ground with 1 mL extracting solution. The homogenates were centrifuged at 10,000× g for 15 min at 4 °C to obtain the supernatant. For the O2− assay, the supernatant was reacted with hydroxylamine hydrochloride, followed by sulfanilic acid and α-naphthylamine. The absorbance was measured at 530 nm. For the H2O2 assay, the supernatant was mixed with titanium reagent to form a yellow peroxide–titanium complex. After centrifugation, the sediment was dissolved in 2 M sulfuric acid, and its absorbance was read at 405 nm. These steps were informed by the manufacturer’s instructions (Solarbio, Beijing, China).
The contents of malondialdehyde (MDA) was determined by thiobarbituric acid (TBA) assay [20]. Leaves (0.1 g) were added into 1 mL extracting solution and ground thoroughly. The homogenates were centrifuged at 10,000× g for 15 min at 4 °C to obtain the supernatant. Then, 0.5 mL of the supernatant was mixed with 1.5 mL of TBA reagent in a test tube. The mixture was heated in a boiling water bath for 30 min, quickly cooled on ice, and centrifuged again. The absorbance of the supernatant was measured at 532 nm and 600 nm. These steps were guided by the manufacturer’s instructions (Solarbio, Beijing, China).
The anthocyanin content was measured using the method established by Qin et al. [21]. Leaves (0.3 g) were added into 300 mL extract consisting of methanol and hydrochloric acid (99:1, v/v) and ground thoroughly. The homogenate was incubated at 4 °C for 24 h, and then centrifuged at 10,000× g for 10 min to obtain the supernatant. The absorbance of the supernatant was measured at 530 nm and 657 nm.
2.7. qRT-PCR Analysis of Related Genes
Total RNA was isolated from Arabidopsis thaliana tissues using the FastPure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China), followed by cDNA synthesis with HiScript III RT SuperMix for qRT-PCR (+gDNA wiper). We utilized ChamQ Universal SYBR qRT-PCR Master Mix (Vazyme, Nanjing, China) to set up a 20 μL reaction system with cDNA as the template and Arabidopsis Actin gene (primers AtActin-F/R) as the internal reference. Each sample was analyzed with three biological replicates and three technical replicates. The qRT-PCR experiments were conducted using the 7500 Software v2.0.5, and gene expression levels were calculated and analyzed utilizing the 2−ΔΔCT method. The primers of anthocyanin synthesis-related genes and salt tolerance-related genes used in this paper are shown in Table S1.
2.8. Morphological Analysis of Arabidopsis Root Cells
After 10 days of 0 and 100 mM NaCl treatment on 1/2 MS solid medium, the root maturation zone of WT and overexpression Arabidopsis seedlings were taken out and placed in FAA (Formalin-Aceto-Alcohol) fixative solution for 4~12 h at 4 °C. Then, samples were removed from the FAA fixative and placed on slides. HCG transparent solution (80 g chloral hydrate, 10 mL 100% glycerol and 30 mL H2O) was added to the slides, and the samples were covered with cover slips for transparent treatment for 24 h. The root epidermal cells were viewed and captured using a DIC microscope (Nikon, Tokyo, Japan). A total of 15 plants were selected from each line, and three independent replicates were conducted. The area of the same part (1–2 cm from the root tip) was selected, and the number and area of root cells were calculated using ImageJ1 software.
2.9. Statistical Analysis
A minimum of three biological and three technical replicates were included in all experiments. Statistical analyses were conducted using IBM SPSS Statistics 25.0. Significant differences (p < 0.05) between groups are indicated by different lowercase letters based on multiple comparison tests.
3. Results
3.1. Characterization of ZmEXPA6
ZmEXPA6 consists of an 837 bp open reading frame (ORF) encoding a 278 amino acid with the relative molecular weight of the protein 30,051.26 Da (C1320H2079N391O386S14) (Figure S2A). The secondary structure contains three secondary structure elements, namely, alpha helix (29.14%), extended strand (17.63%), and random coil (53.24%), among which random coil accounts for the largest proportion (Figure S2B). The protein encoded by ZmEXPA6 is a hydrophilic protein (Figure S2C) without a transmembrane domain (Figure S2E). There is a signal peptide at amino acids 2–29 and a protein domain of DPBB_1 at amino acids 68–144 (Figure S2D). In order to study the evolutionary relationship of the ZmEXPA6 protein, we constructed a phylogenetic tree using homologous amino acid sequences. ZmEXPA6 has the highest evolutionary relationship with sorghum (Sorghum bicolor L.) protein (Figure 1A). The amino acid sequence alignment results of ZmEXPA6 and its homologous proteins showed that ZmEXPA6 had the highest homology with Sorghum bicolor expansin-like A1, which was 90.48% (Figure 1B).

Figure 1.
