Abstract
Cassava (Manihot esculenta Crantz) is essential for global food security, especially in tropical regions. As an important genetic resource, its genetics plays a key role in crop breeding, enabling the development of more productive and pest- and disease-resistant varieties. Scientometrics, which quantitatively analyzes the production and impact of scientific research, is crucial for understanding trends in cassava genetics. This study aimed to apply bibliometric methods to conduct a scientific mapping analysis based on yearly publication trends, paper classification, author productivity, journal impact factor, keywords occurrences, and omic approaches to investigate the application of genetics to the species from 1960 to 2022. From the quantitative data analyzed, 3246 articles were retrieved from the Web of Science platform, of which 654 met the inclusion criteria. A significant increase in scientific production was observed from 1993, peaking in 2018. The first article focused on genetics was published in 1969. Among the most relevant journals, Euphytica stood out with 36 articles, followed by Genetics and Molecular Research (n = 30) and Frontiers in Plant Science (n = 25). Brazil leads in the number of papers on cassava genetics (n = 143), followed by China (n = 110) and the United States (n = 75). The analysis of major methodologies (n = 185) reveals a diversified panorama during the study period. Morpho-agronomic descriptors persisted from 1978 to 2022; however, microsatellite markers were the most widely used, with 102 records. Genomics was addressed in 87 articles, and transcriptomics in 65. By clarifying the current landscape, this study supports cassava conservation and breeding, assists in public policy formulation, and guides future research in the field.
1. Introduction
Cassava (Manihot esculenta Crantz) is one of the most globally relevant crops, especially in tropical and subtropical regions [1]. Its ability to adapt to poor soils and adverse climatic conditions makes it an important food source in developing countries [2]. Approximately 50% of the world’s production is concentrated in African nations, with Brazil as one of the leading producers [3]. Cassava cultivation not only meets food needs but is also a source of raw materials for various products, such as flour and biofuels [4].
Given its agronomic relevance, cassava breeding programs face challenges in releasing cultivars with desirable traits, including resistance to pests and diseases, high productivity, high technological and sensory qualities, and tolerance to climate stress [5]. The use of molecular markers has proven to be an effective approach to enhancing the development of these cultivars, allowing for better selection and adaptation to climate variations [6]. Cassava breeding depends on understanding genetic variation, which is essential for the efficient management of available resources. In this scenario, germplasm banks play a crucial role, directly contributing to the conservation and development of new cultivars [7]. The diversity maintained in these banks enables the selection of important traits, such as disease resistance, adaptation to adverse conditions, and yield breeding, promoting significant advances in sustainable cassava cultivation [8].
Cassava breeding programs face significant challenges in developing cultivars that combine essential traits such as resistance to pests and diseases (e.g., cassava mosaic disease, cassava brown streak disease), high productivity, superior technological (e.g., dry matter, starch yield) and sensory qualities, and tolerance to climate stress [5]. While molecular markers are a powerful tool for enhancing selection efficiency and developing climate-resilient varieties [6], key research gaps hinder their broader application. These include the polygenic nature and low heritability of critical traits like fresh root yield and drought tolerance, complicating robust marker-trait association studies [9]; significant genotype-by-environment (G×E) interactions reducing marker predictive accuracy across diverse target regions [10]; and the rapid evolution of pathogens leading to ‘allele decay’ of resistance genes [11]. Effective breeding fundamentally depends on understanding and utilizing genetic variation. Germplasm banks play a crucial role here by conserving diversity [7], enabling the identification and introgression of valuable alleles for disease resistance, stress adaptation, and yield improvement [8]. However, fully leveraging this diversity requires overcoming the molecular tool challenges and integrating genomic resources with precise phenotyping for efficient resource management and sustainable cultivar development [12].
Scientometrics, which quantifies and measures scientific progress, is crucial for understanding trends and advances in cassava breeding. This analytical approach helps identify promising areas, knowledge gaps, and assess scientific productivity [13]. Although advances in breeding and conservation, with the aid of genetic and genomic approaches, have generated a significant amount of research focused on breeding programs, there is still a lack of studies that synthesize the current state of genetics applied to the crop. This study aims to fill that gap by providing a comprehensive analysis of scientific publications by country and year, identifying the main research groups, relevant journals, and the most frequent genetic approaches.
Scientometrics, the quantitative analysis of scientific literature, provides a powerful and objective approach to mapping the evolution, structure, and impact of research fields [13]. While traditional narrative and systematic reviews offer valuable qualitative syntheses of findings within cassava genetics and breeding [14,15], they are inherently limited in their ability to (1) quantify large-scale trends across decades and thousands of publications; (2) objectively map global collaboration networks and institutional contributions; (3) identify emerging topics and knowledge gaps through keyword evolution and co-occurrence analysis; and (4) trace the adoption and impact of specific technologies (e.g., molecular markers, genomics, gene editing) across the research landscape.
Although significant advances in cassava breeding and conservation, driven by genetic and genomic approaches, have generated a vast body of literature [16], no comprehensive bibliometric analysis exists to quantify and visualize this progress. Existing reviews focus primarily on biological insights (e.g., trait genetics, breeding methods, disease resistance mechanisms [14,15,17]) or specific technologies (e.g., genomic resources) [18], but lack the macro-level, data-driven perspective essential for strategic research planning and resource allocation.
This study addresses this critical gap by conducting the first dedicated bibliometric analysis of cassava genetic research (1960–2022). We leverage scientometric methods to (1) quantify publication growth, geographic distribution, and key contributing institutions/journals; (2) map co-authorship and collaboration networks to identify research hubs and partnerships; (3) analyze keyword and thematic evolution to pinpoint research trends, emerging foci, and underexplored areas; (4) objectively assess the influence of technological milestones on research output. This analysis provides an unprecedented, evidence-based overview of the field’s trajectory, offering actionable insights for researchers, funders, and policymakers to guide future investment and collaboration in cassava genetic improvement.
Through scientific mapping using a scientometric approach, this study will provide insights for researchers to more efficiently direct their efforts in cassava conservation genetics and breeding. The results will also support the formulation of public policies aimed at managing germplasm banks and breeding programs, as well as guide future research. Overall, the research characterizes the current state of scientific knowledge on cassava through a systematic review and mapping analysis of the scientific production on the genetic studies of the species.
2. Materials and Methods
2.1. Data Collection and Filtering
The research data were obtained through an automatic search in the Web of Science (WoS) database (https://www.webofknowledge.com, accessed on 17 February 2024). We chose this platform based on its multidisciplinary nature [17] and its frequent use in studies investigating scientific production. The search period covers the years from 1960 to 2022, using the keywords “Manihot esculenta” and “genetic*,” combined via the Boolean operator “AND”; the use of wild card “*” allowed for capturing morphological variations in the keyword. Given that our interest was in genetic research associated with cassava, we did not add specific keywords (e.g., molecular markers, genetic markers) so as to not lose potential data that related to genetic research but did not specify any tools or approaches.
To filter papers for the bibliometric analysis, we defined the following criteria: only articles published in peer-reviewed journals in English, with a focus on genetic research related to cassava. We opted to use English as the only language for publication retrieval given that studies published in this language are more likely to be cited [18]. Data were independently tabulated by two researchers working in pairs, aiming to maximize consistency and inter-rater agreement.
We based our filtering and selection of papers for the bibliometric and scientometric analysis on the following criteria: duplicated results, papers that fall outside the scope of the research, in other languages, and other types of publications (e.g., books, abstracts, conference proceedings) were removed from the analysis. We accessed the titles, abstracts, and keywords of the articles to verify whether the studies specifically focused on cassava research in the context of genetics, to filter data for descriptive analyses, and for the classification of the papers. Papers, where the abstracts, titles, or keywords did not provide sufficient data for selection and classification, were directly accessed from the publishing journal.
The results of the automatic search were stored as an electronic spreadsheet, using Microsoft Excel software 2019. We divided the dataset into three sheets: the unfiltered raw data from the database, the filtered results that satisfied the selection criteria, and the classification data, respectively. From the accepted papers, the following parameters were extracted for the descriptive analysis: article IDs, first author’s name, publication title, keywords defined by the authors, abstract, year of publication, language, publishing journals, research areas, classification categories defined by the WoS, author affiliations, and DOI. The third sheet (i.e., classification data) represents the most specific analyses, containing the classification and subclassification of the selected papers (Figure 1).
