Transcriptome-Based Phylogenomics and Adaptive Divergence Across Environmental Gradients in Epimedium brevicornu
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Climate Data Acquisition
2.2. RNA Extraction, Transcriptome Library Construction, and High-Throughput Sequencing
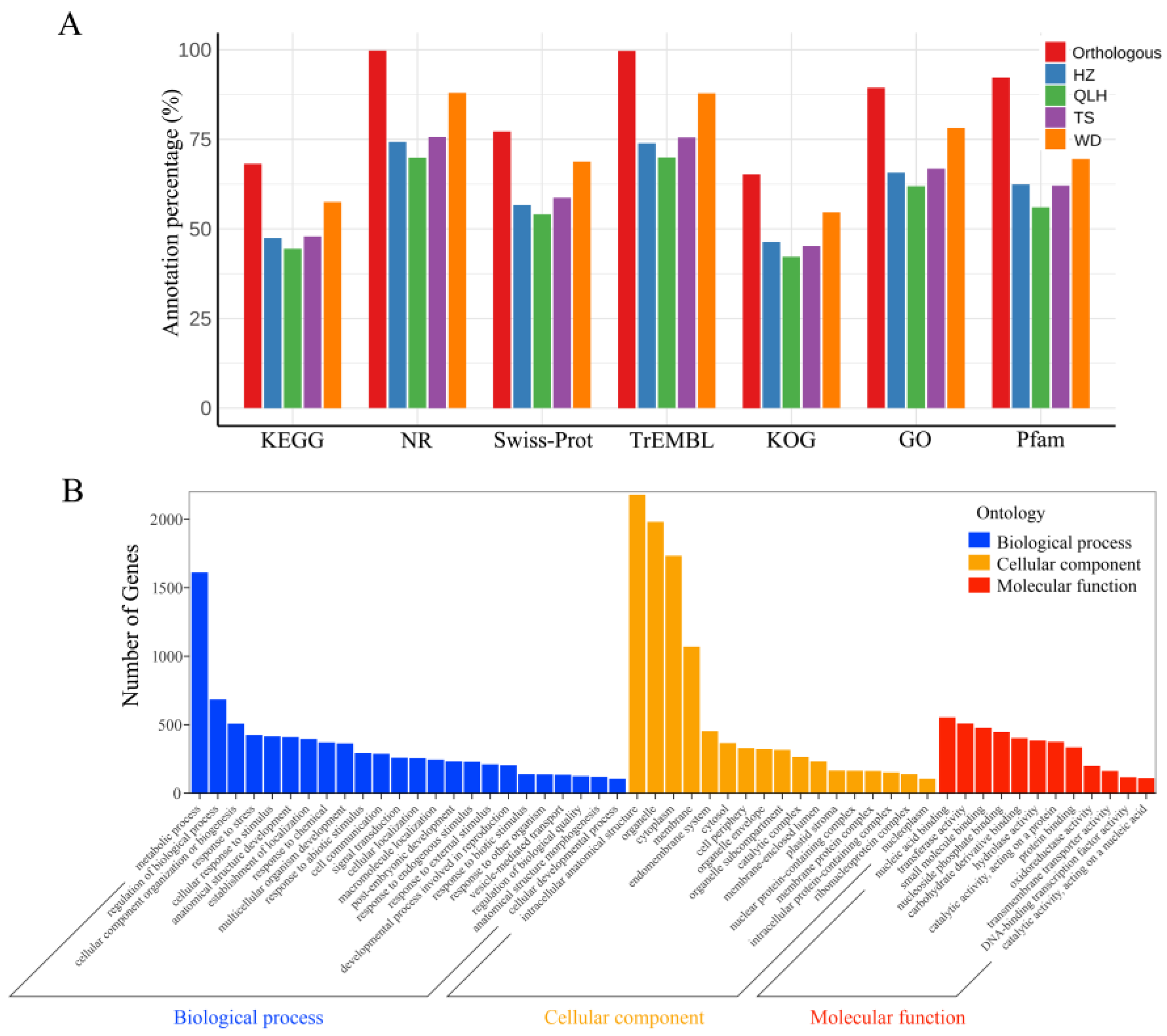
2.3. Data Preprocessing, Transcriptome Assembly, and Functional Annotation
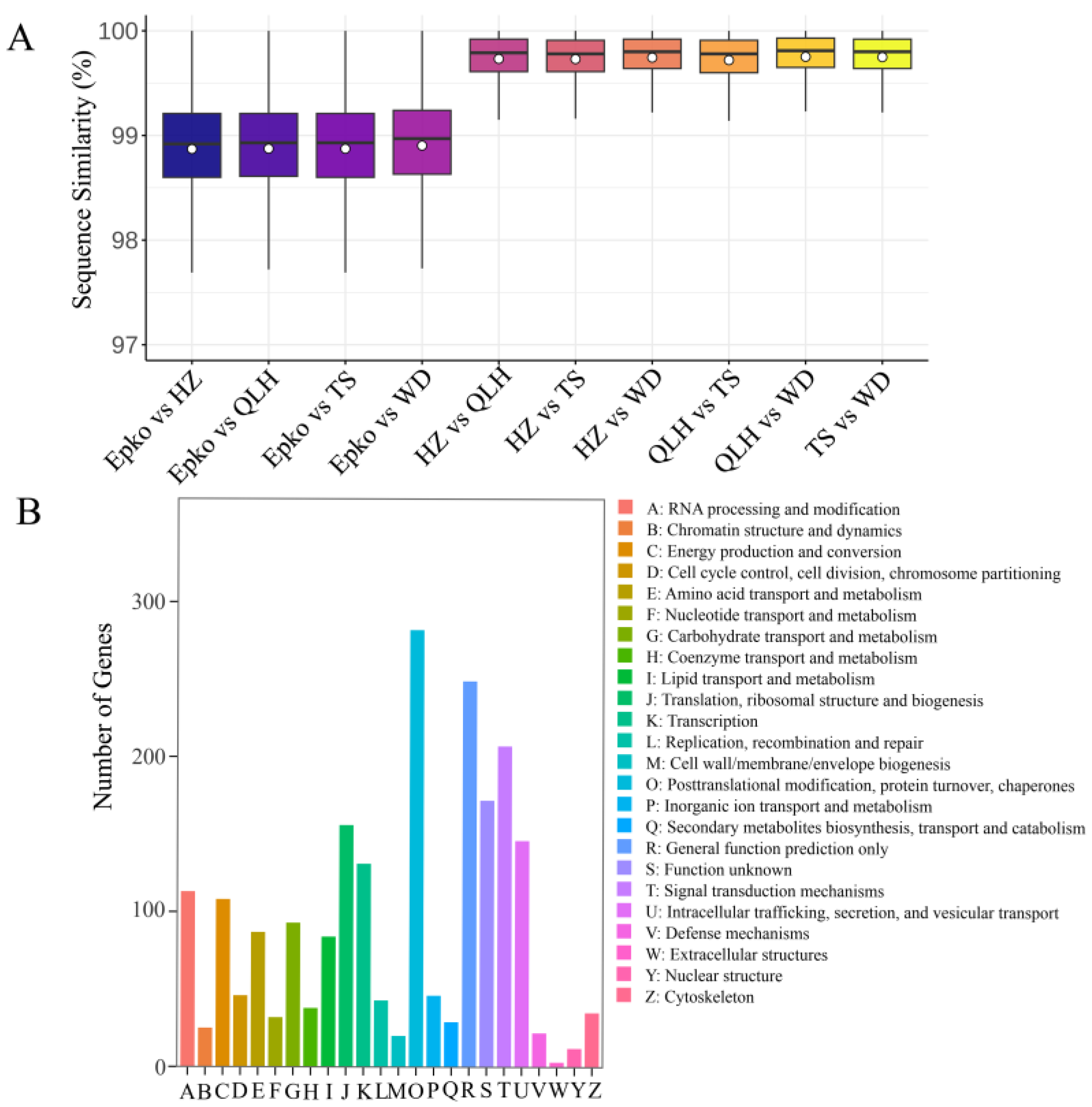
2.4. Orthologous Gene Identification and Sequence Alignment
2.5. Phylogenetic Analysis and Divergence Time Estimation
2.6. Lineage-Specific Evolutionary Rate Acceleration Analysis
2.7. GO Enrichment Analysis of Accelerated-Evolving Genes
2.8. Correlation Analysis Between Accelerated-Evolving Genes and Environmental Factors
2.9. Identification of Candidate Genes Under Positive Selection
2.10. Protein–Protein Interaction Network Construction and Core Gene Identification
3. Results
3.1. Transcriptome Sequencing and De Novo Assembly
3.2. Transcript Annotation and Functional Characterization
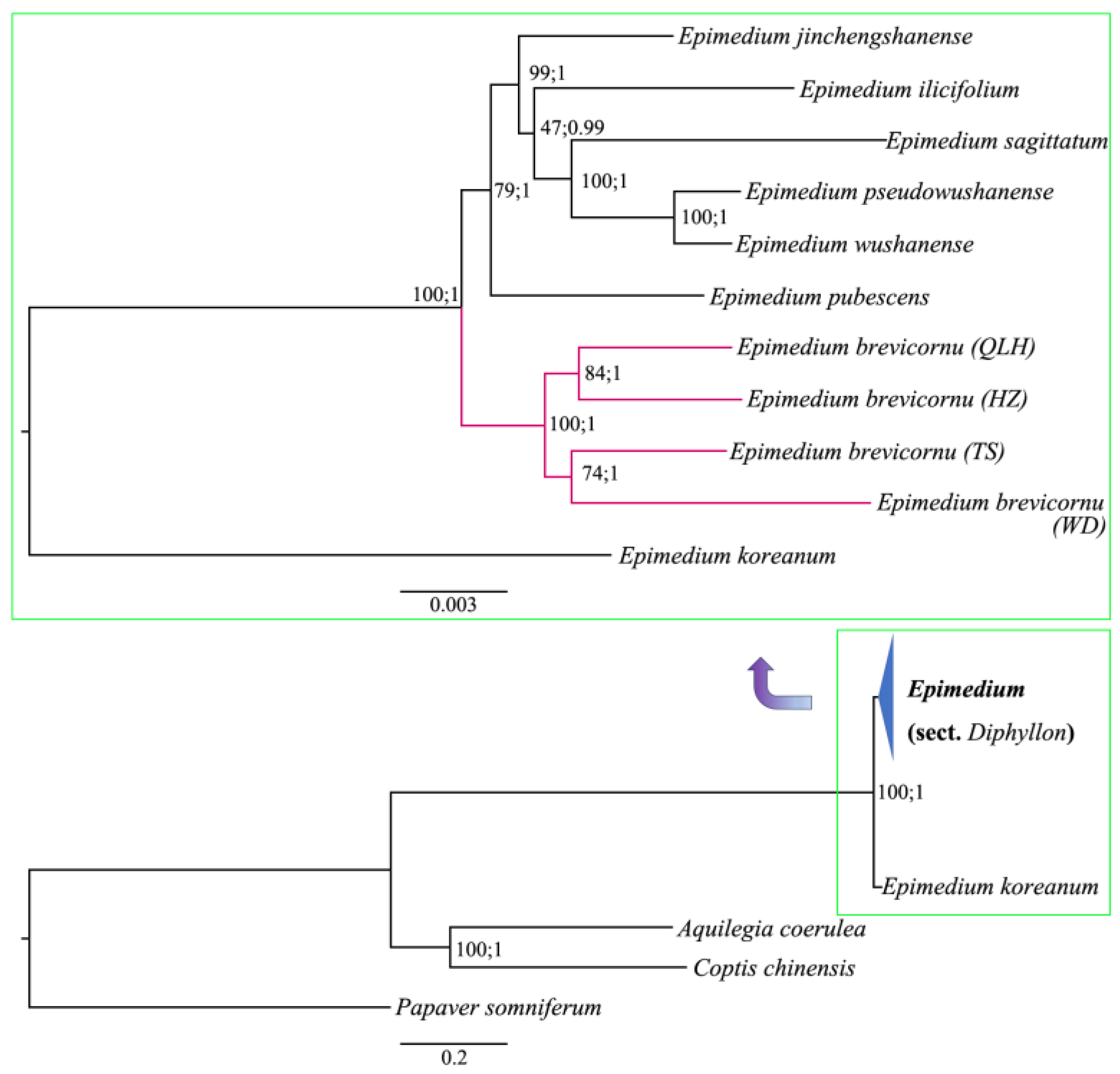
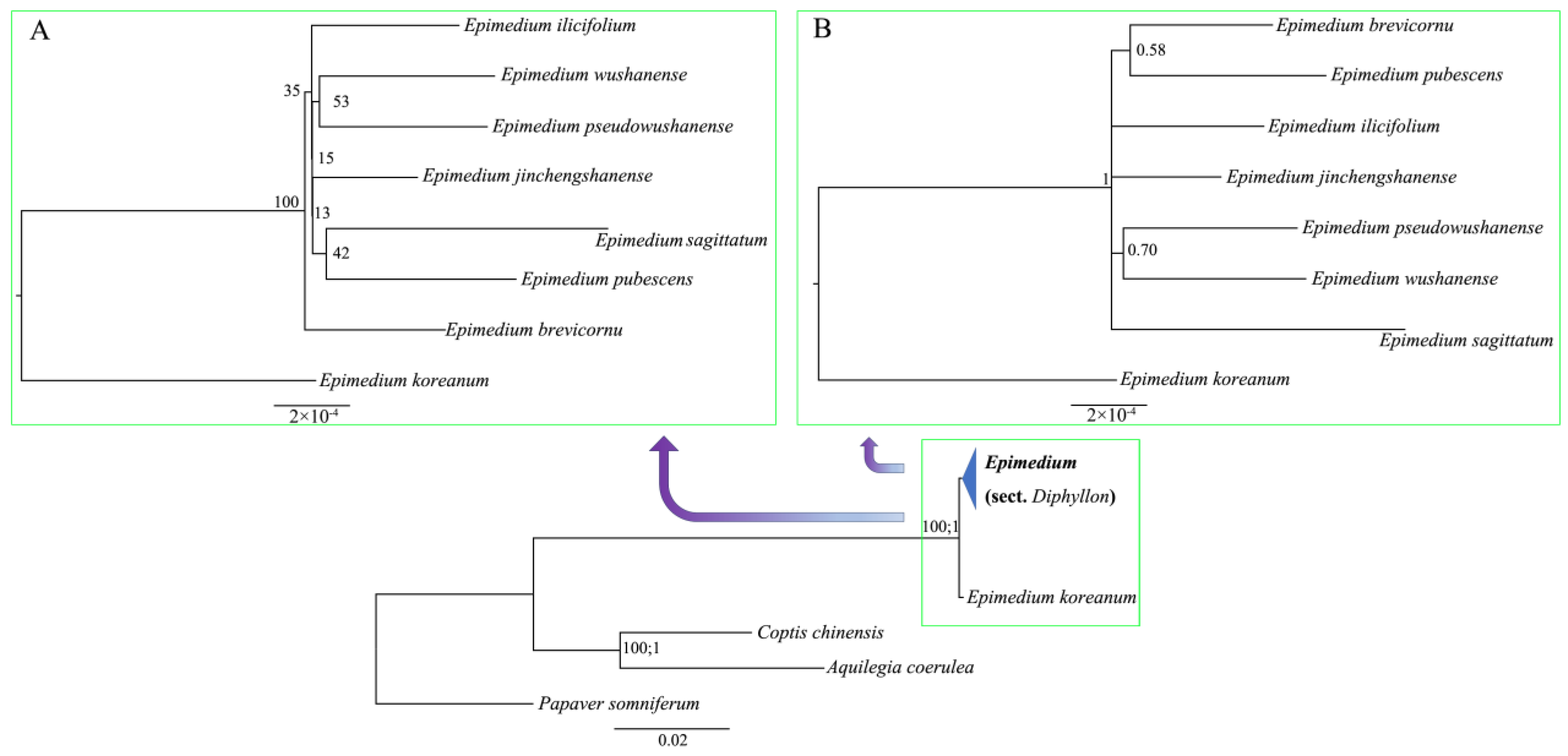
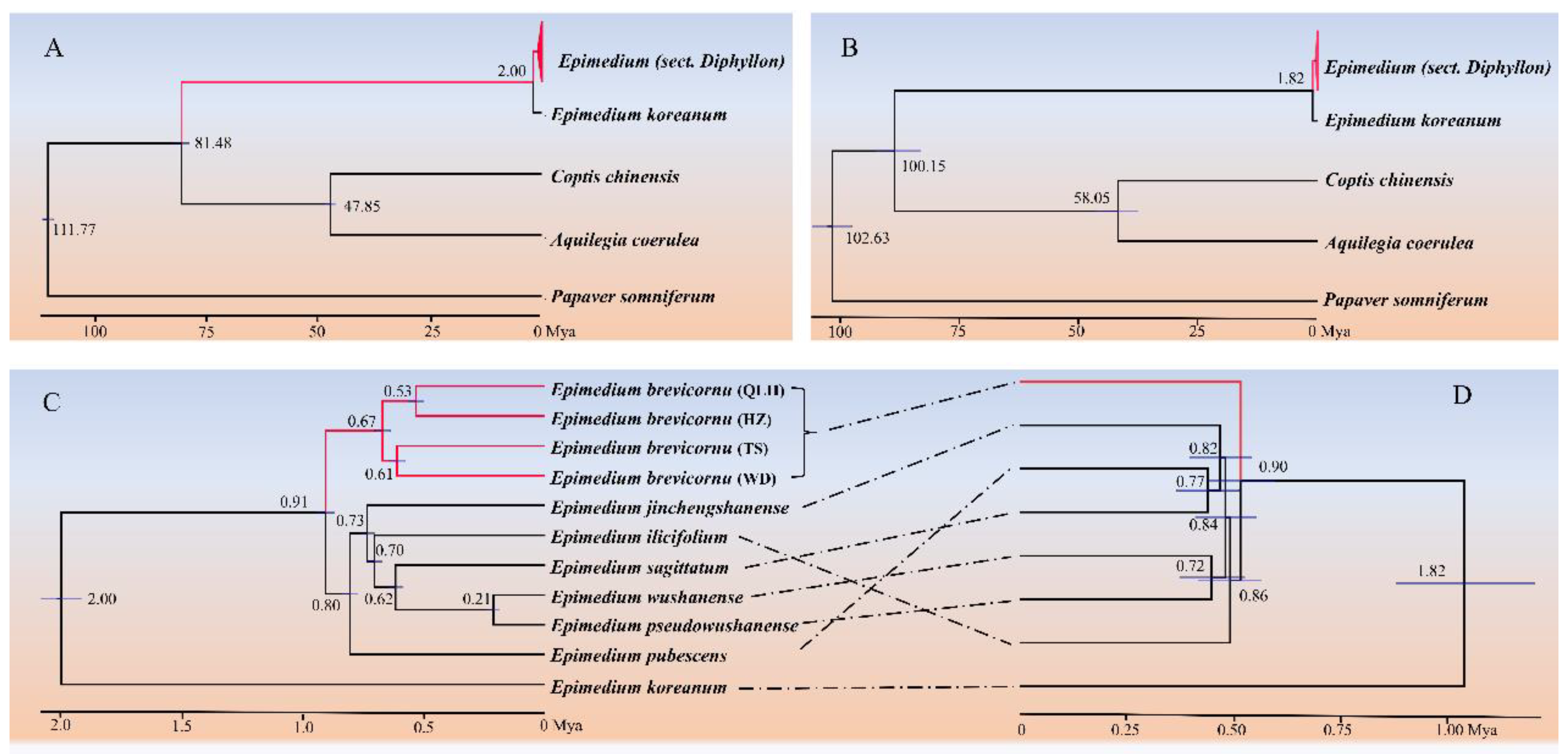
3.3. Phylogenomic Reconstruction of Epimedium and Nuclear–Plastid Topological Discordance
3.4. Divergence Time Estimation of Epimedium Lineages
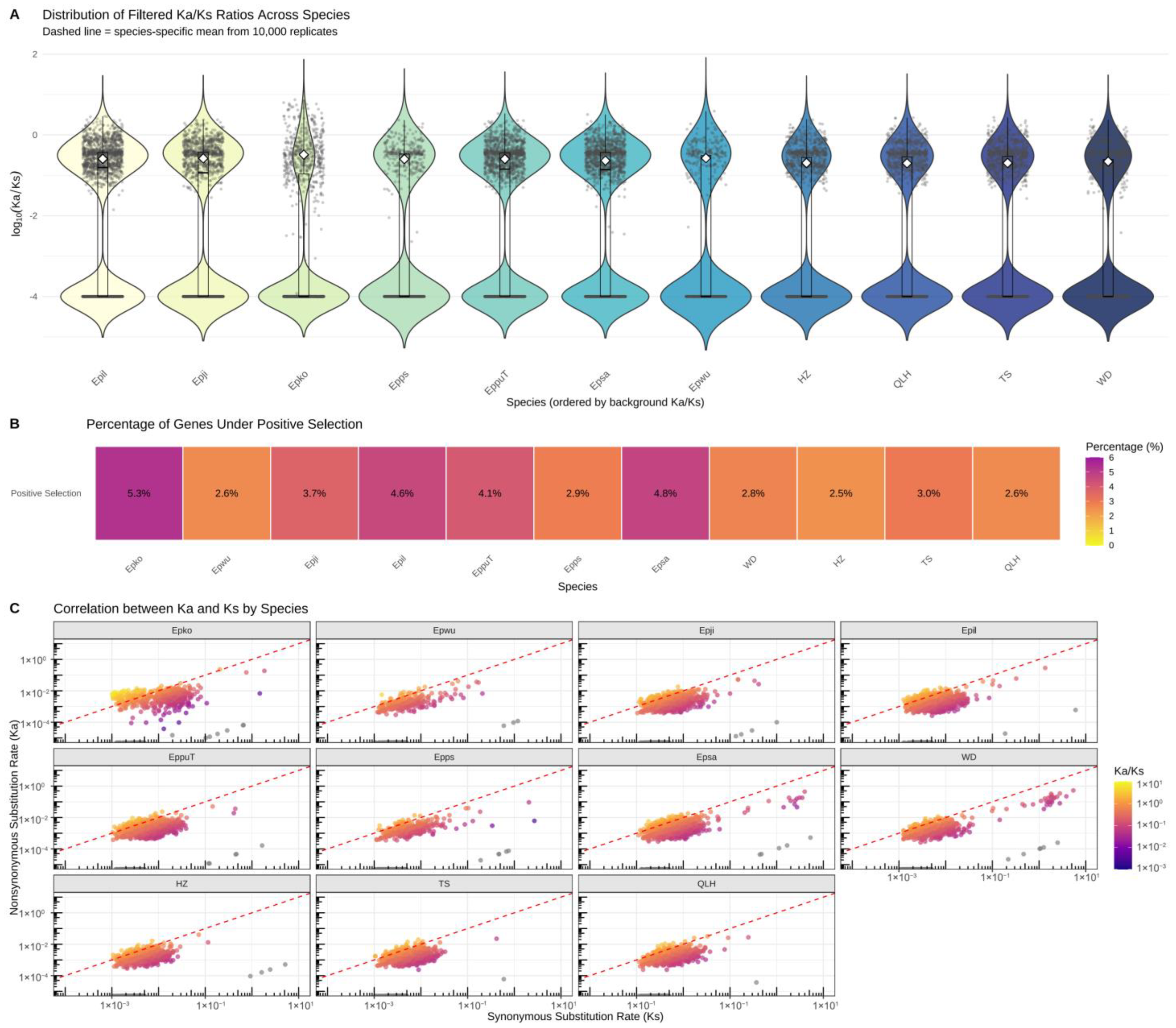
3.5. Molecular Evolutionary Rate Analysis of Chinese Epimedium Lineages
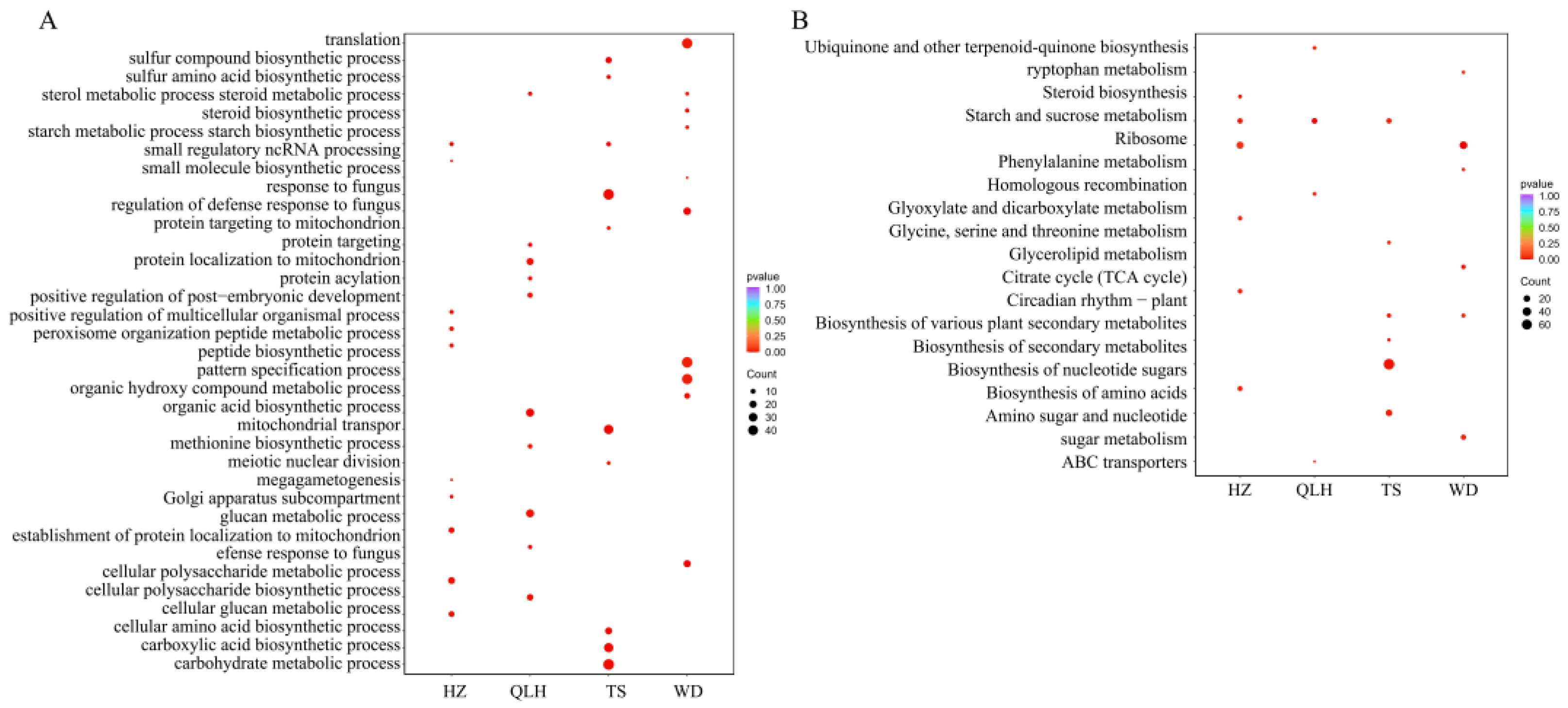
3.6. Identification of Accelerated Evolution Candidate Genes and Functional Enrichment Analysis in Four E. brevicornu Geographic Populations
3.7. Regional Environmental Correlates of Accelerated Gene Evolution in E. brevicornu Populations
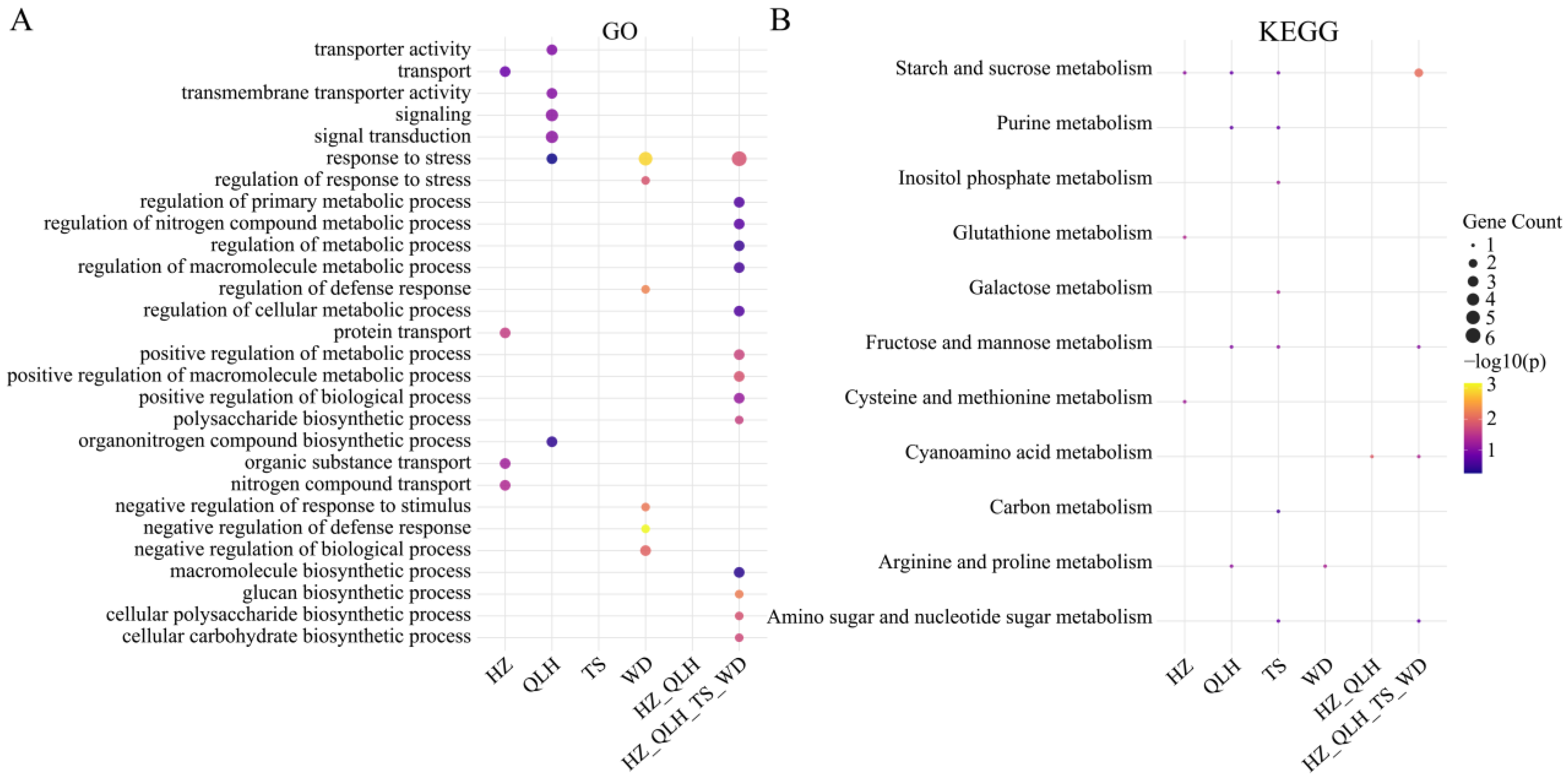
3.8. PSGs and Functional Adaptations in E. brevicornu Populations
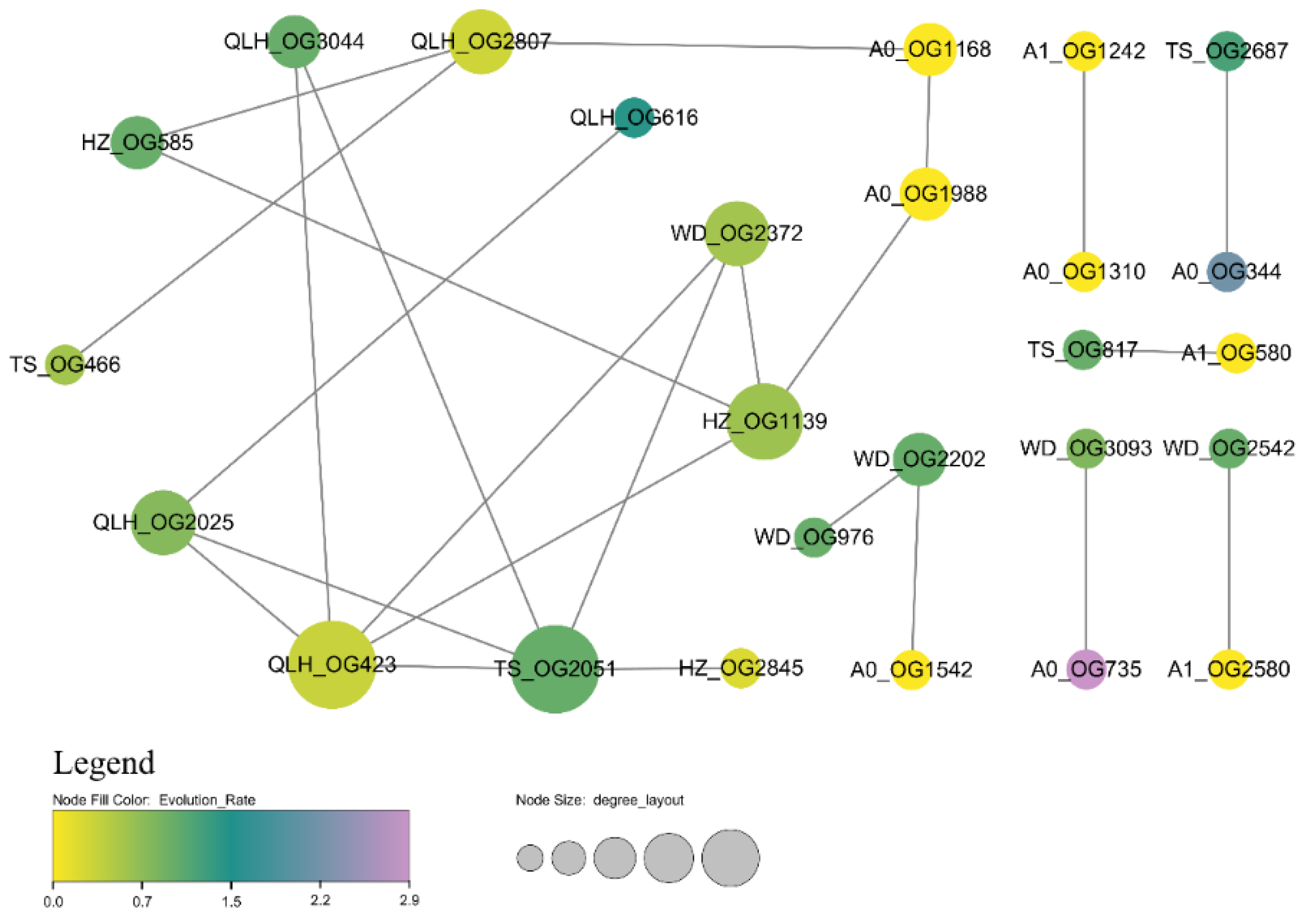
3.9. Protein Interaction Networks Reveal Core Regulators of Adaptive Evolution in E. brevicornu
4. Discussion
4.1. Systematic Classification and Evolutionary Dynamics of Sect. Diphyllon
4.2. Ecological and Historical Interpretation of Divergence Times in Chinese Epimedium Lineages
4.3. Ecological Drivers of Accelerated Population Evolution: Divergent Selective Pressures Shape Gene Function in E. brevicornu
4.4. Positively Selected Genes and Phylogenetic Architecture: Ancestral Node Regulatory Network Remodeling and Population Specialization Trends