Phylogenetic analysis and homologous sequence alignment of ZmEXPA6. (A) Phylogenetic tree of ZmEXPA6. The branch length represents the evolutionary distance between species, and the bar represents 0.02 substitutions per amino acid site. (B) Sequence alignment of homologous amino acids between ZmEXPA6 and different species. The DPBB_1 domain sequence is in the red box.
3.2. Overexpression of ZmEXPA6 Enhanced the Growth of Arabidopsis thaliana Under Salt Stress
By genomic DNA PCR and qRT-PCR (Figure 2A), we selected OE1 and OE2, which had the highest and second-highest expression levels of ZmEXPA6, for the experiments. In order to analyze the function of ZmEXPA6 under salt stress, we treated WT and overexpression Arabidopsis lines with varying NaCl concentrations (0, 50, 100, and 150 mM) using 1/2 MS medium. On the 10th day, the root length was measured. In the standard 1/2 MS, the root length of OE1 and OE2 increased by 16.98% and 14.01% compared with WT (Figure 2C). Under salt stress, the root length of WT was significantly lower than that of OE1 and OE2, and the inhibition degree was significantly higher than that of OE1 and OE2. Compared with normal conditions, the relative reduction ratio of WT under 100 mM NaCl conditions was 56.06%, while the relative reduction ratios of OE1 and OE2 were 36.16% and 39.09%, respectively (Figure S3). This indicates that heterologous overexpression of ZmEXPA6 promoted root elongation in Arabidopsis under salt stress.
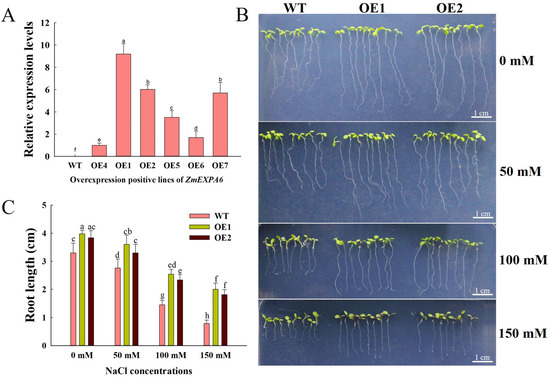
Figure 2.
Root growth under different concentrations of NaCl. (A) The relative expression level of the ZmEXPA6 gene in overexpression lines was detected by qRT-PCR. (B) Root length phenotypes of WT and overexpression lines treated with varying concentrations of NaCl (0, 50, 100, and 150 mM) for 10 days. (C) The root length of seedlings cultured on 1/2 MS with different concentrations of NaCl for 10 days. The data is the mean ± SD of the three replicates, and the significant differences of multiple comparisons at the p < 0.05 level between the groups are represented by different letter labels.
The growth of all lines declined markedly with increasing salt concentrations. However, OE1 and OE2 seedlings exhibited a more robust growth phenotype compared to WT (Figure 3A). Specifically, under normal conditions, the fresh weight of OE1 and OE2 increased by 18.83% and 14.11%, respectively, relative to WT (Figure 3B), while their dry weight rose by 20.69% and 18.58%, respectively (Figure 3C). After salt stress, the growth of WT, OE1, and OE2 was inhibited, but OE1 and OE2 still had higher growth and biomass than WT (Figure 3A). The results show that overexpression of ZmEXPA6 promoted the growth and development of Arabidopsis thaliana and increased its tolerance to salt stress.
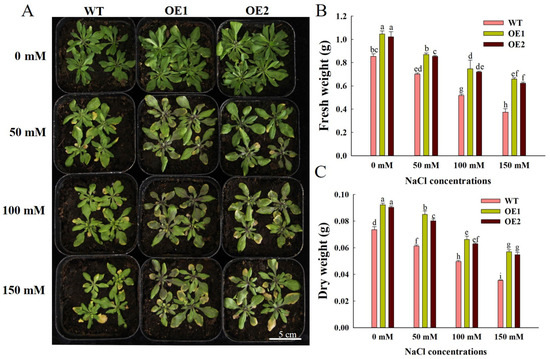
Figure 3.
The phenotypic characterization and biomass statistics of Arabidopsis under different concentrations of NaCl treatment. (A) Phenotypes treated with different concentrations of NaCl for 15 days. (B) Fresh weight. (C) Dry weight. The data of fresh weight and dry weight were taken as the average value ± SD of 10 plants for each line, and the significant differences of multiple comparisons at the p < 0.05 level between the groups were represented by different letter labels.
