Time-Course Transcriptome Analysis Reveals Dynamic Nitrogen Response Mechanisms and Key Regulatory Networks in Sugarcane
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Methods
2.2. RNA-Seq Analysis
2.3. Differentially Expressed Gene (DEG) Analysis
2.4. Functional Annotation
2.5. WGCNA
3. Results
3.1. RNA-Seq Analysis of Sugarcane Leaves Under Nitrogen Treatment
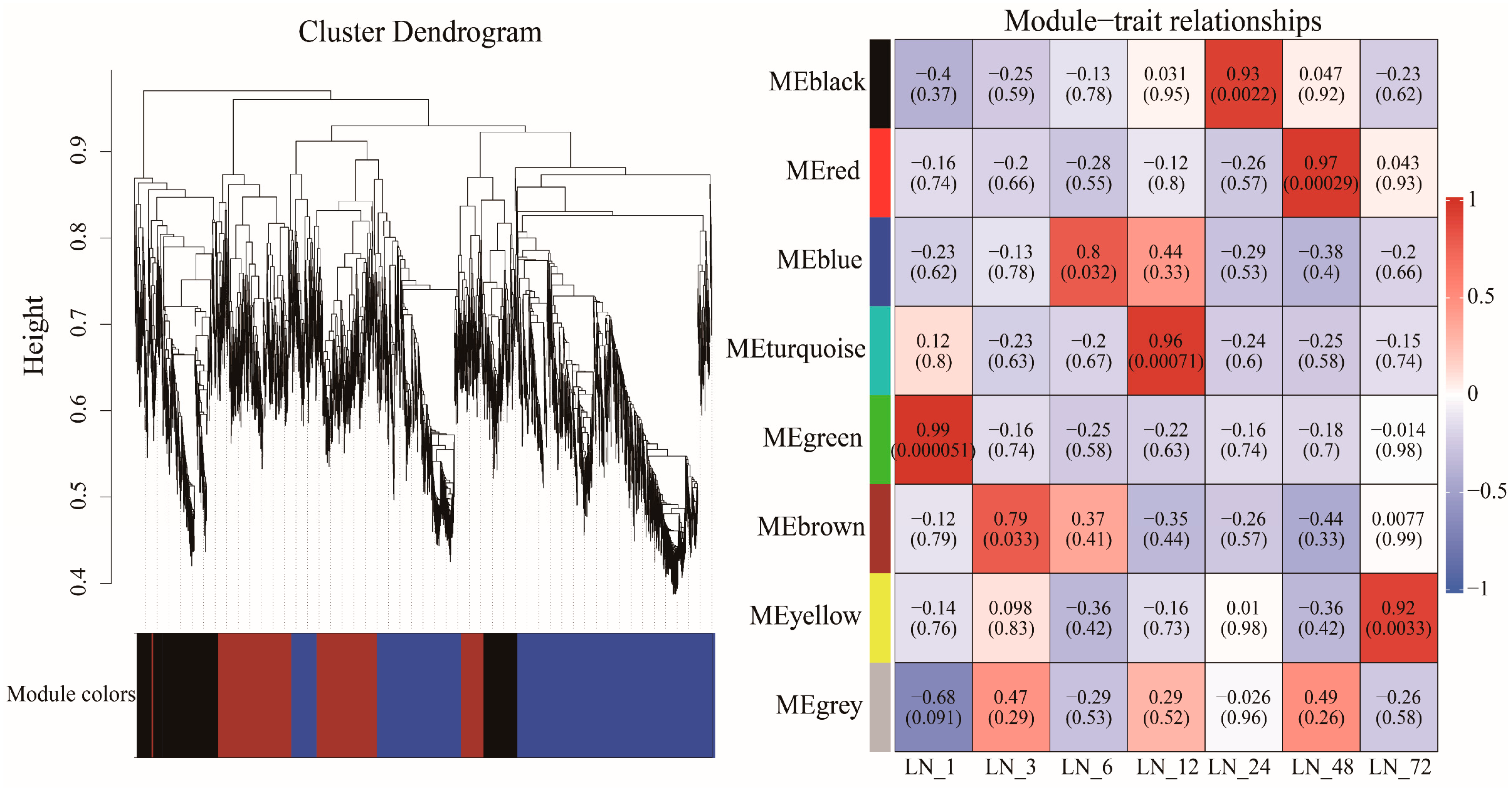
3.2. WGCNA of Sugarcane Under Low Nitrogen Conditions
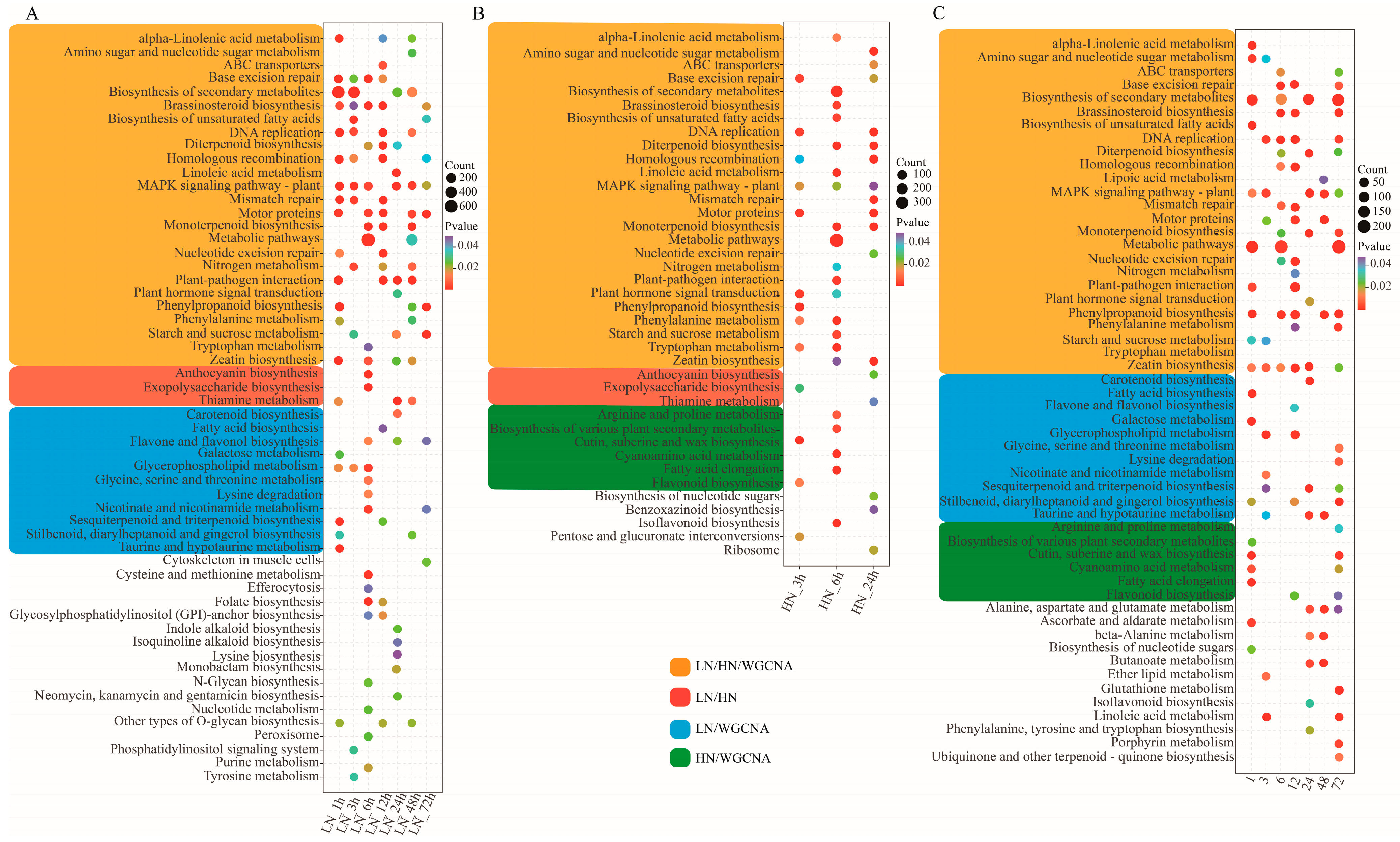
3.3. Functional Annotation and Enrichment Analysis of Differentially Expressed Genes
3.4. Dynamic Response of Nitrogen Metabolism-Related Genes in Sugarcane Leaves to Differential Nitrogen Treatments
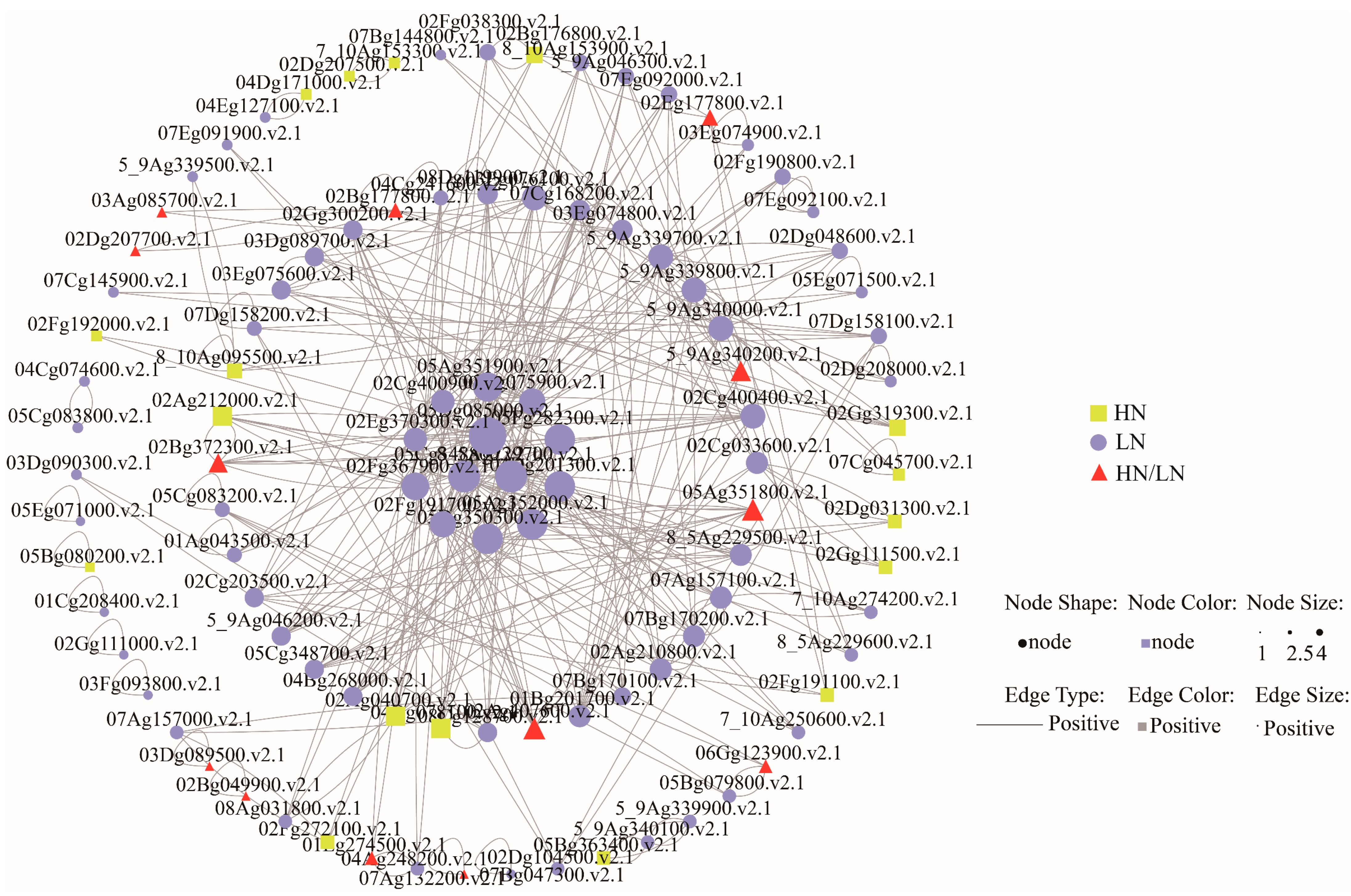
3.5. Dynamic Regulatory Mechanisms of Nitrogen Response in Sugarcane Leaves Mediated by the Zeatin Biosynthesis Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Arencibia, A.D. Gene Transfer in Sugarcane. In Biotechnology of Food Crops in Developing Countries; Hohn, T., Leisinger, K.M., Eds.; Plant Gene Research; Springer: Vienna, Austria, 1999. [Google Scholar] [CrossRef]
- Heinrichs, R.; Otto, R.; Magalhães, A.; Meirelles, G.C. Importance of Sugarcane in Brazilian and World Bioeconomy. In Knowledge-Driven Developments in the Bioeconomy; Dabbert, S., Lewandowski, I., Weiss, J., Pyka, A., Eds.; Economic Complexity and Evolution; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Kishorekumar, R.; Bulle, M.; Wany, A.; Gupta, K.J. An Overview of Important Enzymes Involved in Nitrogen Assimilation of Plants. Methods Mol. Biol. 2020, 2057, 1–13. [Google Scholar] [CrossRef]
