A Genome-Wide Association Study of Sugarcane Smut Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sugarcane Smut Inoculation and Evaluation
2.3. Genome Resequencing and Genotyping
2.4. Linkage Disequilibrium Analysis
2.5. Genome-Wide Association Study (GWAS)
2.6. Candidate Gene Identification for Associated SNPs
3. Results
3.1. Phenotypic Analysis of Smut Resistance in the Sugarcane Natural Population
3.2. Genome-Wide Association Study of Smut Resistance
3.3. Candidate Gene Analysis
3.4. Core Parents and Their Derivatives with SNP-Tagged Smut Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, M.; Zhu, G.; Lin, S.; Zhang, L.H. The mating-type locus b of the sugarcane smut Sporisorium scitamineum is essential for mating, filamentous growth and pathogenicity. Fungal Genet. Biol. 2015, 86, 1–8. [Google Scholar] [CrossRef]
- Wang, D.; Gou, Y.; Yi, C.; Li, Z.; Wang, W.; Lin, P.; Wang, W.; Sun, T.; Wang, T.; Zhao, W.; et al. ScWRKY2: A key regulator for smut resistance in sugarcane. Plant Biotechnol. J. 2025, 1–15. [Google Scholar] [CrossRef]
- Jing, Y.; Zhou, H.; Liu, X.; Tan, F.; Zhang, X.; Zhang, R.; Song, X.; Li, Y.; Yan, M.; Lei, J.; et al. Smut Resistant Identification and Analysis of New Sugarcane Clones of Guitang. Chin. J. Trop. Crops 2020, 41, 333–338. (In Chinese) [Google Scholar]
- Edwards, A.; Ritter, R.; Abel, K.; Manning, A.; Panhuysen, C.; Farrer, L.A. Complement factor polymorphism and age-related macular degeneration. Science 2005, 308, 421–424. [Google Scholar] [CrossRef]
- Hu, C.; Kuang, T.; Shaw, R.K.; Zhang, Y.; Fan, J.; Bi, Y.; Jiang, F.; Guo, R.; Fan, X. Genetic dissection of resistance to gray leaf spot by genome-wide association study in a multi-parent maize population. BMC Plant Biol. 2024, 24, 10. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, A.; Steffenson, B.; Smith, K.; Smith, M.; Dill-Macky, R. Identification of quantitative trait loci for net form net blotch resistance in contemporary barley breeding germplasm from the USA using genome-wide association mapping. Theor. Appl. Genet. 2020, 133, 1019–1037. [Google Scholar] [CrossRef]
- Noh, J.; Do, S.; Kim, G.; Choi, C. A genome-wide association study for the detection of genes related to apple Marssonina Blotch disease resistance in apples. Sci. Hortic. 2020, 262, 108986. [Google Scholar] [CrossRef]
- Zhao, X.; Tang, Q.; Xing, Y.; Wei, L.; Chao, Q.; Zuo, W.; Lubberstedt, T.; Xu, M. Marker-assisted introgression of QhSR1 to improve maize resistance to head smut. Mol. Breed. 2012, 30, 1077–1088. [Google Scholar] [CrossRef]