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stearn, W.T.; Shaw, J. The Genus Epimedium and Other Herbaceous Berberidaceae; Timber Press: Oregon, Portland, 2002. [Google Scholar]
- Shen, P.; Guo, B.L.; Gong, Y.; Hong, D.Y.; Hong, Y.; Yong, E.L. Taxonomic, genetic, chemical and estrogenic characteristics of Epimedium species. Phytochemistry 2007, 68, 1448–1458. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, B.; Wen, S.; Wang, Y.; Pan, C.; Qu, L.; Yin, Y.; Wei, Y. Advancements in the Biotransformation and Biosynthesis of the Primary Active Flavonoids Derived from Epimedium. Molecules 2023, 28, 7173. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, R.; Meng, Y.; Guo, B.L.; Liu, Q.R.; Nie, Z.L. Parallel evolution, atavism, and extensive introgression explain the radiation of Epimedium sect. Diphyllon (Berberidaceae) in southern East Asia. Front. Plant Sci. 2023, 14, 1234148. [Google Scholar] [CrossRef] [PubMed]
- Tsun-Shen, Y. Petal evolution and distribution patterns of Epimedium Linn (Berberidaceae). J. Syst. Evol. 2002, 40, 481. (In Chinese) [Google Scholar]
- Zhang, Y.; Yang, L.; Chen, J.; Sun, W.; Wang, Y. Taxonomic and phylogenetic analysis of Epimedium L. based on amplified fragment length polymorphisms. Sci. Hortic. 2014, 170, 284–292. [Google Scholar] [CrossRef]
- Chen, Y.P.; Sunojkumar, P.; Spicer, R.A.; Hodel, R.G.; Soltis, D.E.; Soltis, P.S.; Paton, A.J.; Sun, M.; Drew, B.T.; Xiang, C.L. Rapid radiation of a plant lineage sheds light on the assembly of dry valley biomes. Mol. Biol. Evol. 2025, 42, msaf011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, L.; Liu, A.; Chen, J.; Wu, L.; Hu, W.; Zhang, W.; Kim, K.; Lee, S.C.; Yang, T.; et al. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 2016, 7, 306. [Google Scholar] [CrossRef]
- Guo, M.; Pang, X.; Xu, Y.; Jiang, W.; Liao, B.; Yu, J.; Xu, J.; Song, J.; Chen, S. Plastid genome data provide new insights into the phylogeny and evolution of the genus Epimedium. J. Adv. Res. 2022, 36, 175–185. [Google Scholar] [CrossRef]
- Yanqin, X.; Wanzhen, C.; Shengfu, H.; Xiaohu, H.; Fei, G.; Ying, W. Morphological variation of non-glandular hairs in cultivated Epimedium sagittatum (Berberidaceae) populations and implications for taxonomy. Biodivers. Sci. 2013, 21, 185. [Google Scholar] [CrossRef]