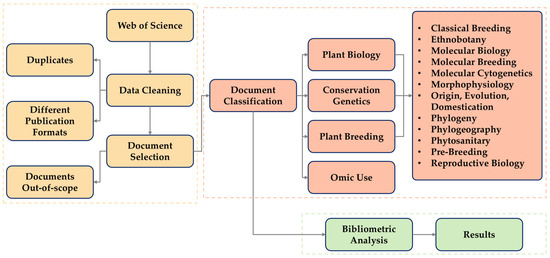
Figure 1.
Stages of selection, exclusion, and classification of scientific articles in our cassava genetics scientometric analysis.
2.2. Qualitative and Quantitative Classification
To establish the state of the art in publications related to genetics in cassava research, we identified temporal trends in publication using a linear model to investigate if the number of papers published per year represented a positive trend, coupled with graphic visualizations to track variations in scientific production. We also mapped the authors and their contributions to the field, highlighting those with the most publications and citations associated. Additionally, we investigated the most relevant institutions involved with cassava genetics research.
The selected articles were classified into three major areas (Figure 1) and twelve sub-areas (Table 1) within the theme of cassava genetic research. To be classified on one of the major areas with their respective subareas, the research needed to align with the following criteria:

Table 1.
Most cited articles on cassava genetics extracted from the Web of Science database for the period 1960–2022.
Plant Biology encompasses the study of physiological, anatomical, and molecular processes that govern the growth and functioning of cassava plants. Within this area, morphophysiology investigates the form and function of plant structures—such as roots, stems, and leaves—offering practical insights for agronomic practices. Taxonomy focuses on the classification and naming of species, contributing to biodiversity documentation and comparative studies. Reproductive Biology explores sexual and asexual reproduction, pollination mechanisms, and gene flow, which are essential for understanding genetic variability. Classical and Molecular Cytogenetics examine chromosome structure and behavior through karyotyping and techniques like FISH, supporting genetic mapping and evolutionary analyses. Molecular biology, using DNA and RNA tools, provides the foundation for gene discovery, expression profiling, and functional genomics.
Conservation Genetics aims to preserve the genetic diversity of cassava and understand its evolutionary history. Studies in this area explore the crop’s origin, domestication, and diversification across regions. Population Genetics and Population Genomics analyze the structure, variability, and dynamics of natural and cultivated populations over time, revealing patterns shaped by selection, drift, and migration. Ethnobotany contributes by documenting traditional knowledge, cultural uses, and local naming systems, strengthening the link between biodiversity and community practices. Phylogenetic analyses reconstruct evolutionary relationships between cassava and related species, while phylogeography integrates genetics with geography to trace lineage dispersal and historical biogeographic events.
Plant Breeding focuses on developing improved cassava cultivars that meet agronomic, nutritional, and environmental demands. Pre-breeding involves identifying and incorporating beneficial alleles from wild relatives or underutilized varieties to broaden the genetic base. Classical breeding applies conventional methods such as hybridization and phenotypic selection to combine desirable traits. In contrast, molecular breeding integrates modern tools like marker-assisted selection and genomic prediction to accelerate breeding cycles and increase precision. Together, these subfields support the development of cassava varieties that are more productive, resilient, and adapted to diverse agroecological conditions.
Information about the methodological tools used in the research was also collected. Articles that presented an omics approach were evaluated for classification into one of the four approaches proposed for this research based on the type of omic employed in the research: genomics, transcriptomics, proteomics, and metabolomics (Figure 1).
2.3. Impact Factor Analysis
We used the 2022 InCites Journal Citation Reports (JCR) report to calculate the impact factor (IF) of the scientific journals retrieved in the search. The JCR is a bibliometric index aimed at evaluating the scientific output of authors, the quality of publications, and the ranking of scientific journals [29].
To analyze the impact factor and the quality of the studies retrieved in the automated search, the titles of the publishing journals were extracted, and a search was conducted in the JCR tool to obtain the impact factor for the year 2022. The impact factor analysis aimed to highlight the research of greater relevance in the global context within the theme of cassava genetics, as well as to obtain descriptive data (e.g., countries with the highest number of high-impact publications).
2.4. Statistical Analysis
We analyzed the data using descriptive statistical techniques (e.g., summarization and frequency counts), complemented by visualization methods to outline the landscape of cassava genetics research publications for the analyzed period. All analyses were performed in the R 4.4.1 environment [30] using the tidyverse package [31].
3. Results
3.1. Temporal Trends
In total, 3246 studies were retrieved from the WoS database, of which only 654 met the inclusion criteria for the analysis. The first article focusing on genetics was published in 1969, with a notable increase in publications since 1993 (n = 6) (Figure 2). A linear regression analysis demonstrated that the number of papers significantly increased by nearly one paper per year (p < 0.05), with the model explaining 66.4% of the variation in the data (R2 = 0.664).
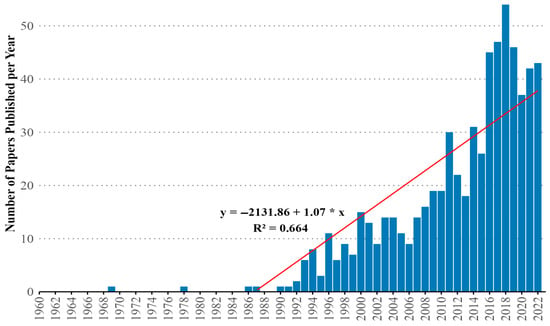
Figure 2.
Annual scientific production in cassava genetics from 1969 to 2022. The red line represents the fitted linear regression.
Our analysis identified 3246 studies from the Web of Science, with 654 meeting inclusion criteria. Publications in cassava genetics began in 1969, but a significant linear increase (nearly one additional paper per year, p < 0.05, R2 = 0.664) commenced notably after 1993 (Figure 2). This sustained growth trajectory strongly correlates with the adoption of pivotal molecular technologies. The initial uptick aligns with the widespread application of PCR-based markers (e.g., RFLPs, SSRs) in the early 1990s, enabling foundational genetic mapping and diversity studies in cassava. The subsequent acceleration mirrors the broader ‘genomics revolution’: the advent of affordable high-throughput sequencing (NGS) around 2009 facilitated the first cassava genome draft, unlocking genome-wide association studies (GWAS), genomic selection (GS), and comprehensive germplasm characterization. Furthermore, the development of CRISPR-Cas9 gene editing after 2013 provided precise tools for functional validation and trait engineering, contributing to sustained publication output. These technological breakthroughs progressively lowered barriers to genetic analysis, transforming cassava from an ‘orphan crop’ with limited tools into a tractable system for modern genetic research, directly driving the observed publication trend.
The average number of papers published per year, by decade, was 5.4 ± 3.5 in the 1990s, 13.4 ± 3.1 in the 2000s, and 31.1 ± 16.4 in the 2010s, peaking in 2018 with 54 papers published (Figure 2). This represents a productivity increase of 475.93% when comparing publications from the 1990s to the last decade.
3.2. Paper Citation History
Based on the data provided by the WoS database, the 654 articles analyzed were cited 15,505 times, averaging 23.7 ± 34.4 citations per document. The 10 most cited articles were published between 1997 and 2016 (Table 1), with an average citation of 195.5 ± 56.1 per document.
The article by Olsen published in 1999 in the Proceedings of the National Academy of Sciences of the United States of America is the most cited, with 338 citations, standing out for its significant contribution to understanding the geographic and genetic origins of cassava, revealing its domestication patterns and ancestral wild relatives. In 1996, Verdaguer published in Plant Molecular Biology, accumulating 229 citations. In 2016, Patanun published in Molecular Genetics and Genomics, receiving 214 citations. De Vetten, with an article in Nature Biotechnology in 2003, accumulated 190 citations.
Xu, published in 2013 in Plant Physiology, was cited 178 times, while Jørgensen, in 2005 in the same journal, received 172 citations, underscoring the relevance of research in this area. Mba, in 2001, published in Theoretical and Applied Genetics, with 167 citations. Olsen published another study in 2001 in the American Journal of Botany, which was cited 161 times, reinforcing his continuous influence in the field. Zeng, in 2010, published in Nucleic Acids Research, with 157 citations.