- Chen, X.; Jia, Y.; Jia, H.; Pinson, R.; Wang, X.; Wu, B. Functional Interactions between Major Rice Blast Resistance Genes Pi-ta and Pi-b, and Minor Blast Resistance Quantitative Trait Loci. Phytopathology 2018, 108, 1095–1103. [Google Scholar] [CrossRef]
- Xu, Z.; Hua, J.; Wang, F.; Cheng, Z.; Meng, Q.; Chen, Y.; Han, X.; Tie, S.; Liu, C.; Li, X.; et al. Marker-assisted selection of qMrdd8 to improve maize resistance to rough dwarf disease. Breed. Sci. 2020, 70, 183–192. [Google Scholar] [CrossRef]
- Gouy, M.; Rousselle, Y.; Thong, A.; Anglade, A.; Royaert, S.; Nibouche, S.; Costet, L. Genome wide association mapping of agro-morphological and disease resistance traits in sugarcane. Euphytica 2015, 202, 269–284. [Google Scholar] [CrossRef]
- Barreto, F.; Rosa, J.; Balsalobre, T.; Pastina, M.; Silva, R.; Hoffmann, H.; Souza, A.; Garcia, A.; Carneiro, M. A genome-wide association study identified loci for yield component traits in sugarcane (Saccharum spp.). PLoS ONE 2019, 14, e0219843. [Google Scholar] [CrossRef]
- Fickett, N.; Gutierrez, A.; Verma, M.; Pontif, M.; Hale, A.; Kimbeng, C.; Baisakh, N. Genome-wide association mapping identifies markers associated with cane yield components and sucrose traits in the Louisiana sugarcane core collection. Genomics 2019, 111, 1794–1801. [Google Scholar] [CrossRef]
- Yang, X.; Luo, Z.; Todd, J.; Sood, S.; Wang, J. Genome-wide association study of multiple yield traits in a diversity panel of polyploid sugarcane (Saccharum spp.). Plant Genome 2020, 13, e20006. [Google Scholar] [CrossRef]
- Dijoux, J.; Rio, S.; Hervouet, C.; Garsmeur, O.; Barau, L.; Dumont, T.; Rott, P.; D’Hont, A.; Hoarau, J. Unveiling the predominance of Saccharum spontaneum alleles for resistance to orange rust in sugarcane using genome-wide association. Theor. Appl. Genet. 2024, 137, 81. [Google Scholar] [CrossRef] [PubMed]
- Mccord, P.; Glynn, N.; Comstock, J. Identifying markers for resistance to sugarcane orange rust (Puccinia kuehnii) via selective genotyping and capture sequencing. Euphytica 2019, 215, 150. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Z.; Fu, D.; Fang, J.; Feng, X.; Xie, J.; Wu, B.; Luo, Y.; Zhu, M.; Qi, Y. Identification of Genetic Loci for Sugarcane Leaf Angle at Different Developmental Stages by Genome-Wide Association Study. Front. Plant Sci. 2022, 13, 841693. [Google Scholar] [CrossRef]
- Cai, Q.; Fan, Y. Descriptors and Data Standard for Sugarcane (Saccharum officinarum L.); China Agricultural Publishing House: Beijing, China, 2006; pp. 45–46. (In Chinese) [Google Scholar]
- Zhang, J.; Qi, Y.; Hua, X.; Wang, Y.; Wang, B.; Qi, Y.; Huang, Y.; Yu, Z.; Gao, R.; Zhang, Y.; et al. The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum. Nat. Genet. 2025, 57, 242–253. [Google Scholar] [CrossRef]