- da Silva, N.F.; da Silva, E.C.; Muraoka, T.; Teixeira, M.B.; Soares, F.A.L.; Cunha, F.N.; Adu-Gyamfi, J.; Cavalcante, W.S.d.S. Nitrogen Utilization from Ammonium Nitrate and Urea Fertilizer by Irrigated Sugarcane in Brazilian Cerrado Oxisol. Agriculture 2020, 10, 323. [Google Scholar] [CrossRef]
- Trivelin, P.C.O.; Vitti, A.C.; Oliveira, M.W.; Gava, G.J.C.; Sarriés, G.A. Nitrogen utilization and sugarcane (plant cane) yield on a sandy soil with incorporated crop residues. Rev. Bras. Cienc. Solo 2002, 26, 637–646. [Google Scholar] [CrossRef]
- Alvarez, J.M.; Vidal, E.A.; Gutiérrez, R.A. Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 2012, 15, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Rennenberg, H.; Dannenmann, M.; Gessler, A.; Kreuzwieser, J.; Simon, J.; Papen, H. Nitrogen balance in forest soils: Nutritional limitation of plants under climate change treatmentes. Plant Biol. 2009, 11 (Suppl. S1), 4–23. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, P.; Gautam, V.; Rengasamy, B.; Adhikari, B.; Udayakumar, M.; Sarkar, A.K. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci. Rep. 2016, 6, 39266. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. Cell Mol. Biol. 2021, 105, 421–430. [Google Scholar] [CrossRef]
- Takei, K.; Takahashi, T.; Sugiyama, T.; Yamaya, T.; Sakakibara, H. Multiple routes communicating nitrogen availability from roots to shoots: A signal transduction pathway mediated by cytokinin. J. Exp. Bot. 2002, 53, 971–977. [Google Scholar] [CrossRef]
- Que, Y.; Su, Y.; Guo, J.; Wu, Q.; Xu, L. A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-Seq. PLoS ONE 2014, 9, e106476. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Lv, Y.; Xu, L.; Dossa, K.; Zhou, K.; Zhu, M.; Xie, H.; Tang, S.; Yu, Y.; Guo, X.; Zhou, B. Identification of putative drought-responsive genes in rice using gene co-expression analysis. Bioinformation 2019, 15, 480–489. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, S.; Li, W.; Chen, B.; Li, W. Gene-coexpression network analysis identifies specific modules and hub genes related to cold treatment in rice. BMC Genom. 2022, 23, 251. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xin, W.; Chen, N.; Yang, F.; Li, J.; Qu, G.; Jiang, X.; Xu, L.; Zhao, S.; Liu, H.; et al. Transcriptome and co-expression network analysis reveals the molecular mechanism of rice root systems in response to low-nitrogen conditions. Int. J. Mol. Sci. 2023, 24, 5290. [Google Scholar] [CrossRef]
- Wei, Q.; Yin, Y.; Deng, B.; Song, X.; Gong, Z.; Shi, Y. Transcriptome analysis of nitrogen-deficiency-responsive genes in two potato cultivars. Agronomy 2023, 13, 2164. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Waern, K.; Snyder, M. RNA-Seq: A method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 2010, 89, 4.11.1–4.11.13. [Google Scholar] [CrossRef]
- Healey, A.L.; Lovell, J.T.; Garsmeur, O.; Jenkins, J.W.; Grimwood, J.; Aitken, K.S.; Henry, R.J.; D’Hont, A.; Schmutz, J. Leveraging Multiple Sequencing Technologies to Generate a Haplotype Specific Assembly of Sugarcane R570; CIRAD: San Diego, CA, USA, 2020. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Abrams, Z.B.; Johnson, T.S.; Huang, K.; Payne, P.R.O.; Coombes, K. A protocol to evaluate RNA sequencing normalization methods. BMC Bioinform. 2019, 20 (Suppl. S24), 679. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Rosati, D.; Palmieri, M.; Brunelli, G.; Morrione, A.; Iannelli, F.; Frullanti, E.; Giordano, A. Differential gene expression analysis pipelines and bioinformatic tools for the identification of specific biomarkers: A review. Comput. Struct. Biotechnol. J. 2024, 23, 1154–1168. [Google Scholar] [CrossRef]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Li, H.; Wang, Q.; Huang, T.; Liu, J.; Zhang, P.; Li, L.; Xie, H.; Wang, H.; Liu, C.; Qin, P. Transcriptome and Metabolome Analyses Reveal Mechanisms Underlying the Response of Quinoa Seedlings to Nitrogen Fertilizers. Int. J. Mol. Sci. 2023, 24, 11580. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Y.; Zhou, Y.; Wang, J.; Geng, X.; Shi, H. Transcriptome analysis of tobacco root response to different concentrations of nitrate. Agron. J. 2020, 112, 1111–1125. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative transcriptomics of rice genotypes with contrasting responses to nitrogen treatment reveals genes influencing nitrogen uptake through the regulation of root architecture. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef]
- Wang, R.; Guegler, K.; LaBrie, S.T.; Crawford, N.M. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 2000, 12, 1491–1509. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Dua, V.K.; Chakrabarti, S.K. Transcriptome analysis of potato shoots, roots and stolons under nitrogen treatment. Sci. Rep. 2020, 10, 1152. [Google Scholar] [CrossRef]
- Sinha, S.K.; Sevanthi V., A.M.; Chaudhary, S.; Tyagi, P.; Venkadesan, S.; Rani, M.; Mandal, P.K. Transcriptome analysis of two rice varieties contrasting for nitrogen use efficiency under chronic n starvation reveals differences in chloroplast and starch metabolism-related genes. Genes 2018, 9, 206. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, S.; Nigro, D.; Lasorella, C.; Marcotuli, I.; Gadaleta, A.; de Pinto, M.C. The Role of Glutamine Synthetase (GS) and Glutamate Synthase (GOGAT) in the Improvement of Nitrogen Use Efficiency in Cereals. Biomolecules 2023, 13, 1771. [Google Scholar] [CrossRef] [PubMed]