- De Smet, Y.; Goetghebeur, P.; Wanke, S.; Asselman, P.; Samain, M.S. Additional evidence for recent divergence of Chinese Epimedium (Berberidaceae) derived from AFLP, chloroplast and nuclear data supplemented with characterisation of leaflet pubescence. Plant Ecol. Evol. 2012, 145, 73–87. [Google Scholar] [CrossRef]
- Zhang, M.L.; Uhink, C.H.; Kadereit, J.W. Phylogeny and biogeography of Epimedium/Vancouveria (Berberidaceae): Western North American-East Asian disjunctions, the origin of European mountain plant taxa, and East Asian species diversity. Syst. Bot. 2007, 32, 81–92. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, D.; Guo, K.; Zhao, L.; Meng, F.; Xiao, J.; Niu, Y.; Sun, Y. Comparative analysis of codon usage patterns in chloroplast genomes of ten Epimedium species. BMC Genom. Data 2023, 24, 3. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, H.; Zhang, S.; Jin, J.; Liang, X.; Huang, B.; Wang, L. The complete chloroplast genome of the rare species Epimedium tianmenshanensis and comparative analysis with related species. Physiol. Mol. Biol. Plants 2020, 26, 2075–2083. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Z.; Su, Y.; Wang, T. Population transcriptomic sequencing reveals allopatric divergence and local adaptation in Pseudotaxus chienii (Taxaceae). BMC Genom. 2021, 22, 388. [Google Scholar] [CrossRef]
- Xu, W.; Ren, C.; Zhang, X.; Comes, H.P.; Liu, X.; Li, Y.; Kettle, C.J.; Jalonen, R.; Gaisberger, H.; Qiu, Y. Genome sequences and population genomics reveal climatic adaptation and genomic divergence between two closely related sweetgum species. Plant J. 2024, 118, 1372–1387. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, L.; Shu, J.; Jin, D.; Shen, H.; Chen, H.; Zhang, R.; Yan, Y. Transcriptomic evidence of adaptive evolution of the epiphytic fern Asplenium nidus. Int. J. Genom. 2019, 1, 1429316. [Google Scholar] [CrossRef]
- Zhu, X.; Wen, S.; Gul, H.; Xu, P.; Yang, Y.; Liao, X.; Wu, L. Exploring regulatory network of icariin synthesis in Herba Epimedii through integrated omics analysis. Front. Plant Sci. 2024, 15, 1409601. [Google Scholar] [CrossRef]
- Xu, C.; Liu, X.; Shen, G.; Fan, X.; Zhang, Y.; Sun, C.; Suo, F.; Guo, B. Time-series transcriptome provides insights into the gene regulation network involved in the icariin-flavonoid metabolism during the leaf development of Epimedium pubescens. Front. Plant Sci. 2023, 14, 1183481. [Google Scholar] [CrossRef]