- Sadowski, M.; Kraft, A.; Szalaj, P.; Wlasnowolski, M.; Tang, Z.; Ruan, Y.; Plewczynski, D. Spatial chromatin architecture alteration by structural variations in human genomes at the population scale. Genome Biol. 2019, 20, 148. [Google Scholar] [CrossRef]
- Khvorykh, G.V.; Khrunin, A.V. Imputeqc: An R package for assessing imputation quality of genotypes and optimizing imputation parameters. BMC Bioinform. 2020, 21, 304. [Google Scholar] [CrossRef] [PubMed]
- Pino, D.C.D.; Lozano, R.; Wolfe, M.D.; Jannink, J.L. Genome-Wide Association Studies and Heritability Estimation in the Functional Genomics Era. In Population Genomics; Springer: Cham, Switzerland, 2018; pp. 361–425. [Google Scholar]
- Chen, S.; Feng, X.; Zhang, Z.; Hua, X.; Zhang, Q.; Chen, C.; Li, J.; Liu, X.; Weng, C.; Chen, B.; et al. ScDB: A comprehensive database dedicated to Saccharum, facilitating functional genomics and molecular biology studies in sugarcane. Plant Biotechnol. J. 2024, 22, 14457. [Google Scholar] [CrossRef]
- Bhuiyan, S.; Croft, B.J. Incidence versus severity—What difference would it make to smut screening. In Proceedings of the 33rd Annual Conference of the Australian Society of Sugar Cane Technologists 2011, Mackay, QLD, Australia, 4–6 May 2011; pp. 118–125. [Google Scholar]
- Marques, J.R.P.; Hoy, W.J.; Beatriz, A.; Viveros, F.G.; Vieira, M.L.C.; Baisakh, N. Sugarcane Cell Wall-Associated Defense Responses to Infection by Sporisorium scitamineum. Front. Plant Sci. 2018, 9, 9698. [Google Scholar] [CrossRef] [PubMed]
- Orellana, P.P.A.; Perez, P.J. Evaluation of the resistance in sugar cane plantlets (Saccharum spp. hybrids) against Ustilago scitaminea Syd. Jpn. J. Appl. Phys. 1987, 17, 733–734. [Google Scholar]
- Deng, Q.Q.; Xu, G.H.; Dou, Z.M.; Shen, W.K. Identification of three Sporisorium scitamineum pathogenic races in mainland China. Int. J. Agric. Biol. 2018, 20, 799–802. [Google Scholar]
- Shen, W.; Deng, H. Analysis of Results from Smut Resistant Identification in Sugarcane Varieties Introduced. Chin. Agric. Sci. Bull. 2011, 27, 234–238. (In Chinese) [Google Scholar]
- Zhang, Y.Y.; Huang, N.; Xiao, X.H.; Huang, L.; Liu, F.; Su, W.H.; Que, Y.X. Molecular variation of Sporisorium scitamineum in Mainland China revealed by internal transcribed spacers. Genet. Mol. Res. GMR 2015, 14, 7894–7909. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef]
- Giulietti, S.; Bigini, V.; Savatin, D.V. ROS and RNS production, subcellular localization and signaling triggered by immunogenic danger signals. J. Exp. Bot. 2024, 75, 4512–4534. [Google Scholar] [CrossRef]