Finally, Fregene, with a 1997 publication in Theoretical and Applied Genetics, obtained 149 citations. Although Fregene is the author with the highest number of associated publications (n = 33), in many of them (28 articles), he appears as a co-author, highlighting his extensive collaboration in the field.
3.3. Journals
Research papers were published across 212 journals, averaging 3.1 ± 4.7 publications per journal. Of these, 200 published less than 10 papers (94.3%), with the remaining 12 (5.7%) publishing between 10 and 36 publications for the period. The journals with the highest number of publications (Figure 3) are Euphytica (n = 36), Genetics and Molecular Research (n = 30), Frontiers in Plant Science (n = 25), Theoretical and Applied Genetics (n = 24), Plant Molecular Biology (n = 22), PLoS One (n = 21), Scientific Reports (n = 19), Crop Science (n = 12), Genetic Resources and Crop Evolution (n = 12), BMC Genomics (n = 11), African Journal of Biotechnology (n = 11), and Plant Molecular Biology Reporter (n = 10). These journals accounted for 233 of the 654 (35.6%) publications analyzed, thus being considered the main journals related to cassava genetics for the period.
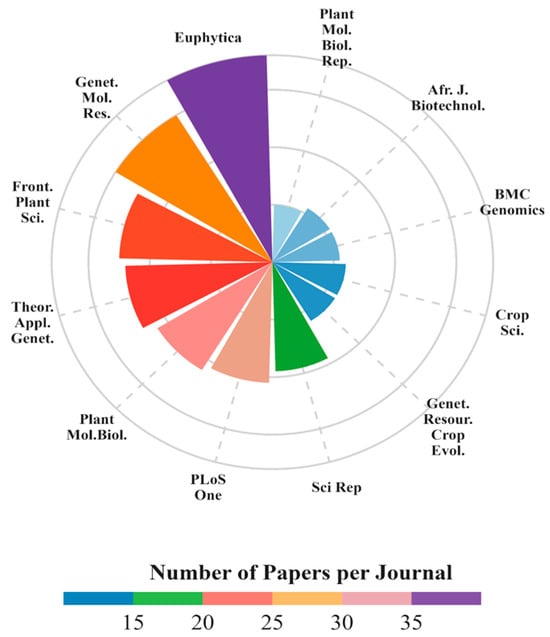
Figure 3.
Top journals with 10 or more publications related to cassava genetics articles from 1969 to 2022. Journal names were abbreviated following ISO4 convention: Euphytica = Euphytica; Plant Mol. Biol. Rep. = Plant Molecular Biology Reporter; Afr. J. Biotechnol. = African Journal of Biotechnology; BMC Genomics = BMC Genomics; Crop Sci. = Crop Science, Genet Resour Crop Evol = Genetic Resources and Crop Evolution; Sci. Rep. = Scientific Reports; PLoS One = PLoS One; Plant Mol. Biol. = Plant Molecular Biology; Theor. Appl. Genet. = Theoretical and Applied Genetics; FPLS = Frontiers in Plant Science; GMR = Genetics and Molecular Research.
The impact factors (IF) for journals with 10 or more publications associated were obtained from the 2022 JCR report (Figure 4), with an average IF of 3.2 ± 1.8. To assess a potential relationship between the number of papers published and the journals’ IF, we performed a linear regression analysis. The results showed no significant correlation between a higher IF and the number of papers published (p > 0.05; R2 = 0.007712).
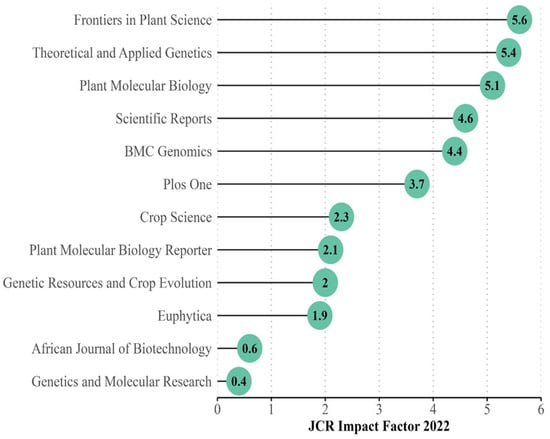
Figure 4.
Impact factor of the top 10 journals with the most publications related to cassava genetics articles from 1969–2022 based on the JCR Report 2022.
3.4. Authors and Co-Authors
We identified a total of 2050 authors and co-authors involved in cassava genetics research between 1960 and 2022. Of these, 1919 (93.6%) published between 1–5 papers, 93 (4.5%) 5–10 papers, 28 (1.4%) 10–20 papers, and only 10 authors (0.5%) are associated with more than 20 articles, with Fregene, Rabbi, and Tohme the only authors showing the highest number of publications (Figure 5). In contrast, when first authorship was analyzed, except for Tohme, the same authors published only 5 papers, with the highest number of papers associated with a first author being 21, authored by Nassar.
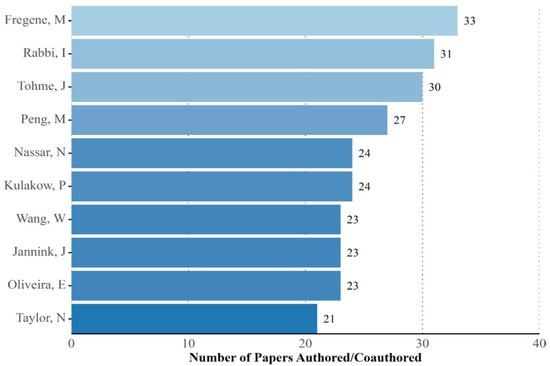
Figure 5.
Top 10 authors associated with cassava genetics publications. The numbers were based on how many papers the author was associated with, not distinguishing between first authorship or co-authorship.
3.5. Countries and Institutions
The 654 articles were published by authors from 54 different countries (Figure 6), with an average of 12.1 ± 27 papers per country. The top 10 countries with the highest number of publications for the period were Brazil (n = 143), China (n = 110), United States (n = 75), Colombia (n = 67), Thailand (n = 35), Nigeria (n = 31), France (n = 22), England (n = 19), South Africa (n = 13), and Switzerland (n = 13).
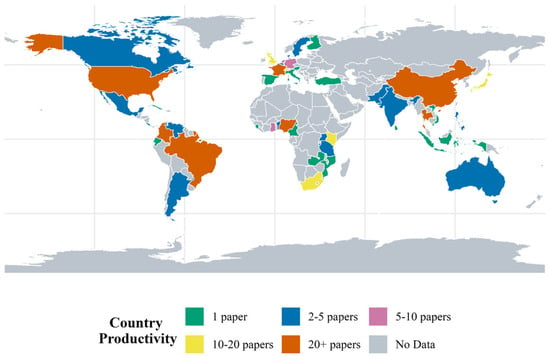
Figure 6.
Country productivity for the period 1960–2022 based on the number of papers. Countries in gray did not have studies involved in cassava genetics research.
With 143 papers, Brazil had the highest productivity for the period of 1960–2022, corresponding to 21.9% of the total production among all 54 countries, and 27.1% of the production among the 10 most productive countries for the period. Brazil also holds the most productive institution for the period, the Brazilian Agricultural Research Corporation (Embrapa), associated with authors of 117 papers. Regarding authors associations, Embrapa is followed by other nine institutions: Alliance (n = 96), the Chinese Academy of Tropical Agricultural Sciences (n = 95), the International Center for Tropical Agriculture (CIAT) (n = 95), the Consultative Group on International Agricultural Research (CGIAR) (n = 75), Cornell University (n = 67), the International Institute of Tropical Agriculture (IITA) (n = 59), Hainan University (n = 56), Mahidol University (n = 42), and the Institut de Recherche pour le Developpement (IRD) (n = 37).