- Chen, J.; Xu, Y.; Wei, G.; Liao, S.; Zhang, Y.; Huang, W.; Yuan, L.; Wang, Y. Chemotypic and genetic diversity in Epimedium sagittatum from different geographical regions of China. Phytochemistry 2015, 116, 180–187. [Google Scholar] [CrossRef]
- Qin, W.; Yang, Y.; Wang, Y.; Zhang, X.; Liu, X. Transcriptomic and metabolomic analysis reveals the difference between large and small flower taxa of Herba Epimedii during flavonoid accumulation. Sci. Rep. 2022, 12, 2762. [Google Scholar] [CrossRef] [PubMed]
- Savolainen, O.; Lascoux, M.; Merilä, J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013, 14, 807–820. [Google Scholar] [CrossRef]
- Ma, H.; He, X.; Yang, Y.; Li, M.; Hao, D.; Jia, Z. The genus Epimedium: An ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2011, 134, 519–541. [Google Scholar] [CrossRef]
- Xu, N.; Zhou, G.; Li, X.; Lu, H.; Meng, F.; Zhai, H. Geographical classification of Epimedium based on HPLC fingerprint analysis combined with multi-ingredients quantitative analysis. Biomed. Chromatogr. 2017, 31, e3871. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Du, X.; Zhao, C.; He, P.; Meng, F. Spatial distribution dynamics for Epimedium brevicornum Maxim. from 1970 to 2020. Ecol. Evol. 2024, 14, e11010. [Google Scholar] [CrossRef] [PubMed]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Regev, A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Emms, D.M.; OrthoFinder, S.K. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Löytynoja, A.; Goldman, N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 2005, 102, 10557–10562. [Google Scholar] [CrossRef]
- Sun, Y. FasParser2: A graphical platform for batch manipulation of tremendous amount of sequence data. Bioinformatics 2018, 34, 2493–2495. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Guo, M.; Ren, L.; Xu, Y.; Liao, B.; Song, J.; Li, Y.; Mantri, N.; Guo, B.; Chen, S.; Pang, X. Development of plastid genomic resources for discrimination and classification of Epimedium wushanense (Berberidaceae). Int. J. Mol. Sci. 2019, 20, 4003. [Google Scholar] [CrossRef]
- Joly, S.; McLenachan, P.A.; Lockhart, P.J. A Statistical Approach for Distinguishing Hybridization and Incomplete Lineage Sorting. Am. Nat. 2009, 174, E54–E70. [Google Scholar] [CrossRef] [PubMed]