3.3. Overexpression of ZmEXPA6 Promoted the Elongation of Arabidopsis Root Cells
Overexpression of ZmEXPA6 promoted root elongation of Arabidopsis. To investigate the functional role of ZmEXPA6 in root development, we observed the root cells of each line. Under normal conditions, the average root cell area was 5480 μm2 in OE1 and 4104 μm2 in OE2. The average root cell length was 144.45 μm in OE1 and 126.63 μm in OE2. However, the cell area (2763 μm2) and cell length (82.06 μm) of the WT were significantly smaller than those of the OE lines (Figure 4B,C). Under salt stress, root cell development in each line was significantly inhibited. However, the length and area of root cells in OE1 and OE2 were still significantly greater than those in WT. Under salt stress, the relative reduction rates of root cell length of OE1 and OE2 were 13.8% and 14.1%, respectively, and that of WT was 15.2%. The results show that the overexpression of ZmEXPA6 increased the size of root cells, thus promoting the elongation of roots and improving the salt tolerance of overexpression lines.
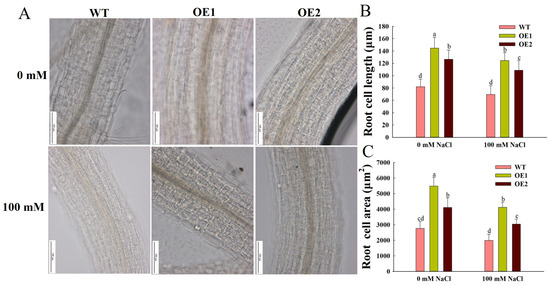
Figure 4.
The development of root cells under salt stress. (A) The development of root cells treated with varying NaCl concentrations (0 and 100 mM) for 10 days. (B) Root cell length. (C) Root cell area. The root cell length and area data were from the same part of the region (1–2 cm from the root tip), and the average ± SD of 15 plants in each line was taken. Statistically significant differences between groups (p < 0.05) are indicated by different lowercase letters.
3.4. Overexpression of ZmEXPA6 Promoted Anthocyanin Synthesis in Arabidopsis Under Salt Stress
Anthocyanin is a natural plant pigment that gives various colors to plant tissues when plants are stressed. The accumulation of anthocyanins can significantly enhance the salt tolerance of Arabidopsis [22]. Under normal conditions, there was no color difference between WT, OE1, and OE2. After 10 days of treatment on 1/2 MS with 100 mM NaCl, the leaves and stems of OE1 and OE2 became purple, while WT had no significant color change (Figure 5A). Under control growth conditions, there was no difference between anthocyanin levels in WT and overexpression Arabidopsis lines. After salt treatment, the anthocyanin levels in OE1 and OE2 increased by about 61.7% and 59.7% compared with WT (Figure 5B). This is in agreement with the observed phenotypic results.
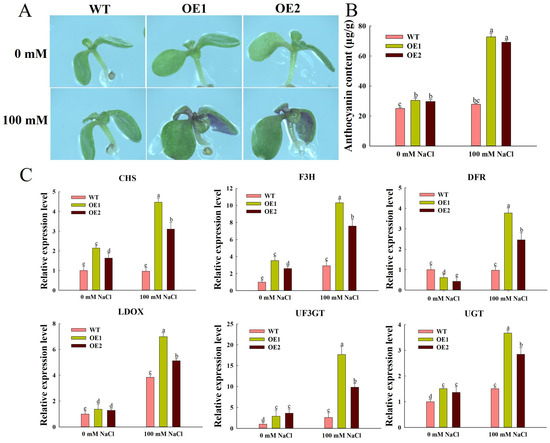
Figure 5.
Anthocyanin synthesis under normal and salt stress. (A) The color changes of WT and ZmEXPA6 overexpression lines treated with 0 and 100 mM NaCl for 10 days. (B) Anthocyanin content. Seedlings treated with 0 and 100 mM NaCl for 10 days were measured for anthocyanin content. (C) Transcription levels of CHS, F3H, DFR, LDOX, UF3GT, and UGT in seedlings treated with 0 and 100 mM NaCl. The data were the mean ± SD of 3 replicates, and different letters were used to indicate the significant difference of multiple comparisons between groups at the p < 0.05 level.
We employed qRT-PCR to analyze the expression levels of key anthocyanin biosynthesis-related genes in Arabidopsis under salt stress conditions [23]. After salt stress, the transcription levels of AtCHS, AtF3H, and AtDFR in OE1 were 4.61-fold, 3.53-fold, and 3.88-fold higher than those in WT, respectively. In OE2, the transcription levels of AtCHS, AtF3H, and AtDFR were elevated by 3.20-, 2.60-, and 2.54-fold relative to WT. The expression levels of AtLDOX and AtUF3GH were also significantly higher than those of the WT. This indicates that overexpression of ZmEXPA6 promoted anthocyanin synthesis in Arabidopsis under salt stress (Figure 5C).