3.6. Keyword Occurrences
Keywords are the foundation of data analysis, allowing researchers to identify patterns, trends, and gaps in academic knowledge [15]. They also facilitate communication and collaboration among researchers by providing a common language to describe and categorize research topics [16]. Understanding the central role of keywords in data analysis is essential for rigorous and meaningful research in any field of study [17]. We analyzed the keywords defined by the authors, revealing a total of 1437 unique terms and 1390 unique words used between 1960 and 2022 to index cassava research. To visualize the relationship between terms and the frequency of keyword occurrence, we constructed a word cloud (Figure 7).

Figure 7.
Word cloud of selected keywords from cassava genetics articles for the period 1960–2022.
The keywords “Cassava”, “Manihot esculenta”, and “genetic diversity” were the most commonly used, appearing 234, 126, and 38 times, respectively, reflecting the research focus for the period. When analyzing unique words used to construct keywords, “genetic”, “gene”, and “diversity” appeared 144, 64, and 52 times, respectively. The analysis of the keywords allowed us to better understand the research focus for the period, and it demonstrated that our search was able to capture the most significant part of the research related to cassava genetics.
3.7. Temporal Trends of the Main Molecular Tools
The analysis of the main molecular tools and methodologies used in cassava genetics research over the past four decades reveals a clear evolution in the available techniques, reflecting continuous advancements in technology and genetic analysis capabilities (Figure 8). A total of 185 tools and methodologies were detected for the reviewed period.
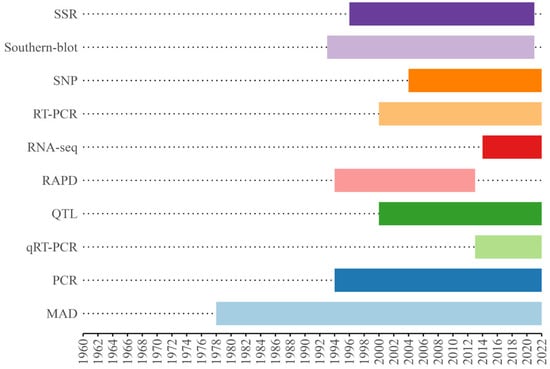
Figure 8.
Temporal trend of the 10 main tools in cassava genetics articles. SSR—Simple Sequence Repeats, SNP—Single Nucleotide Polymorphism, RT-PCR—Reverse Transcription Polymerase Chain Reaction, RNA-seq—RNA Sequencing, RAPD—Random Amplified Polymorphic DNA, QTL—Quantitative Trait Loci, qRT-PCR—Quantitative Reverse Transcription Polymerase Chain, PCR—Polymerase Chain Reaction, MAD—Agro Morphological Descriptors.
The use of morpho-agronomic descriptors (MAD) began in 1978 and was recorded up until 2022 (Figure 8), demonstrating its relevance over time. The Polymerase Chain Reaction (PCR), since its introduction in 1986, was also used until 2022, remaining a central tool in genetic research. Random Amplified Polymorphic DNA (RAPD), used from 1994 to 2013, had a specific period of intensive use.
Microsatellite markers (SSR), on the other hand, were widely employed from 1996 to 2021. The Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) technique was recorded between 2000 and 2022, with Quantitative Trait Loci (QTL) applied to identifying specific genomic regions associated with complex traits from 2000 to 2022. Single Nucleotide Polymorphism (SNP) markers and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) also had records up to 2022, since their introduction in 2004 and 2013, respectively. More recent technologies, such as RNA sequencing (RNA-seq) and Genotyping by Sequencing (GBS), began to be used in 2014, with GBS records extending to 2022. This continued use highlights that the continued use of some of these techniques may occur as specific research applications demand them.
The number of applications of the 10 main molecular tools varied over the years, reflecting different demands and technological advances. These variations can be observed in the frequency of use of each technique, which changed during distinct periods.
The analysis of the applications of the 10 main molecular tools in genetic research reveals that SSR markers lead in usage, with 102 records. RT-PCR also had a significant presence, being applied in 62 studies. Other widely used techniques include MAD and PCR, with 71 and 77 applications, respectively.
Although SNP markers were recorded 64 times and qRT-PCR 31 times, these and other techniques, such as RNA-seq, used on 37 occasions, remain relevant and adaptable to new needs. Isoenzymes and GBS had lower application frequencies, with 15 and 18 records, respectively. These numbers reflect the current scenario in cassava genetics studies, but it is important to highlight that these tools may be revisited in new contexts as research demands evolve.
3.8. Analysis of Article Distribution by Class and Subclass
Figure 9 illustrates the distribution of research papers across various subclasses. Molecular Biology dominates with 235 papers, followed by Phytosanitary studies (n = 110) and Molecular Breeding (n = 47). Pre-Breeding (n = 42) and Phylogeny (n = 17) occupy intermediate positions, while subclasses like Reproductive Biology (n = 14), Classical Breeding (n = 11), and Molecular Cytogenetics (n = 8) have significantly fewer contributions. Ethnobotany (6), Morphophysiology (n = 5), Phylogeography (n = 4), and Origin, Evolution, and Domestication (n = 2) are minimally represented. This distribution highlights a strong research focus on molecular and phytosanitary areas, with traditional and evolutionary topics receiving comparatively less attention.
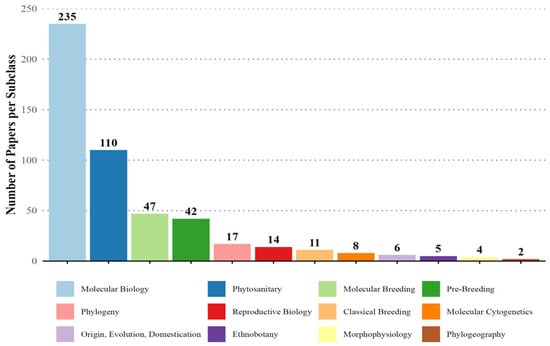
Figure 9.
Distribution of papers associated with cassava genetics by subclasses for the 1960–2022 period.
Research focus was categorized across three thematic classes: Plant Biology, Breeding, and Conservation. Plant Biology (Figure 10A) shows a predominant emphasis on Molecular Biology (75%), with a lesser focus on Reproductive Biology, Molecular Cytogenetics, Morphophysiology, and Ethnobotany. Breeding (Figure 10B) highlights a shift from Classical Breeding to Molecular Breeding and Phytosanitary studies, the latter dominating over 50% of the focus for the 1960–2022 period. Conservation (Figure 10C) underscores a high interest in Phylogeny, followed by Origin, Evolution, and Domestication, with Ethnobotany being underrepresented. Overall, the data reflects a clear trend toward molecular and phytosanitary approaches, with traditional and ethnobotanical research receiving limited attention.
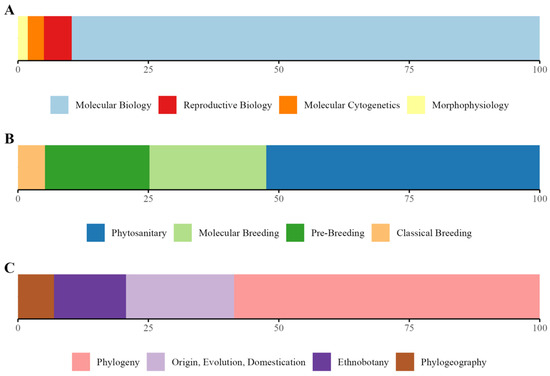
Figure 10.
Percentage of the most representative subclasses distributed by class ((A) Plant Biology, (B) Plant Breeding, and (C) Conservation Genetics) for the 1960–2022 period.
Our data revealed that some papers may be classified in more than one class and also be included in more than one subclass. The data demonstrated the association between different subclass combinations and the number of articles. The most prominent focus is on Population Genetics/Population Genomics, with 104 articles, showcasing its central role in interdisciplinary studies. Other combinations, such as Reproductive Biology with Classic Cytogenetics (5 articles), Population Genetics with Pre-Breeding (4 articles), and Reproductive Biology with Molecular Biology (3 articles), reflect emerging but less dominant integrations. This distribution underscores the prominence of Population Genetics in multidisciplinary research, with smaller contributions from other combined approaches.
The thematic analysis of articles on cassava genetics demonstrated a wide and diverse distribution among the main research class and their respective subclasses. This approach provides a comprehensive overview of the areas of interest and the predominant methodologies in the field.