- Sebastin, R.; Kim, J.; Jo, I.H.; Yu, J.K.; Jang, W.; Han, S.; Park, H.S.; AlGarawi, A.M.; Hatamleh, A.A.; So, Y.S.; et al. Comparative chloroplast genome analyses of cultivated and wild Capsicum species shed light on evolution and phylogeny. BMC Plant Biol. 2024, 24, 797. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Koester, T.; Staiger, D. Pre-mRNA Splicing in Plants: In Vivo Functions of RNA-Binding Proteins Implicated in the Splicing Process. Biomolecules 2015, 5, 1717–1740. [Google Scholar] [CrossRef]
- Xin, R.; Zhu, L.; Salomé, P.A.; Mancini, E.; Marshall, C.M.; Harmon, F.G.; Yanovsky, M.J.; Weigei, D.; Huq, E. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E7018–E7027. [Google Scholar] [CrossRef]
- Yang, C.; Georgiou, M.; Atkinson, R.; Collin, J.; Al-Aama, J.; Nagaraja-Grellscheid, S.; Johnson, C.; Ali, R.; Armstrong, L.; Mozaffari-Jovin, S.; et al. Pre-mRNA processing factors and retinitis pigmentosa: RNA splicing and beyond. Front. Cell Dev. Biol. 2021, 9, 700276. [Google Scholar] [CrossRef]
- Shang, X.; Cao, Y.; Ma, L. Alternative splicing in plant genes: A means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Zhurov, V.; Navarro, M.; Bruinsma, K.A.; Arbona, V.; Santamaria, M.E.; Cazaux, M.; Wybouw, N.; Osborne, E.J.; Ens, C.; Rioja, C.; et al. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiol. 2014, 164, 384–399. [Google Scholar] [CrossRef]
- Hardie, D.G. Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Biol. 1999, 50, 97–131. [Google Scholar] [CrossRef]
- McIntosh, K.B.; Bonham-Smith, P.C. Ribosomal protein gene regulation: What about plants? Can. J. Bot. 2006, 84, 342–362. [Google Scholar] [CrossRef]
- Veith, T.; Martin, R.; Wurm, J.P.; Weis, B.L.; Duchardt-Ferner, E.; Safferthal, C.; Hennig, R.; Mirus, O.; Bohnsack, M.T.; Wöhnert, J.; et al. Structural and functional analysis of the archaeal endonuclease Nob1. Nucleic Acids Res. 2012, 40, 3259–3274. [Google Scholar] [CrossRef]
- Macovei, A.; Vaid, N.; Tula, S.; Tuteja, N. A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal. Behav. 2012, 7, 1138–1143. [Google Scholar] [CrossRef]
- Quan, Q.; Li, Y. Effect of morphs on reproductive biology of Epimedium pubescens (Berberidaceae): A species endemic to China. Int. J. Agric. Biol. 2018, 20, 945–950. [Google Scholar] [CrossRef]