- Jiang, G.; Yin, D.; Zhao, J.; Chen, H.; Guo, L.; Zhu, L.; Zhai, W. The rice thylakoid membranebound ascorbate peroxidase OsAPX8 functions in tolerance to bacterial blight. Sci. Rep. 2016, 6, 26104. [Google Scholar]
- Zhang, J.; Jia, X.; Wang, G.; Ma, S.; Wang, S.; Yang, Q.; Chen, X.; Zhang, Y.; Lyu, Y.; Wang, X. Ascorbate peroxidase 1 confers resistance to southern corn leaf blight in maize. J. Integr. Plant Biol. 2022, 64, 1196–1211. [Google Scholar] [CrossRef]
- You, X.; Zhang, F.; Liu, Z.; Wang, M.; Xu, X.; He, F.; Wang, D.; Wang, R.; Wang, Y.; Wang, G.; et al. Rice catalase OsCATC is degraded by E3 ligase APIP6 to negatively regulate immunity. Plant Physiol. 2022, 190, 1095–1099. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, L.; Zhu, R.; Zhou, X.; Zhang, J.; Li, D.; He, S.; Qiao, Y. Phytophthora sojae effector PsAvh113 associates with the soybean transcription factor GmDPB to inhibit catalasemediated immunity. Plant Biotechnol. J. 2023, 21, 1393–1407. [Google Scholar] [CrossRef]
- Su, Y.; Guo, J.; Ling, H.; Chen, S.; Wang, S.; Xu, L.; Allan, A.C.; Que, Y. Isolation of a Novel Peroxisomal Catalase Gene from Sugarcane, Which is Responsive to Biotic and Abiotic Stresses. PLoS ONE 2014, 9, e84426. [Google Scholar] [CrossRef]
- Casu, R.E.; Dimmock, C.M.; Chapman, S.C.; Grof, C.P.L.; McIntyre, C.L.; Bonnett, G.D.; Manners, J.M. Identification of differentially expressed transcripts frommaturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Mol. Biol. 2004, 54, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Guo, J.; Li, G.; Que, Y.; Xu, L. Electronic cloning and characterization of CAT gene from Saccharum officinarum using bioinformatics tool. Chin. J. Bioinform. 2012, 10, 65–70. (In Chinese) [Google Scholar]
- Liu, Y.; Yao, Y.; Hu, X.; Xu, L.; Xing, S.; Zhang, S. Isolation and Characterization of Catalase (SsCAT-1) Gene in Saccharum spontaneum L. Mol. Plant Breed. 2014, 12, 1251–1258. (In Chinese) [Google Scholar]
- Liu, Y.; Yao, Y.; Hu, X.; Xu, L.; Xing, S.; Zhang, S. Isolation and Characterization of Catalase Gene (EaCAT-1a) in Erianthus arundinaceus (Retz.) Jesw. Southwest China J. Agric. Sci. 2015, 4, 1535–1541. (In Chinese) [Google Scholar]
- Su, Y.; Wang, Z.; Li, Z.; Liu, F.; Xu, L.; Que, Y.; Dai, M.; Chen, Y. Molecular Cloning and Functional Identification of Peroxidase Gene ScPOD02 in Sugarcane. ACTA Agron. Sin. 2017, 43, 510–521. [Google Scholar] [CrossRef]