As for the omics techniques, 488 articles do not present any omic approach. In contrast, 166 papers utilized at least one type of omic. Genomics is addressed in 87 articles, and transcriptomics in 65 articles, with a combination of both approaches being detected in 8 articles. The presence of combinations of genomics and transcriptomics suggests a growing interest in integrated approaches, although these techniques are less frequent compared to genomics alone. From the four omic classes detected, only two omics presented less than 6 papers: proteomics (n = 6) and metabolomics, which were detected in only one article (Figure 11).
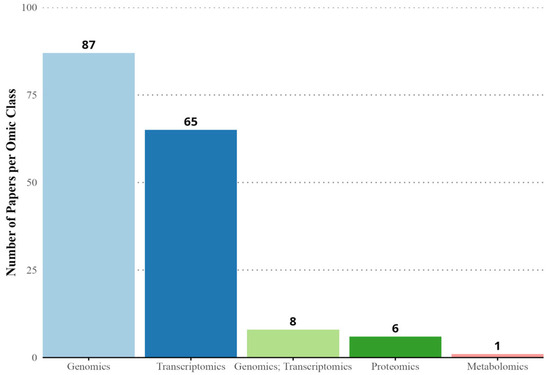
Figure 11.
Distribution of papers related to cassava genetics with an omic approach for the period 1960–2022.
4. Discussion
4.1. Temporal Trends
The number of publications involving methodological approaches in cassava genetics increased significantly after 1993 (Figure 2). One plausible explanation is the development of new technologies, such as the emergence of PCR and all classes of molecular markers, including RAPD, RFLP, SSR, AFLP, ISSR, and SNP, which have been used for a wide range of research, whether in Conservation [32,33], Breeding [34], or basic research (i.e., plant biology) [35].
After a decline in the number of publications involving genetic studies on cassava in 1995 (Figure 2), there was a notable increase from three publications in 1995 to 11 in 1996. Between 1978 and 1995, Brazil contributed only three articles authored or co-authored by Nassar [36,37,38]. After 1997, there was an increase in Brazilian publications involving cassava genetics, likely driven by three factors: the implementation of the Real Plan, the growth of investments in research grants and funding through the National Council for Scientific and Technological Development (CNPq), and the dissemination of internet access in Brazil starting in 1996 [39,40].
The period of stability between 1997 and 1999 was followed by a continuous rise in publications starting in 2000 (Figure 2). This increase coincided with the introduction of next-generation sequencing technologies, which revolutionized genomic research by making it more accessible and efficient [41]. These advancements marked a new era for cassava genetics, particularly with the commercialization of these technologies beginning in 2005.
Starting in 2010, China emerged as a major contributor to cassava genetics research, with a publication peak in 2018, totaling 54 articles. In that year, China led with 17 publications, followed by Brazil with 10 and the United States with seven (Figure 6). The surge in scientific output from China reflects substantial investment in research and development, as well as a growing global interest in cassava genetics due to its economic and agricultural importance.
Thus, the analysis of temporal trends not only reveals the evolution of cassava genetics research but also underscores the influence of technological, economic, and social factors on this trajectory.
4.2. Direct Citation History
Despite the growth in cassava genetics studies, there has been a noticeable decrease in citation frequency over time, which indicates the evolving impact of research on the scientific community [42]. Highly cited works often offer long-term value, such as species descriptions or hybridization insights [43], while more specialized studies receive fewer citations [44]. Older studies tend to be cited more frequently, as observed by [45], reflecting their foundational role.
Among the top-cited authors (Figure 5), United States researchers, especially Olsen, who elucidated cassava’s evolutionary origin, are prominent, with two pivotal studies on phylogeography and genetic variation cited 338 and 161 times, respectively [46,47]. Phylogeographic research, essential for understanding historical impacts on genetic distributions, integrates principles of molecular ecology, underscoring biological diversity [48]. Macroevolutionary studies complement this by addressing isolation and phylogenetic processes, driven by molecular advances like PCR [49].
The spread of systemic diseases through vegetative propagation remains a concern, as pathogens accumulate over cycles, as seen with the cassava common mosaic virus [50]. Verdaguer’s study on the cassava vein mosaic virus promoter, with 229 citations, furthered knowledge of virus dynamics [51]. Recent years have also seen rising interest in non-coding RNAs, such as miRNAs, critical in stress response. Patanun’s 2016 study, identifying cassava miRNAs, has been widely cited and has propelled research on resilient cultivars [18]. Public concern over antibiotic and herbicide resistance genes in transgenic plants has led to alternative transgenic methods. De Vetten’s 2003 study introduced a marker-free selection process, widely adopted in plant biotechnology [52].
China’s contribution to cassava genetics is notable, with key publications by Xu on delaying postharvest deterioration and Zeng on miRNAs in Euphorbiaceae, advancing research on stress responses in cassava [53,54]. Cassava’s cyanogenic glycosides, primarily linamarin, pose detoxification challenges. Jørgensen’s 2005 study demonstrated reduced cyanide levels in cassava, guiding detoxification research [24].
Among the top-cited scientists, Mba and Fregene have contributed significantly to cassava genetic mapping, emphasizing SSR markers’ role in characterizing germplasm and analyzing genetic variability, aiding cassava breeding efforts [55]. This body of research underscores the impact and continued advancement of molecular techniques in cassava genetics.
4.3. Journals
Within the scope of journal diversity, the 10 journals with the highest number of publications stand out (Figure 3). The evaluation of the quality and relevance of scientific journals is commonly conducted through the IF, provided by JCR, available in the WoS database [56].
According to [57], this aspect has been a topic of considerable debate and discussion in the literature. He highlights that some researchers defend its positive characteristics. Similarly, ref. [58] affirm that the issue has been widely debated, with many researchers acknowledging its advantages. Consequently, a trend observed is that articles published in high-impact scientific journals are more likely to be read and subsequently cited [59]. This assertion is corroborated by analyzing the most cited article, where Olsen published a study in Proceedings of the National Academy of Sciences of the United States of America (IF = 11.1).
However, among the 10 journals with the highest number of publications, as shown in Figure 3, only Plant Molecular Biology (IF = 5.1) and Theoretical and Applied Genetics (IF = 5.4) are among those with the most cited articles. On the other hand, Genetics and Molecular Research, ranked second in terms of the number of publications, has the lowest IF among the top 10 journals (IF = 0.4). It is important to note that these metrics are developed by Clarivate Analytics, the entity responsible for evaluating journals indexed in WoS, providing an international perspective on the impact of scientific journals [60]. These metrics include the IF and other citation-related analyses of articles published in specific journals [61].
4.4. Authors and Co-Authors
The productivity of authors is not necessarily correlated with the number of citations received, as an author can publish many articles and receive few citations, while another may receive a high number of citations for a smaller number of articles [62]. Among the 10 authors and co-authors who stood out for the number of articles produced, as shown in Figure 5, Fregene, director of Agriculture and Agro-industry at the African Development Bank, is notable. Fregene stands out as the most prolific researcher in cassava genetics, with his first study published in 1994 in Theoretical and Applied Genetics. His prominent position is evidenced by the fact that he is the author of the tenth most cited study, as shown in Table 1. Fregene is the only researcher among the most productive to also be one of the most cited, reflecting his long career and extensive collaboration with institutions and researchers. Other authors with significant publications are Peng (n = 27), Wang (n = 22), Hu (n = 16), and Zhiqiang (n = 16), all from Chinese institutions that emerged after 2014. This growth is driven by China’s substantial investment in research, contrasting with Brazil, which has faced challenges due to funding cuts since 2016 [63,64].
Despite difficulties in including Chinese and peripheral journals in international databases [65]. China’s investment in research has promoted a significant increase in international scientific production [66]. In Brazil, Oliveira and Nassar are the only researchers among the most prolific (Figure 5); Oliveira works at Embrapa Cassava and Fruits, focusing on molecular genetics and plant breeding, with notable international collaborations. On the other hand, Nassar is a professor at the University of Brasília, with a long research history related to germplasm conservation and, also, the identification of new varieties and species of Manihot.