- Sun, Y.; Fung, K.P.; Leung, P.C.; Shaw, P.C. A phylogenetic analysis of Epimedium (Berberidaceae) based on nuclear ribosomal DNA sequences. Mol. Phylogenet. Evol. 2005, 35, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, F.; Yan, S.; Zhu, Z.; Gao, X.; Zhao, X. Phylogenomics and plastome evolution of Indigofera (Fabaceae). Front. Plant Sci. 2023, 14, 1186598. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zheng, L.; Song, H.; Yu, X. Genome-wide identification and expression patterns of the aspartic protease gene family in Epimedium pubescens. BMC Genom. 2025, 26, 1–16. [Google Scholar] [CrossRef]
- Cui, Z.; Chen, Y.; Zhang, W.; Zhou, S.; Zhou, L.; Zhang, M.; Li, C. Research history, glacial chronology and origins of Quaternary glaciations in China. Quat. Sci. 2011, 31, 749–764. (In Chinese) [Google Scholar]
- Wen, J.; Zhang, J.Q.; Nie, Z.L.; Zhong, Y.; Sun, H. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 2014, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Shi, T.; Mo, Z.; Zhao, C. Across a phylogeographic break in the Qinling Mountains-Huaihe River Line: Quaternary evolutionary history of a medicinal and edible homologous plant (Allium macrostemon) in China. BMC Ecol. Evol. 2024, 24, 107. [Google Scholar] [CrossRef]
- Qiu, Y.; Fu, C.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Wang, Y.; Comes, H.; Cao, Y.; Guo, R.; Mao, Y.; Qiu, Y. Quaternary climate change drives allo-peripatric speciation and refugial divergence in the Dysosma versipellis-pleiantha complex from different forest types in China. Sci. Rep. 2017, 7, 40261. [Google Scholar] [CrossRef]
- Zhang, K.; Leng, Y.; Hao, R.; Zhang, W.; Li, H.; Chen, M.; Zhu, F. Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques. Int. J. Mol. Sci. 2024, 25, 12666. [Google Scholar] [CrossRef]
- Andrew, S.C.; Simonsen, A.K.; Coppin, C.W.; Arnold, P.A.; Briceño, V.F.; McLay, T.G.; Jackson, C.J.; Gallagher, R.V.; Mokany, K. Expression–environment associations in transcriptomic heat stress responses for a global plant lineage. Mol. Ecol. 2024, 33, e17473. [Google Scholar] [CrossRef]
- Gurung, P.D.; Upadhyay, A.K.; Bhardwaj, P.K.; Sowdhamini, R.; Ramakrishnan, U. Transcriptome analysis reveals plasticity in gene regulation due to environmental cues in Primula sikkimensis, a high altitude plant species. BMC Genom. 2019, 20, 989. [Google Scholar] [CrossRef]