3.5. Overexpression of ZmEXPA6 Increased the Accumulation of Osmotic Adjustment Substances Under Salt Stress
Under salt stress, excessive salinity in soil will reduce water potential in soil and cause osmotic stress [24]. In response, plants accumulate a large amount of osmotic adjustment substances to resist this osmotic stress [25]. Therefore, we measured the contents of proline, soluble sugar, and soluble protein in WT, OE1, and OE2. Under normal growth conditions, there was no significant difference between WT, OE1, and OE2 (Figure 6A–C). Under salt stress, the contents of proline, soluble sugar, and soluble protein in each line increased with the increase in salt concentration. However, compared with WT, the proline content of OE1 and OE2 increased by 16.39% and 15.11% on average, the soluble sugar content increased by 11.97% and 8.68% on average, and the soluble protein content increased by 14.83% and 13.74% on average under the 100 mM NaCl treatment (Figure 6A–C).
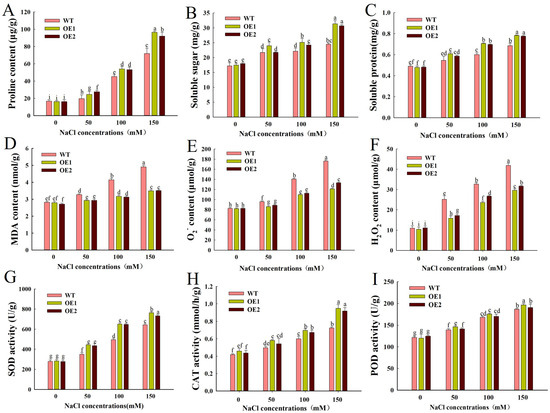
Figure 6.
Physiological indexes under normal and salt stress. (A) Proline content. (B) Soluble sugar content. (C) Soluble protein content. (D) MDA content. (E) O2− content. (F) H2O2 content. (G) SOD activity. (H) CAT activity. (I) POD activity. The data were the average value of 3 replicates ± SD, and different letters were used to indicate the significant difference of multiple comparisons between groups at the level of p < 0.05.
3.6. Overexpression of ZmEXPA6 Enhanced the Antioxidant Capacity of Arabidopsis Under Salt Stress
Excessive accumulation of reactive oxygen species such as O2− and H2O2 under salt stress can cause oxidative damage in plants. MDA serves as a critical biomarker for assessing the extent of cell membrane lipid peroxidation. Therefore, we examined the content of MDA, O2−, and H2O2 in each line under salt stress. Under normal conditions, there was no significant difference in the content of MDA, O2−, and H2O2 in each line, but the content of each line increased significantly after salt stress. Under the 100 mM NaCl treatment, OE1 and OE2 exhibited significantly lower levels of MDA, O2−, and H2O2 compared with WT. Specifically, MDA content decreased by 23.47% and 24.48% (Figure 6D), O2− levels decreased by 21.9% and 19.8% (Figure 6E), and H2O2 content decreased by 27.61% and 18.07% (Figure 6F). This indicates that overexpression of ZmEXPA6 caused less damage to Arabidopsis under salt stress.
Under control conditions, SOD, CAT, and POD activities showed no significant differences among WT, OE1, and OE2 lines. After salt stress treatment, both OE1 and OE2 showed significantly higher SOD and CAT activities compared to WT. Under the 100 mM NaCl treatment, compared with WT, SOD activity in OE1 and OE2 increased by 23.81% and 23.51% (Figure 6G); CAT activity increased by 13.86% and 10.93% (Figure 6H). Overexpression of ZmEXPA6 enhanced antioxidant enzyme activity, thereby increasing the antioxidant capacity of overexpression Arabidopsis under salt stress.
3.7. The Expression Level of Salt Stress-Related Genes
To elucidate the molecular mechanisms underlying ZmEXPA6-mediated salt stress tolerance, we analyzed the expression profiles of five key stress-responsive genes involved in ion homeostasis (AtSOS1, AtSOS2, and AtSOS3), antioxidant defense (AtCAT1), and proline metabolism (AtP5CS1). Quantitative analysis revealed that salt stress significantly up-regulated the expression of all five stress-responsive genes in both ZmEXPA6 overexpression lines and WT lines. Under control conditions, there was no significant difference between the transcription levels of AtSOS1, AtSOS2, AtSOS3, and AtCAT1 (Figure 7A–C), while the transcription level of AtP5CS1 was significantly higher than that of WT—the levels in OE1 and OE2 were 3.60 and 3.24 times higher, respectively, than in WT (Figure 7D). Notably, under salt stress conditions, the ZmEXPA6 overexpression lines exhibited significantly higher transcription levels of all examined genes compared to WT lines.