- Zhang, B.; Shao, M.; Huang, Y.; Huang, X.; Song, X.; Chen, H.; Wang, S.; Tan, Q.; Yang, L.; Li, Y. Cloning and Expression Analysis of Peroxidase Gene (ScAPX1) from Sugarcane. Biotechnol. Bull. 2019, 35, 31–37. (In Chinese) [Google Scholar]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Sun, B.Y.; Yang, L.Y.; Dong, S.M. Advances in regulatory mechanisms of plant resistance genes. Chin. Bull. Life Sci. 2025, 37, 477–489. (In Chinese) [Google Scholar]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Reddy, A.C.; Venkat, S.; Sing, T.H.; Aswath, C.; Reddy, K.M.; Reddy, D.C.L. Isolation, characterization and evolution of NBS-LRR encoding disease-resistance gene analogs in eggplant against bacterial wilt. Eur. J. Plant Pathol. 2015, 143, 417–426. [Google Scholar] [CrossRef]
- Xu, Y.J.; Liu, F.; Zhu, S.W.; Li, X.Y. The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and Arabidopsis. Front. Plant Sci. 2018, 9, 1033. [Google Scholar] [CrossRef]
- Hassan, Z.; Rahim, M.A.; Jung, H.J.; Park, J.I.; Kim, H.T.; Nou, I.S. Genome-wide characterization of NBS-encoding genes in watermelon and their potential association with gummy stem blight resistance. Int. J. Mol. Sci. 2019, 20, 902. [Google Scholar] [CrossRef] [PubMed]
- Ge, R.C.; Chen, G.P.; Zhao, B.C.; Shen, Y.Z.; Huang, Z.J. Cloning and functional characterization of a wheat serine/threonine kinase gene (TaSTK) related to salt-resistance. Plant Sci. 2007, 173, 55–60. [Google Scholar] [CrossRef]
- Gao, L.; Wang, S.; Li, X.Y.; Wei, X.J.; Zhang, Y.J.; Wang, H.Y.; Liu, D.Q. Expression and functional analysis of a pathogenesis-related protein 1 gene, TcLr19PR1, involved in wheat resistance against leaf rust fungus. Plant Mol. Biol. Report. 2015, 33, 797–805. [Google Scholar] [CrossRef]
- Bundo, M.; Coca, M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 2016, 14, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.J.; Qin, L.O.; Wu, M.X.; Zou, W.H.; Zang, S.J.; Zhao, Z.N.; Lin, P.X.; Guo, J.L.; Wang, H.B.; Que, Y.X. Identification and characterization of WAK gene family in Saccharum and the negative roles of ScWAK1 under the pathogen stress. Int. J. Biol. Macromol. 2023, 224, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, F.; Zhang, X.; Wang, W.; Sun, T.; Chen, Y.; Dai, M.; Yu, S.; Xu, L.; Su, Y.; et al. Expression characteristics and functional analysis of the ScWRKY3 gene from sugarcane. Int. J. Mol. Sci. 2018, 19, 4059. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, X.; He, Q.; Guo, D.; Liu, C.; Cao, J.; Wu, Z.; Kang, Z.; Wang, X. TaMYB29: A novel R2R3-MYB transcription factor involved in wheat defense against stripe rust. Front. Plant Sci. 2021, 12, 783388. [Google Scholar] [CrossRef] [PubMed]