- Dahal, K.; Vanlerberghe, G.C. Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 2017, 213, 560–571. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.P.; Romero-Rodríguez, M.C.; Sánchez-Lucas, R.; Navarro-Cerrillo, R.M.; Medina-Aunon, J.A.; Jorrín-Novo, J.V. 2-DE proteomics analysis of drought treated seedlings of Quercus ilex supports a root active strategy for metabolic adaptation in response to water shortage. Front. Plant Sci. 2015, 6, 627. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Ding, G. Response mechanism of carbon metabolism of Pinus massoniana to gradient high temperature and drought stress. BMC Genom. 2024, 25, 166. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.J.; Kawagoe, T.; Sugisaka, J.; Honjo, M.N.; Iwayama, K.; Kudoh, H. Annual transcriptome dynamics in natural environments reveals plant seasonal adaptation. Nat. Plants 2019, 5, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Rathore, N.; Kumar, P.; Mehta, N.; Swarnkar, M.K.; Shankar, R.; Chawla, A. Time-series RNA-Seq transcriptome profiling reveals novel insights about cold acclimation and de-acclimation processes in an evergreen shrub of high altitude. Sci. Rep. 2022, 12, 15553. [Google Scholar] [CrossRef]
- Brachi, B.; Filiault, D.; Whitehurst, H.; Darme, P.; Le Gars, P.; Le Mentec, M.; Morton, T.C.; Kerdaffrec, E.; Rabanal, F.; Anastasio, A.; et al. Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proc. Natl. Acad. Sci. USA 2022, 119, e2201285119. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, H.; Gao, H.; Yu, S.; Liu, C.; Wang, Y.; Sun, Y.; Zhang, D. Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. Int. J. Mol. Sci. 2025, 26, 5864. [Google Scholar] [CrossRef]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Wang, L.; Lo, C.; Zhu, F. Alternative splicing and its roles in plant metabolism. Int. J. Mol. Sci. 2022, 23, 7355. [Google Scholar] [CrossRef]
- Anderson, J.T.; Willis, J.H.; Mitchell-Olds, T. Evolutionary genetics of plant adaptation. Trends Genet. 2011, 27, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, S.; Xu, X.; Zhou, W.; Wu, L. Alternative splicing: An efficient regulatory approach towards plant developmental plasticity. Wiley Interdiscip. Rev. RNA 2023, 14, e1758. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Lamanna, A.C.; Karbstein, K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc. Natl. Acad. Sci. USA 2009, 106, 14259–14264. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. The roles of reactive oxygen metabolism in drought: Not so cut and dried. Plant Physiol. 2014, 164, 1636–1648. [Google Scholar] [CrossRef]