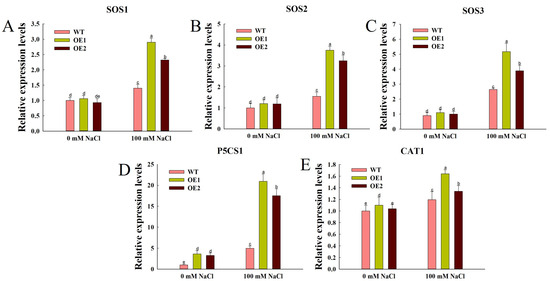
Figure 7.
Transcription level of salt stress response genes under normal and salt stress treatment. (A) SOS1. (B) SOS2. (C) SOS3. (D) P5CS1. (E) CAT1. The data were the mean ± SD of 3 replicates, and different letters were used to indicate the significant difference of multiple comparisons between groups at the p < 0.05 level.
4. Discussion
As a strong and highly dynamic cell structure, the cell wall usually mediates its own morphological and structural changes with cell wall modifiers to promote plant growth and development and adapt to stress [9,26]. Stress-mediated cell wall modification is considered to be an essential way for plants to respond to stress [27,28]. Expansin has been identified as a key cell wall-modifying protein that regulates both plant developmental processes and stress adaptation responses [29,30,31]. The expansin gene was first isolated from cucumber hypocotyls [32], and then a large number of genes were found in rice, wheat, soybean, maize, and tomato [33,34,35,36]. So far, the biological functions of expansin genes have been explored in many species, but there are few reports on the salt tolerance function of maize expansin genes. In previous work, we screened a highly expressed expansin gene, ZmEXPA6, obtained from a maize root transcriptome library, under 200 Mm NaCl treatment (Figure S1). The gene contains a DPBB domain similar to the expansins of other species (Double Psi Beta-Barrel) (Figure S2D). This domain exhibits polysaccharide-binding capacity and disrupts inter-polysaccharide hydrogen bonding within the cell wall matrix, mediating cell wall relaxation and mechanical softening [6,32].
Salt stress, one of the most detrimental abiotic stresses limiting global crop productivity, severely impairs multiple aspects of plant growth and development, mainly in plant height, root length, seed germination, and plant growth status [4]. Roots are important organs for plants to absorb water and nutrients, and their growth and development have an important impact on plant salt tolerance. Numerous expansins, including soybean GmEXLB1 [37] and GmEXPB2 [38] and rice OsEXPB2 [39], OsEXPA8 [40], and OsEXPA10 [41], have been demonstrated to be associated with root development and plant stress tolerance. In this study, overexpression ZmEXPA6 Arabidopsis lines showed better growth phenotype (Figure 3) and longer root length (Figure 2) after salt treatment. These findings demonstrate that ZmEXPA6 overexpression positively regulates root development and elongation, and improves the salt tolerance of overexpression lines. As previously reported, plant growth and development is inseparable from the increase in cell size and number, and cell expansion depends on cell wall loosening [6]. Expansin is considered to be the most important cell wall modifier for cell elongation. It mediates cell wall extension and relaxation through pH-dependent, non-enzymatic mechanisms, thereby facilitating cellular expansion [8]. In this study, similar results were obtained when examining root cells of ZmEXPA6 overexpression lines. Under salt stress, the root cell length and area of ZmEXPA6 overexpression Arabidopsis lines were significantly higher than those of WT lines (Figure 4). This may explain how ZmEXPA6 overexpression enhances root elongation. Cosgrove ‘s loosing theory tells us that expansin-mediated cell wall adaptive response enables cells to continue to grow with reduced expansion pressure, increasing the ratio of root to stem and allowing the root system to find water in the soil while limiting the loss of water in the leaves [42]. Thus, it can resist the damage suffered under salt stress and adapt to salt stress.
Anthocyanins are water-soluble flavonoids widely distributed in plants [43,44]. In recent years, studies have found that they play an important role in plants’ response to abiotic stress (such as salt stress) [45]. On the one hand, anthocyanins, as potent antioxidants, can scavenge ROS produced under salt stress and reduce oxidative damage [46,47,48]. On the other hand, they cooperate with osmotic regulators such as proline and soluble sugar to regulate cell osmotic potential and maintain cell turgor [49,50]. Under salt stress, ROS signaling induces the expression of anthocyanin synthesis genes, which makes anthocyanins play an important role in resisting stress. For example, Arabidopsis overexpression AtPAP1 exhibits significantly higher anthocyanin levels and greater salt tolerance than WT lines [51]. One study revealed that the Arabidopsis AtDFR gene improves salt and drought tolerance in Brassica napus through the modulation of anthocyanin biosynthesis [52]. Anthocyanins and flavonoids are involved in the signal mechanism of salt stress response in anthocyanin synthesis-deficient mutants [53]. In this study, after salt treatment, the transcription levels of anthocyanin synthesis genes such as CHS, F3H, DFR, LDOX, UF3GF, and UGT in ZmEXPA6 overexpression lines were significantly up-regulated, and the anthocyanin content significantly increased. These results indicate that ZmEXPA6 overexpression could positively regulate anthocyanin synthesis and accumulation under salt stress, and improve salt tolerance of overexpression lines.