In the United States, Jannink of the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) and Kulakow, of Kansas State University, are noted for their contributions to genetic breeding and phytoremediation. Tohme from CIAT, and Rabbi from IITA, are also notable, with significant contributions to cassava genetics and the development of varieties for West Africa, respectively.
4.5. Countries and Institutions
Regarding the leading countries and institutions in cassava genetics research, China stands out as the country with the highest scientific production (Figure 6). Institutions such as the Chinese Academy of Tropical Agricultural Sciences and Hainan University are prominent in this context. China has substantially invested in training researchers, which, according to [67], contributes to its advancement in agricultural research and innovation.
Brazil ranks second in scientific production, with Embrapa being the country’s leading institution. Embrapa has played a crucial role in cassava genetics research, as evidenced by [68]. This collaboration between Embrapa and other Brazilian institutions has driven the development of new cultivars and applied modern methodologies tailored to local needs.
France and Italy also stand out (Figure 6), with institutions such as CGIAR and IRD in France, and Alliance in Italy, contributing significantly to research. CGIAR, founded in 1972, continues to address global challenges related to food security, while Alliance Bioversity International and IRD focus on solutions for poverty and climate change. Colombia, with CIAT, and Thailand, with Mahidol University, have also gained prominence. CIAT, now part of Alliance, and Mahidol University’s Institute of Molecular Biosciences are examples of institutions committed to research and innovation in cassava genetics and other areas.
International collaboration, exemplified by the NextGen Cassava Project, an initiative involving the Bill and Melinda Gates Foundation and DFID, demonstrates how cooperation between institutions and countries can accelerate progress in cassava genetics research. This project not only contributes to food security but also promotes sustainable development with several results [69]. The integration of resources and technical knowledge through this collaborative network is crucial for turning scientific discoveries into practical solutions, positively impacting communities that depend on cassava.
4.6. Keyword Occurrences
Careful selection of keywords is essential in any scientific data analysis, as they play a crucial role in indexing and discovering academic works [70]. Systematic strategies to identify the most relevant keywords are necessary, as discussed by [71]. Transparency and rigor in the selection and reporting of keywords are vital to ensure the replicability and reliability of results [72].
Examining the keyword cloud presented in Figure 7, it is observed that “Cassava” emerges as the most frequent term. This indicates a focus on topics related to cassava, encompassing its biology, cultivation, genetics, or applications. “Manihot esculenta” also appears prominently, signifying a specific emphasis on the botanical species of cassava. The inclusion of the term “Genetic” suggests the exploration of genetic issues relevant to cassava, such as genetic variation, genetic breeding, or genomics.
4.7. Temporal Trends of Key Molecular Tools
The evolution of molecular techniques used in cassava genetics research (Figure 8) reflects significant advancements in scientific technology over recent decades. Since the introduction of morphological and agronomic descriptors (MAD) in 1978, which enabled data collection on the physical and agronomic traits of plants, there has been a shift toward more sophisticated molecular methods to address the increasing complexity of genetic studies [73]. In 1996, SSRs became a critical tool for analyzing genetic variability, offering a detailed resolution of genetic differences and enhancing the understanding of genetic diversity and evolutionary relationships within cassava populations [74,75].
Isoenzymes, introduced in 1987, represented progress in genetic differentiation based on enzyme activity variations. However, limitations in resolving genetic variants led to their gradual replacement by more advanced methods [69]. The advent of RAPD in 1994 allowed genetic variation detection without prior DNA sequence information, playing an essential role in early cassava molecular genetics but eventually being supplanted by more precise techniques like SNPs and qRT-PCR.
PCR, introduced in 1994, revolutionized DNA amplification with high specificity and sensitivity, remaining fundamental for various molecular studies, as evidenced by its continued application in 36 studies [76]. Since 2004, SNPs marked a breakthrough in detecting genetic variations, offering high-resolution analysis, critical for genotyping and genome-wide association studies [77,78], with 28 records highlighting their relevance.
Starting in 2013, qRT-PCR brought a new approach to gene expression quantification, enabling precise RNA expression measurement and insights into regulatory mechanisms under different conditions, reflected in 24 studies [75]. RNA-seq, introduced in 2014, revolutionized transcriptomic analysis by enabling comprehensive quantification and novel transcript identification, allowing for a detailed understanding of gene expression across physiological and developmental contexts [79]. Simultaneously, GBS, also introduced in 2014, combined next-generation sequencing with genotyping methods, facilitating large genetic dataset analysis cost-effectively. Its continued use until 2022, with 12 studies, underscores its importance in high-density genomic analysis and identifying genetic variants associated with phenotypic traits [73,74].
QTL analysis, in use since 2000, remains a crucial tool for identifying genomic regions linked to quantitative traits like productivity and disease resistance, with 11 studies confirming its relevance in understanding complex trait genetics and supporting desirable trait selection [80,81,82]. The evolution of these molecular tools demonstrates continuous progress in genetic analysis capabilities, with newer methodologies progressively replacing older techniques, each contributing to cassava genetics and adapting to new technological and scientific demands [83].
In addition to the progressive refinement of individual molecular tools, recent cassava research has moved toward integrated multi-omics approaches. By combining genomics, transcriptomics, proteomics, and metabolomics, researchers can obtain a more comprehensive understanding of the molecular networks underlying key agronomic traits. These integrative strategies have enhanced the discovery of candidate genes related to drought tolerance, root quality, and disease resistance. Advanced techniques such as genome-wide association studies (GWAS), enabled by high-density SNP datasets from GBS and resequencing, are increasingly used to map complex traits. Furthermore, the application of RNA-seq has expanded functional genomics by revealing regulatory pathways activated under stress conditions. These tools have had practical implications in cassava breeding programs, supporting marker-assisted selection and guiding parental selection with greater precision.
Emerging technologies such as CRISPR/Cas genome editing and third-generation sequencing platforms (e.g., PacBio, Nanopore) are beginning to transform cassava genetic research by enabling targeted gene modification and the resolution of complex genomic regions. At the same time, the rise of computational biology and artificial intelligence is facilitating the analysis of large-scale genomic datasets, improving trait prediction models and accelerating genomic selection. Despite these advances, challenges remain, including limited access to high-throughput infrastructure in low-income regions, difficulties in tissue culture and genetic transformation, and the need for better-characterized mapping populations. Addressing these bottlenecks will be essential to fully harness the potential of molecular tools in cassava improvement and conservation efforts.
4.8. Distribution of Articles by Topic
The thematic distribution analysis of cassava genetics articles revealed a broad range of research interests and approaches (Figure 9 and Figure 10). Plant biology leads scientific production with 279 articles, notably in the molecular subclass (236 articles), reflecting a strong focus on high-resolution techniques to explore genetic and biochemical processes [84]. In plant breeding, which has 212 articles, the subclass of plant health dominates with 108 publications, underscoring the priority of developing disease and stress-resistant cultivars [85]. Pre-breeding and molecular breeding approaches, totaling 42 and 47 articles, respectively, illustrate an integration of traditional and modern methods for genetic breeding [86,87].
Conservation genetics records 147 articles, led by Population Genetics/Genomic studies (103 articles), emphasizing the importance of genetic diversity and integrating areas like Phylogeny, Origin, Evolution, Domestication, and Ethnobotany [88]. Omics techniques show genomics (88 articles) and transcriptomics (66 articles) as the most common approach, although integrated studies are limited (eight articles), indicating a gradual but expanding interest in combined approaches [89]. Proteomics and Metabolomics appear minimally (six and one article, respectively), likely due to methodological constraints, but their integration holds potential for future research [90,91,92].
This study highlights six decades of cassava genetics research evolution through scientometric analysis, showcasing key trends and technological advancements. It underscores the value of international collaboration, especially between China and Brazil, and emphasizes molecular techniques and diverse methodologies. These findings provide a foundation for future research and the development of innovative practices to enhance cassava production, crucial for food security and tropical economies.
5. Conclusions
This scientometric and bibliometric analysis provides a comprehensive overview of six decades of cassava genetic research. Our findings show a clear increase in scientific output since the 1990s, driven by technological advances such as SSR markers, SNP genotyping, and omics-based approaches like genomics and transcriptomics. Despite this growth, there remains a limited exploration of integrative biological themes—such as the functional role of genes in agronomic traits, the genetic basis of disease resistance, and the application of emerging biotechnologies to improve cassava.