- Hamel, L.P.; Nicole, M.C.; Sritubtim, S.; Morency, M.J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J.; et al. Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Barak, S.; Singh Yadav, N.; Khan, A. DEAD-box RNA helicases and epigenetic control of abiotic stress-responsive gene expression. Plant Signal. Behav. 2014, 9, e977729. [Google Scholar] [CrossRef]
- de Miguel, M.; Rodríguez-Quilón, I.; Heuertz, M.; Hurel, A.; Grivet, D.; Jaramillo-Correa, J.P.; Vendramin, G.G.; Plomion, C.; Majada, J.; Alía, R.; et al. Polygenic adaptation and negative selection across traits, years and environments in a long-lived plant species (Pinus pinaster Ait., Pinaceae). Mol. Ecol. 2022, 31, 2089–2105. [Google Scholar] [CrossRef] [PubMed]
- Hämälä, T.; Savolainen, O. Genomic Patterns of Local Adaptation under Gene Flow in Arabidopsis lyrata. Mol. Biol. Evol. 2019, 36, 2557–2571. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, S. Local Adaptation by Alleles of Small Effect. Am. Nat. 2015, 186 (Suppl. S1), S74–S89. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, S.; Qi, J.; Zhao, J.; Song, Q.; Xing, L.; Du, W.; Wang, X.; Zhang, X.; Zhou, X. Transcriptome-Based Phylogenomics and Adaptive Divergence Across Environmental Gradients in Epimedium brevicornu. Agronomy 2025, 15, 2139. https://doi.org/10.3390/agronomy15092139
Lu S, Qi J, Zhao J, Song Q, Xing L, Du W, Wang X, Zhang X, Zhou X. Transcriptome-Based Phylogenomics and Adaptive Divergence Across Environmental Gradients in Epimedium brevicornu. Agronomy. 2025; 15(9):2139. https://doi.org/10.3390/agronomy15092139
Chicago/Turabian StyleLu, Songsong, Jianwei Qi, Jun Zhao, Qianwen Song, Luna Xing, Weibo Du, Xuhu Wang, Xiaowei Zhang, and Xiaolei Zhou. 2025. "Transcriptome-Based Phylogenomics and Adaptive Divergence Across Environmental Gradients in Epimedium brevicornu" Agronomy 15, no. 9: 2139. https://doi.org/10.3390/agronomy15092139
APA StyleLu, S., Qi, J., Zhao, J., Song, Q., Xing, L., Du, W., Wang, X., Zhang, X., & Zhou, X. (2025). Transcriptome-Based Phylogenomics and Adaptive Divergence Across Environmental Gradients in Epimedium brevicornu. Agronomy, 15(9), 2139. https://doi.org/10.3390/agronomy15092139