Under saline conditions, elevated soil salt concentrations reduce water potential, compromising cellular water uptake and ultimately resulting in osmotic stress caused by water shortage [4]. The accumulation of osmoregulatory substances represents a key adaptive mechanism for plants to mitigate salt stress [25]. It has been reported that the proline content of OfEXLA1 overexpression lines in Osmanthus fragrans was significantly higher than that of the WT under salt induction [13]. The content of soluble sugar, soluble protein, and proline in wheat TaEXPB7-B overexpression lines continued to increase after salt treatment and significantly exceeded that observed in WT lines [16]. The content of proline, soluble sugar, and soluble protein in ZmEXPA6 overexpression lines significantly increased after salt stress treatment (Figure 6A–C). Consistent with these findings, ZmEXPA6 overexpression similarly promoted osmolyte biosynthesis, corroborating previous reports.
The increase of ROS under salt stress can be used as a signal for plant stress response, but high concentrations of ROS are ion-toxic and harmful to plants [25]. To mitigate oxidative damage, plants have evolved sophisticated ROS-scavenging systems [54]. In the ROS-scavenging system, SOD can rapidly convert ROS into H2O2, which can be converted into O2 and H2O through the action of POD and CAT [55]. Therefore, SOD, POD, and CAT are used as indicators to determine the antioxidant capacity of plants, reflecting the tolerance of plants to salt stress. MDA, the terminal byproduct of lipid peroxidation, is widely recognized as a sensitive biomarker for oxidative stress-induced membrane damage [56]. In this study, we found that the accumulation of MDA, H2O2, and O2− content in the overexpression lines was less (Figure 6D–F), and the activity of SOD, POD, and CAT in ZmEXPA6 overexpression lines was significantly increased after salt treatment (Figure 6G–I). This indicates that the overexpression of ZmEXPA6 increased the active oxygen-scavenging ability of Arabidopsis thaliana and made the overexpression lines have stronger salt tolerance, which further supports previous reports.
Salt stress induces excessive Na+ accumulation in plants, disrupting both ionic and osmotic homeostasis. We therefore analyzed the expression of five salt tolerance marker genes in the Arabidopsis genome (Figure 7). SOS1 is a Na+/H+ transporter on the plasma membrane that can transport Na+ to the outside of the cell. SOS3 and SOS2 combine to regulate the Na+ efflux of SOS1, thus reducing the intracellular Na+ concentration and reducing the salt stress injury of plants [57,58]. The expression of SOS1, SOS2, and SOS3 was up-regulated in ZmEXPA6 overexpression lines after salt treatment (Figure 7A–C). AtP5CS1 is a key enzyme for proline synthesis and regulates proline synthesis. Compared with the WT, the significant increase of proline content in the overexpression lines was related to the up-regulated expression of the gene (Figure 7D). The high expression of the AtCAT gene also enhanced the antioxidant activity of overexpression lines (Figure 7E).
5. Conclusions
In summary, heterologous overexpression of ZmEXPA6 significantly promoted root elongation and seedling growth in Arabidopsis thaliana, as well as improved its tolerance to salt stress. The expression level of salt stress response genes in ZmEXPA6 overexpression was higher. Under salt stress, ZmEXPA6 overexpression increased the accumulation of anthocyanin, proline, soluble sugar, and soluble protein and the activities of SOD, CAT, and POD, and decreased the contents of MDA, O2−, and H2O2, thereby improving salt tolerance of Arabidopsis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agronomy15092240/s1: Figure S1: The expression level of ZmEXPA6 in maize roots at different days before and after NaCl treatment; Figure S2: Bioinformatics analysis of ZmEXPA6; Figure S3: Compared with 0 mM NaCl, the root length reduction ratio of WT and overexpression lines under different salt concentrations; Table S1: Primers used in this study.
Author Contributions
Conceptualization, Y.S. and B.L.; methodology, Z.Y. and G.Y.; software, Q.L.; validation, Q.L. and X.L.; formal analysis, Y.S.; investigation, Y.S. and Q.L.; resources, X.L.; data curation, Y.S.; writing—original draft preparation, Y.S.; writing—review and editing, Y.S. and B.L.; visualization, S.M.; supervision, B.L.; project administration, S.M.; funding acquisition, C.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China, grant number 2022YFD1201704; the Agricultural Science and Technology Innovation Projects of Shandong Academy of Agricultural Sciences, China, grant numbers CXGC2025C02 and CXGC2025A03; the Science and Technology Demonstration Project of Shandong Province, grant number 2024SFGC0402.