- Zayed, O.; Hewedy, O.A.; Abdelmoteleb, A.; Ali, M.; Youssef, M.S.; Roumia, A.F.; Seymour, D.; Yuan, Z.C. Nitrogen Journey in Plants: From Uptake to Metabolism, Treatment Response, and Microbe Interaction. Biomolecules 2023, 13, 1443. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, S.; Chen, D.; Pan, Y.; Bhanot, D.; Xu, T.; Gupta, S.; Zhang, J.; Huang, Y. WGCNA analysis and identification of key genes in tobacco in response to different nitrogen levels. BMC Plant Biol. 2025, 25, 465. [Google Scholar] [CrossRef]
- Pengpeng, W.; Tan, Z. Ammonia assimilation in rumen bacteria: A review. Anim. Biotechnol. 2013, 24, 107–128. [Google Scholar] [CrossRef]
- Inokuchi, R.; Kuma, K.I.; Miyata, T.; Okada, M. Nitrogen-assimilating enzymes in land plants and algae: Phylogenic and physiological perspectives. Physiol. Plant. 2002, 116, 1–11. [Google Scholar] [CrossRef]
- Machingura, M.; Salomon, E.; Jez, J.M.; Ebbs, S.D. The β-cyanoalanine synthase pathway: Beyond cyanide detoxification. Plant Cell Environ. 2016, 39, 2329–2341. [Google Scholar] [CrossRef]
- Howden, A.J.; Harrison, C.J.; Preston, G.M. A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. Cell Mol. Biol. 2009, 57, 243–253. [Google Scholar] [CrossRef]
- Gu, J.; Li, Z.; Mao, Y.; Struik, P.C.; Zhang, H.; Liu, L.; Wang, Z.; Yang, J. Roles of nitrogen and cytokinin signals in root and shoot communications in maximizing of plant productivity and their agronomic applications. Plant Sci. Int. J. Exp. Plant Biol. 2018, 274, 320–331. [Google Scholar] [CrossRef]