The analysis highlights a strong concentration of studies in molecular genetics and phytosanitary breeding, while areas such as Reproductive Biology, Classical Breeding, and Conservation Genetics remain underrepresented. These gaps suggest opportunities for deeper investigations into genotype–phenotype relationships and the adaptation of cassava to climate change, especially using high-resolution tools.
This study demonstrates the value of bibliometric analysis as a complement to traditional reviews, offering a macro-perspective on research trends, collaborative networks, and knowledge gaps. However, limitations include the exclusive use of the Web of Science database, the restriction to English-language articles, and the focus on metadata rather than full-text analysis.
Moving forward, cassava researchers can benefit from integrating scientometric insights with biological validation to ensure that future studies not only follow trends but also address practical challenges in cassava improvement and food security.
Author Contributions
All authors contributed equally to this project and share equal authorship. M.V.B.M.S., corresponding author: conceptualization, methodology, formal quantitative analysis, writing—original draft, review and editing. V.C.S., G.R.d.B., F.M.N., W.F.d.N., E.A.V. and M.V.B.M.S.: conceptualization, methodology, formal qualitative scientometrics analysis, writing—original draft, review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript. The financial support for publication was provided by the Minas Gerais State University (Edital PROPPG Nº 02/2025—PROPUBLIC/UEMG). We are grateful to the three anonymous reviewers for their constructive feedback, insightful suggestions, and careful corrections, which greatly improved the quality of this manuscript.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors did not receive support from any organization for the submitted work. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript. The authors have no competing interests to declare that are relevant to the content of this article.
References
- Pushpalatha, R.; Gangadharan, B. Is Cassava (Manihot esculenta Crantz) a Climate “Smart” Crop? A Review in the Context of Bridging Future Food Demand Gap. Trop. Plant Biol. 2020, 13, 201–211. [Google Scholar]
- Burns, A.; Gleadow, R.; Cliff, J.; Zacarias, A.; Cavagnaro, T. Cassava: The drought, war and famine crop in a changing world. Sustainability 2010, 2, 3572–3607. [Google Scholar]
- FAO. Food and Agriculture Organization of the United Nations. Available online: http://faostat.fao.org (accessed on 10 May 2024).
- Andrade, L.R.S.; Felisardo, R.J.A.; Cruz, I.A.; Bilal, M.; Iqbal, H.M.N.; Mulla, S.I.; Bharagava, R.N.; Souza, R.L.; Azevedo, L.C.B.; Ferreira, L.F.R. Integrated biorefinery and life cycle assessment of cassava processing residue–From production to sustainable evaluation. Plants 2022, 11, 3577. [Google Scholar]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.; Hershey, C. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: A case study for cassava. Theor. Appl. Genet. 2015, 128, 1647–1667. [Google Scholar]
- Olasanmi, B.; Kyallo, M.; Yao, N. Marker-assisted selection complements phenotypic screening at seedling stage to identify cassava mosaic disease-resistant genotypes in African cassava populations. Sci. Rep. 2021, 11, 2850. [Google Scholar]
- Aguilera, J.G.; Marim, B.G.; Setotaw, T.A.; Zuffo, A.M.; Nick, C.; Silva, D.D. The combination of data as a strategy to determine the diversity of tomato subsamples. Amaz. J. Plant Res. 2019, 3, 276–289. [Google Scholar]
- Rondon, M.J.P.; Tiago, A.V.; Hoogerheide, E.S.S. On-farm conservation of cassava in Cuiabá, Mato Grosso state, Brazil: Ethnobotanical aspects and genetic diversity. Pesqui. Agropecu. Trop. 2023, 53, e73659. [Google Scholar]
- Maziya-Dixon, B.; Dixon, A.G.O.; Townsend, D. The future of cassava in the era of biotechnology in Southern Africa. Int. J. Plant Sci. 2022, 183, 327–338. [Google Scholar]
- Okeke, U.G.; Akdemir, D.; Rabbi, I.Y.; Kulakow, P.; Jannink, J.-L. Accuracies of univariate and multivariate genomic prediction models in African cassava. Genet. Sel. Evol. 2017, 49, 88. [Google Scholar]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; Pino del Carpio, D.; Ramu, P.; Jannink, J.-L. Genome-wide association and prediction reveals genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Nkouaya Mbanjo, E.G.; Rabbi, I.Y.; Ferguson, M.E.; Kayondo, S.I.; Tripathi, L.; Kulakow, P.; Egesi, C. Technological innovations for improving cassava production in Sub-Saharan Africa: Integrating genomic resources with precise phenotyping for sustainable cultivar development. Front. Genet. 2020, 11, 623736. [Google Scholar]
- Chen, C.; Song, M. Visualizing a field of research: A methodology of systematic scientometric reviews. PLoS ONE 2019, 14, e0223994. [Google Scholar]
- Ceballos, H.; Kulakow, P.; Hershey, C. Cassava breeding: Current status, bottlenecks and the potential of biotechnology tools. Trop. Plant Biol. 2015, 8, 127–137. [Google Scholar]
- Ferguson, M.E.; Rabbi, I.Y.; Kim, D.-J.; Gedil, M.; Lopez-Lavalle, L.A.L.; Okogbenin, E. Molecular markers and their application to cassava breeding: Past, present and future. Trop. Plant Biol. 2019, 5, 95–109. [Google Scholar]
- Kawuki, R.S.; Kaweesi, T.; Esuma, W.; Pariyo, A.; Kayondo, I.S.; Ozimati, A.; Kyaligonza, V.; Abaca, A.; Orone, J.; Tumuhimbise, R.; et al. Eleven years of breeding efforts to combat cassava brown streak disease. Breed. Sci. 2016, 66, 560–571. [Google Scholar]
- Singh, V.K.; Singh, P.; Karmakar, M.; Leta, J.; Mayr, P. The journal coverage of Web of Science, Scopus and Dimensions: A comparative analysis. Scientometrics 2021, 126, 5113–5142. [Google Scholar]
- Legg, J.P.; Kumar, P.L.; Makeshkumar, T.; Tripathi, L.; Ferguson, M.; Kanju, E.; Ntawuruhunga, P.; Cuellar, W. Cassava virus diseases: Biology, epidemiology, and management. Adv. Virus Res. 2015, 91, 85–142. [Google Scholar]
- Olsen, K.M.; Schaal, B.A. Evidence on the origin of cassava: Phylogeography of Manihot esculenta. Proc. Natl. Acad. Sci. USA 1999, 96, 5586–5591. [Google Scholar]
- Verdaguer, B.; de Kochko, A.; Beachy, R.N.; Fauquet, C. Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol. 1996, 31, 1129–1139. [Google Scholar]
- Patanun, O.; Lertpanyasampatha, M.; Sojikul, P.; Viboonjun, U.; Narangajavana, J. Computational identification of microRNAs and their targets in cassava (Manihot esculenta Crantz.). Mol. Biotechnol. 2013, 53, 257–269. [Google Scholar]
- de Vetten, N.; Wolters, A.M.; Raemakers, K.; van der Meer, I.; ter Stege, R.; Heeres, E.; Heeres, P.; Visser, R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat. Biotechnol. 2003, 21, 439–442. [Google Scholar]
- Xu, J.; Duan, X.; Yang, J.; Beeching, J.R.; Zhang, P. Enhanced Reactive Oxygen Species Scavenging by Overproduction of Superoxide Dismutase and Catalase Delays Postharvest Physiological Deterioration of Cassava Storage Roots. Plant Physiol. 2013, 161, 1517–1528. [Google Scholar]
- Jørgensen, K.; Bak, S.; Busk, P.K.; Sørensen, C.; Olsen, C.E.; Puonti-Kaerlas, J.; Møller, B.L. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers: Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol 2005, 139, 363–374. [Google Scholar]