Data Availability Statement
Data will be made available on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef]
- Navada, S.; Vadstein, O.; Gaumet, F.; Tveten, A.K.; Spanu, C.; Mikkelsen, Ø.; Kolarevic, J. Biofilms remember: Osmotic stress priming as a microbial management strategy for improving salinity acclimation in nitrifying biofilms. Water Res. 2020, 176, 115732. [Google Scholar] [CrossRef]
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, Y.; Cho, H.T.; Kende, H. Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant cell wall loosening by expansins. Annu. Rev. Cell Dev. Biol. 2024, 40, 329–352. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Cosgrove, D.J. Structure and growth of plant cell walls. Nat. Rev. Mol. Cell Biol. 2024, 25, 340–358. [Google Scholar] [CrossRef]
- Samalova, M.; Gahurova, E.; Hejatko, J. Expansin-mediated developmental and adaptive responses: A matter of cell wall biomechanics? Quant. Plant Biol. 2022, 3, e11. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Xu, Z.; Kong, Y. Overexpression of NtEXPA11 modulates plant growth and development and enhances stress tolerance in tobacco. Plant Physiol. Biochem. 2020, 151, 477–485. [Google Scholar] [CrossRef]
- Dong, B.; Wang, Q.; Zhou, D.; Wang, Y.; Miao, Y.; Zhong, S.; Fang, Q.; Yang, L.; Xiao, Z.; Zhao, H. Abiotic stress treatment reveals expansin like A gene OfEXLA1 improving salt and drought tolerance of Osmanthus fragrans by responding to abscisic acid. Hortic. Plant J. 2024, 10, 573–585. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, G.; An, J.; Li, Q.; Chen, Y.; Zhao, X.; Wu, J.; Wang, Y.; Hao, Q.; Wang, W. Expansin gene TaEXPA2 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 298, 110596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Hao, W.; Zhang, L.; Chen, L. Expression of two α-type expansins from Ammopiptanthus nanus in Arabidopsis thaliana enhance tolerance to cold and drought stresses. Int. J. Mol. Sci. 2019, 20, 5255. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F. TaEXPB7-B, beta-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef] [PubMed]
- Mara, C.; Grigorova, B.; Liu, Z. Floral-dip transformation of Arabidopsis thaliana to examine pTSO2::β-glucuronidase reporter gene expression. J. Vis. Exp. 2010, 40, e1952. [Google Scholar]
- Xu, Y.; Zou, J.; Zheng, H.; Xu, M.; Zong, X.; Wang, L. RNA-Seq transcriptome analysis of rice primary roots reveals the role of flavonoids in regulating the rice primary root growth. Genes 2019, 10, 213. [Google Scholar] [CrossRef]
- Yan, Z.; Li, K.; Li, Y.; Wang, W.; Leng, B.; Yao, G.; Zhang, F.; Mu, C.; Liu, X. The ZmbHLH32-ZmIAA9-ZmARF1 module regulates salt tolerance in maize. Int. J. Biol. Macromol. 2023, 253, 126987. [Google Scholar]
- Han, G.; Yuan, F.; Guo, J.; Zhang, Y.; Wang, B. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 2019, 285, 55–67. [Google Scholar] [CrossRef]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J. Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.W.; Lee, H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V. The role of anthocyanins in plant tolerance to drought and salt stresses. Plants 2023, 12, 2558. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.M.; Dai, H.; Zheng, W.; Cao, F.; Zhang, G.; Sun, D.; Wu, F. Genotypic differences in physiological characteristics in the tolerance to drought and salinity combined stress between Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2013, 63, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Hessini, K.; Martínez, J.P.; Gandour, M.; Albouchi, A.; Soltani, A.; Abdelly, C. Effect of water stress on growth, osmotic adjustment, cell wall elasticity and water-use efficiency in Spartina alterniflora. Environ. Exp. Bot. 2009, 67, 312–319. [Google Scholar] [CrossRef]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef]
- Qiu, D.; Xu, S.; Wang, Y.; Hong, L. Primary cell wall modifying proteins regulate wall mechanics to steer plant morphogenesis. Front. Plant Sci. 2021, 12, 751372. [Google Scholar] [CrossRef]
- Wang, X.; Ma, J.; He, F.; Wang, L.; Zhang, T.; Liu, D.; Xu, Y.; Li, F.; Feng, X. A study on the functional identification of overexpressing winter wheat expansin gene TaEXPA7-B in rice under salt stress. Int. J. Mol. Sci. 2024, 25, 7707. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.; Wu, D.; Zhao, H.; Gong, L.; Xu, J. Regulation of SmEXPA13 expression by SmMYB1R1-L enhances salt tolerance in Salix matsudana Koidz. Int. J. Biol. Macromol. 2024, 270, 132292. [Google Scholar] [CrossRef]