- Singh, S.; Letham, D.S.; Jameson, P.E.; Zhang, R.; Parker, C.W.; Bandenoch-Jones, J.; Noodén, L.D. Cytokinin Biochemistry in Relation to Leaf Senescence: IV. Cytokinin Metabolism in Soybean Explants. Plant Physiol. 1988, 88, 788–794. [Google Scholar] [CrossRef]
- Yong, J.W.; Wong, S.C.; Letham, D.S.; Hocart, C.H.; Farquhar, G.D. Effects of elevated [CO2] and nitrogen nutrition on cytokinins in the xylem sap and leaves of cotton. Plant Physiol. 2000, 124, 767–780. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol. 2001, 42, 85–93. [Google Scholar] [CrossRef]
- Takei, K.; Yamaya, T.; Sakakibara, H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004, 279, 41866–41872. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Zhang, L.; Liu, S.; Chen, M.; Yang, X. Time-Course Transcriptome Analysis Reveals Dynamic Nitrogen Response Mechanisms and Key Regulatory Networks in Sugarcane. Agronomy 2025, 15, 2164. https://doi.org/10.3390/agronomy15092164
Wang W, Zhang L, Liu S, Chen M, Yang X. Time-Course Transcriptome Analysis Reveals Dynamic Nitrogen Response Mechanisms and Key Regulatory Networks in Sugarcane. Agronomy. 2025; 15(9):2164. https://doi.org/10.3390/agronomy15092164
Chicago/Turabian StyleWang, Wanru, Lijun Zhang, Shuai Liu, Meiyan Chen, and Xiping Yang. 2025. "Time-Course Transcriptome Analysis Reveals Dynamic Nitrogen Response Mechanisms and Key Regulatory Networks in Sugarcane" Agronomy 15, no. 9: 2164. https://doi.org/10.3390/agronomy15092164
APA StyleWang, W., Zhang, L., Liu, S., Chen, M., & Yang, X. (2025). Time-Course Transcriptome Analysis Reveals Dynamic Nitrogen Response Mechanisms and Key Regulatory Networks in Sugarcane. Agronomy, 15(9), 2164. https://doi.org/10.3390/agronomy15092164