- Que, Y.; Su, Y.; Guo, J.; Wu, Q.; Xu, L. A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-seq. PLoS ONE 2014, 9, e106476. [Google Scholar] [CrossRef]
- Agisha, V.N.; Ashwin, N.M.R.; Vinodhini, R.T.; Nalayeni, K.; Ramesh, S.A.; Malathi, P.; Viswanathan, R. Transcriptome analysis of sugarcane reveals differential switching of major defense signaling pathways in response to Sporisorium scitamineum isolates with varying virulent attributes. Front. Plant Sci. 2022, 13, 969826. [Google Scholar] [CrossRef] [PubMed]

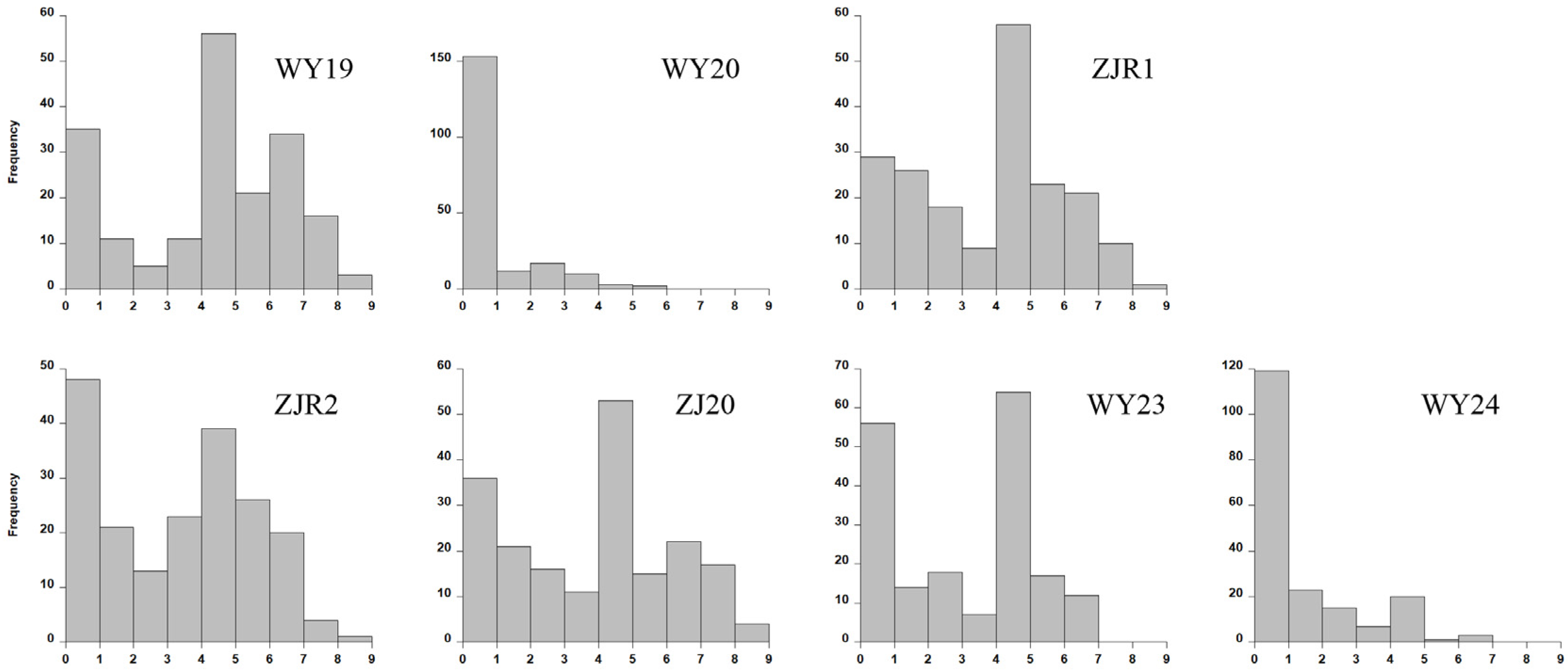

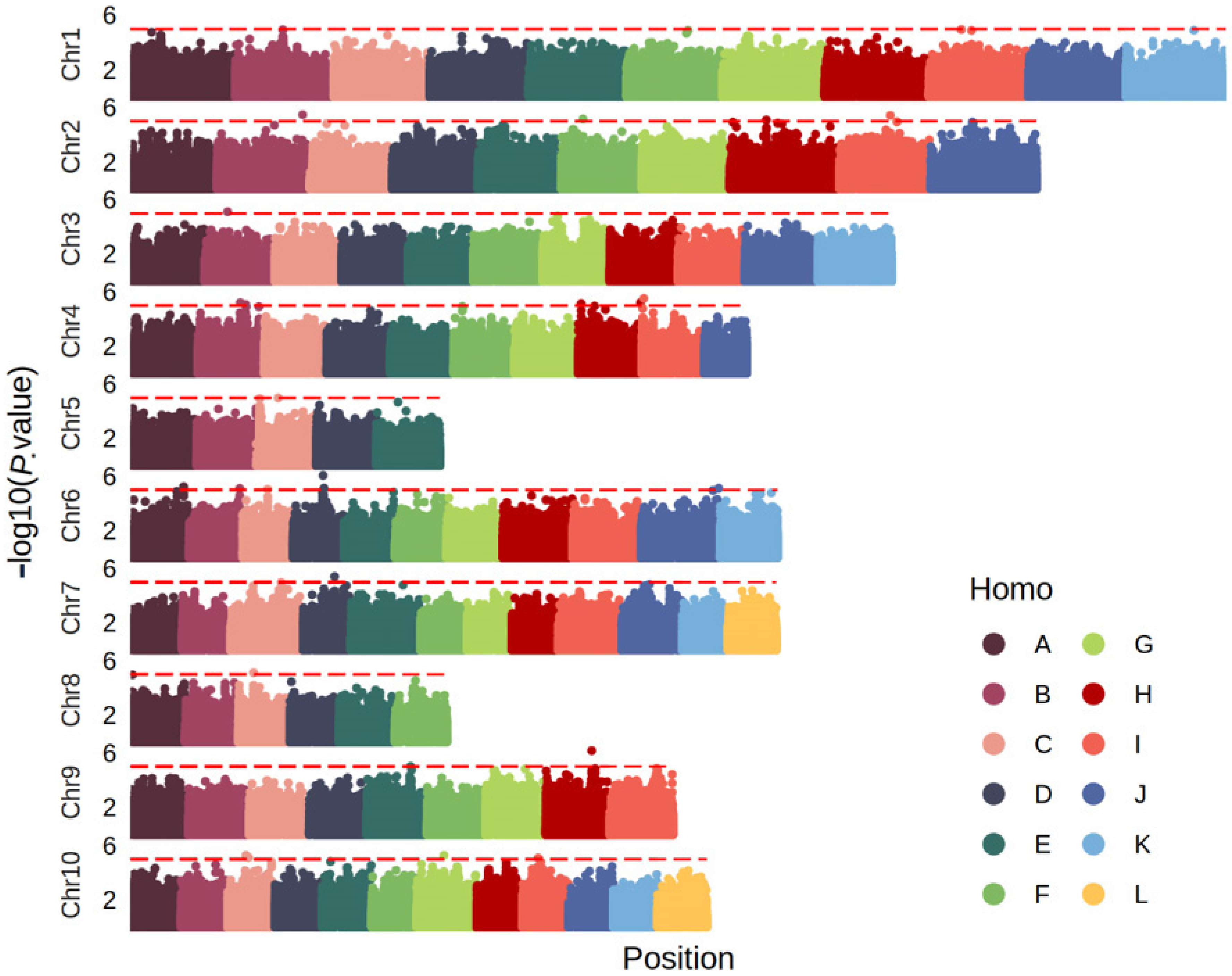
| Name | WY19 1 | WY20 | ZJR1 | ZJR2 | ZJ20 | WY23 | WY24 | Snp1 | Snp2 | Snp3 | Snp4 | Snp5 | Snp6 | Snp7 | Snp8 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C88-380 | 1 3 | 1 | 1 | 1 | 1 | 1 | 1 | CC 4 | CC | GC | TT | AA | CC | GG | AA |
| Fu_nong-07-14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | CC | CC | GC | TT | AA | CC | GG | AA |
| HOCP07-612 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | CC | CC | CC | TT | AA | CC | GG | AA |
| CP84-1198 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | CC | CC | CG | TT | AA | CC | GG | AA |
| GT-03-1453 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | CC | CC | CC | TT | AA | CC | GG | AA |
| CP75-1632 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | TT | CC | CG | CT | AA | CC | GG | AA |
| Q208 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | CC | CC | GC | TT | AA | CC | GG | AA |
| Fu_nong-91-23 | 2 | NA | 1 | 2 | 1 | 1 | 1 | TT | CC | GC | TT | AA | CC | GG | AG |
| CP72-2086 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | CT | CC | CC | TT | AG | CC | GG | AA |
| Yunzhe-89-151 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | CC | CC | CC | CT | AA | CC | GG | AA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Li, X.; Cai, E.; Feng, X.; Fang, J.; Wu, J.; Zhang, N. A Genome-Wide Association Study of Sugarcane Smut Resistance. Agronomy 2025, 15, 2111. https://doi.org/10.3390/agronomy15092111
Chen X, Li X, Cai E, Feng X, Fang J, Wu J, Zhang N. A Genome-Wide Association Study of Sugarcane Smut Resistance. Agronomy. 2025; 15(9):2111. https://doi.org/10.3390/agronomy15092111
Chicago/Turabian StyleChen, Xinglong, Xuhui Li, Enping Cai, Xiaomin Feng, Junteng Fang, Jiayun Wu, and Nannan Zhang. 2025. "A Genome-Wide Association Study of Sugarcane Smut Resistance" Agronomy 15, no. 9: 2111. https://doi.org/10.3390/agronomy15092111
APA StyleChen, X., Li, X., Cai, E., Feng, X., Fang, J., Wu, J., & Zhang, N. (2025). A Genome-Wide Association Study of Sugarcane Smut Resistance. Agronomy, 15(9), 2111. https://doi.org/10.3390/agronomy15092111