- Kuluev, B.R.; Musin, K.G.; Yakupova, A.B. The expansin gene NtEXPA5 increases stress tolerance of tobacco hairy roots through an effect on the antioxidant system. Ecol. Genet. 2021, 19, 5–12. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, P.; Kumar, A.; Chawla, N.; Dhatt, A.S. Multifaceted regulation of anthocyanin biosynthesis in plants: A comprehensive review. J. Plant Growth Regul. 2024, 43, 3048–3062. [Google Scholar] [CrossRef]
- Huang, J.; Takano, T.; Akita, S. Expression of α-expansin genes in young seedlings of rice (Oryza sativa L.). Planta 2000, 211, 467–473. [Google Scholar] [CrossRef]
- Lizana, X.C.; Riegel, R.; Gomez, L.D.; Herrera, J.; Isla, A.; McQueen-Mason, S.J.; Calderini, D.F. Expansins expression is associated with grain size dynamics in wheat (Triticum aestivum L.). J. Exp. Bot. 2010, 61, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Meeley, R.B.; Cosgrove, D.J. Analysis and expression of the α-expansin and β-expansin gene families in maize. Plant Physiol. 2001, 126, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wang, B.; Du, H.; Li, W.; Li, X.H.; Zhang, C. GmEXLB1, a soybean expansin-like B gene, alters root architecture to improve phosphorus acquisition in arabidopsis. Front. Plant Sci. 2019, 10, 00808. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Zou, H.; Wenwen, Y.; Zang, G.; Kang, Z.; Zhang, Z.; Huang, J.; Wang, G. OsEXPB2, a β-expansin gene, is involved in rice root system architecture. Mol. Breed. 2015, 35, 41. [Google Scholar] [CrossRef]
- Ma, N.; Wang, Y.; Qiu, S.; Kang, Z.; Che, S.; Wang, G.; Huang, J.; Bennett, M. Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS ONE 2013, 8, e75997. [Google Scholar] [CrossRef]
- Che, J.; Yamaji, N.; Shen, R.F.; Ma, J.F. An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 2016, 88, 132–142. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Expanding wheat yields with expansin. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Ahmadzai, A.S.; Hu, C.; Zhang, C.; Li, Y. Mechanisms of anthocyanin-mediated salt stress alleviation and cellular homeostasis in plants. Plant Growth Regul. 2025, 105, 566–573. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, Z.; Li, D.; Cai, M.; Liang, Z.; Chen, Q.; Du, X.; Wang, J.; Gu, R.; Li, L. Transcriptome analysis revealed the potential molecular mechanism of anthocyanidins’ improved salt tolerance in maize seedlings. Plants 2023, 12, 2793. [Google Scholar] [CrossRef] [PubMed]
- Htay, N.A.; Kil, K.C. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar]
- Zhang, Q.; Zhai, J.; Shao, L.; Lin, W.; Peng, C. Accumulation of anthocyanins: An adaptation strategy of mikania micrantha to low temperature in winter. Front. Plant Sci. 2019, 10, 1049. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X. NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef]
- Luo, S.; Shu, H.; Miao, Y.; Sun, S.; Zhang, X.; Yi, Y.; Sun, W. The Rhododendron agastum flavonoid 3-O-glycosyltransferase Ra3GT2 contributes to salt and drought stress tolerance through modulating anthocyanin synthesis. Plant Physiol. Biochem. PPB 2025, 228, 110209. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Do anthocyanins function as Osmoregulators in leaf tissues? Adv. Bot. Res. 2002, 37, 103–127. [Google Scholar]
- Hughes, N.M.; Carpenter, K.L.; Cannon, J.G. Estimating contribution of anthocyanin pigments to osmotic adjustment during winter leaf reddening. J. Plant Physiol. 2013, 170, 230–233. [Google Scholar] [CrossRef]
- Oh, J.E.; Kim, Y.H.; Kim, J.H.; Kwon, Y.R.; Lee, H. Enhanced level of anthocyanin leads to increased salt tolerance in Arabidopsis PAP1-D plants upon sucrose treatment. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 79–88. [Google Scholar] [CrossRef]
- Kim, J.H.; Hyun, W.Y.; Nguyen, H.N.; Jeong, C.Y.; Xiong, L.; Hong, S.W.; Lee, H. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2015, 38, 559–571. [Google Scholar] [CrossRef]
- Oosten, M.J.V.; Sharkhuu, A.; Batelli, G.; Bressan, R.A.; Maggio, A. The Arabidopsis thaliana mutant air1 implicates SOS3 in the regulation of anthocyanins under salt stress. Plant Mol. Biol. 2013, 83, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Branicky, R.; Noe, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Comprehensive analysis of carotenoid cleavage dioxygenases gene family and its expression in response to abiotic stress in poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Oosten, M.J.V.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1 as plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