- Mba, R.; Stephenson, P.; Edwards, K.; Melzer, S.; Nkumbira, J.; Gullberg, U.; Apel, K.; Gale, M.; Tohme, J.; Fregene, M. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: Towards an SSR-based molecular genetic map of cassava. Theor. Appl. Genet. 2001, 102, 21–31. [Google Scholar]
- Olsen, K.M.; Schaal, B.A. Microsatellite variation in cassava (Manihot esculenta, Euphorbiaceae) and its wild relatives: Further evidence for a southern Amazonian origin of domestication. Am. J. Bot. 2001, 88, 131–142. [Google Scholar]
- Zeng, C.; Wang, W.; Zheng, Y.; Chen, X.; Bo, W.; Song, S.; Zhang, W.; Peng, M. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2010, 38, 981–995. [Google Scholar]
- Fregene, M.; Angel, F.; Gomez, R.; Rodriguez, F.; Chavarriaga, P.; Roca, W.; Tohme, J.; Bonierbale, M. A molecular genetic map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 1997, 95, 431–441. [Google Scholar]
- Birkle, C.; Pendlebury, D.A.; Schnell, J.; Adams, J. Web of Science as a data source for research on scientific and scholarly activity. Quant. Sci. Stud. 2020, 1, 363–376. [Google Scholar]
- Di Bitetti, M.S.; Ferreras, J.A. Publish (in English) or perish: The effect on citation rate of using languages other than English in scientific publications. Ambio 2017, 46, 121–127. [Google Scholar]
- Garfield, E. The history and meaning of the journal impact factor. J. Am. Med. Assoc. 2006, 295, 90–93. [Google Scholar]
- Oler, J.R.L.; Hoogerheide, E.S.S.; Pinto, J.M.A.; Tiago, A.; Silva, J.F.V.; Veasey, E. Research Article Influence of the use of manioc on its genetic diversity conservation in a quilombo community in Mato Grosso, Brazil. Genet. Mol. Res. 2019, 18, 1–15. [Google Scholar]
- Carrasco, N.F.; Oler, J.R.L.; Marchetti, F.F.; Carniello, M.A.; Amorozo, M.C.M.; Valle, T.L.; Veasey, E.A. Growing Cassava (Manihot esculenta) in Mato Grosso, Brazil: Genetic Diversity Conservation in Small–Scale Agriculture. Econ. Bot. 2016, 70, 15–28. [Google Scholar]
- dos Santos Silva, P.P.; Sousa, M.B.e.; de Oliveira, E.J.; Morgante, C.V.; de Oliveira, C.R.S.; Vieira, S.L.; Borel, J.C. Genome-wide association study of drought tolerance in cassava. Euphytica 2021, 217, 60. [Google Scholar]
- Jette, A.M.; McDonough, C.M.; Haley, S.M.; Ni, P.; Olarsch, S.; Latham, N.; Hunter, D. A computer-adaptive disability instrument for lower extremity osteoarthritis research demonstrated promising breadth, precision, and reliability. J. Clin. Epidemiol. 2009, 62, 807–815. [Google Scholar]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Straus, S.E. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar]
- Alves-Pereira, A.; Clement, C.R.; Picanço-Rodrigues, D.; Veasey, E.A.; Dequigiovanni, G.; Ramos, S.L.F.; Zucchi, M.I. A population genomics appraisal suggests independent dispersals for bitter and sweet manioc in Brazilian Amazonia. Evol. Appl. 2020, 13, 342–361. [Google Scholar]
- Nassar, N.M. Conservation of the genetic resources of cassava (Manihot esculenta) determination of wild species localities with emphasis on probable origin. Econ. Bot. 1978, 32, 311–320. [Google Scholar]
- Nassar, N.M. Production of triploid cassava, Manihot esculenta Crantz by hybrid diploid gametes. Field Crops Res. 1992, 30, 173–182. [Google Scholar]
- Nassar, H.N.; Nassar, N.M.; Vieira, C.; Saraiva, L.S. Cytogenetic behaviour of the interspecific hybrid of Manihot neusana Nassar and cassava, M. esculenta Crantz, and its backcross progeny. Can. J. Plant Sci. 1995, 75, 675–678. [Google Scholar]
- Maynard, D.C.S. An Introduction to the History of the Internet: A Brazilian Perspective. In The Internet and Health in Brazil; Pereira Neto, A., Flynn, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 19–34. [Google Scholar]
- Angel, A. The real plan: The successful struggle to consolidate economic governance. In Consolidating Economic Governance in Latin America. Governance, Development, and Social Inclusion in Latin America; Palgrave Macmillan: Cham, Switzerland, 2021; pp. 87–104. [Google Scholar]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar]
- Liu, Y.; He, H. Scientometrics of Scientometrics Based on Web of Science Core Collection Data between 1992 and 2020. Information 2023, 14, 637. [Google Scholar]
- Bredeson, J.V.; Lyons, J.B.; Prochnik, S.E.; Wu, G.A.; Ha, C.M.; Edsinger-Gonzales, E.; Grimwood, J.; Schmutz, J.; Rabbi, I.Y.; Egesi, C.; et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016, 34, 562–570. [Google Scholar]
- Aksnes, D.W.; Langfeldt, L.; Wouters, P. Citations, citation indicators, and research quality: An overview of basic concepts and theories. Sage Open 2019, 9, 2158244019829575. [Google Scholar]
- Moed, H.F. Citation Analysis in Research Evaluation; Springer: Dordrecht, The Netherlands, 2006; Volume 9. [Google Scholar]
- Albuquerque, H.Y.G.D.; Oliveira, E.J.D.; Brito, A.C.; Andrade, L.R.B.D.; Carmo, C.D.D.; Morgante, C.V.; Vieira, E.A.; Moura, E.F.; Faleiro, F.G. Identification of duplicates in cassava germplasm banks based on single-nucleotide polymorphisms (SNPs). Sci. Agric. 2019, 76, 328–336. [Google Scholar]
- Ocampo, J.; Ovalle, T.; Labarta, R.; Le, D.P.; de Haan, S.; Vu, N.A.; Becerra Lopez-Lavalle, L.A. DNA fingerprinting reveals varietal composition of Vietnamese cassava germplasm (Manihot esculenta Crantz) from farmers’ field and genebank collections. Plant Mol. Biol. 2021, 109, 215–232. [Google Scholar]
- Papadopoulou, A.; Knowles, L.L. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc. Natl. Acad. Sci. USA 2016, 113, 8018–8024. [Google Scholar]
- Hickerson, M.J.; Carstens, B.C.; Cavender-Bares, J.; Crandall, K.A.; Graham, C.H.; Johnson, J.B.; Yoder, A.D. Phylogeography’s past, present, and future: 10 years after. Mol. Phylogenet. Evol. 2010, 54, 291–301. [Google Scholar]
- Sullivan, M.L.; Knollenberg, B.J. Red clover HDT, a BAHD Hydroxycinnamoyl-coenzyme A: L-3,4-Dihydroxyphenylalanine (L-DOPA) hydroxycinnamoyl transferase that synthesizes clovamide and other N-Hydroxycinnamoyl-aromatic amino acid amides. Front. Plant Sci. 2021, 12, 727461. [Google Scholar]
- Sanz-Carbonell, A.; Marques, M.C.; Bustamante, A.; Fares, M.A.; Rodrigo, G.; Gomez, G. Inferring the regulatory network of the miRNA-mediated response to biotic and abiotic stress in melon. BMC Plant Biol. 2019, 19, 78. [Google Scholar]
- Nyaboga, E.N.; Njiru, J.M.; Tripathi, L. Factors influencing somatic embryogenesis, regeneration, and Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14. Front. Plant Sci. 2015, 6, 411. [Google Scholar]
- Li, S.; Yu, X.; Lei, N.; Cheng, Z.; Zhao, P.; He, Y.; Wang, W.; Peng, M. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci. Rep. 2017, 7, 45981. [Google Scholar]
- Steinwand, M.A.; Ronald, P.C. Crop biotechnology and the future of food. Nat. Food 2020, 1, 273–283. [Google Scholar]
- Amelework, A.B.; Bairu, M.W. Advances in genetic analysis and breeding of cassava (Manihot esculenta Crantz): A review. Plants 2022, 11, 1617. [Google Scholar]
- Pendlebury, D.A. The use and misuse of journal metrics and other citation indicators. Arch. Immunol. Ther. Exp. 2020, 57, 1–11. [Google Scholar]
- Bar-Ilan, J. Informetrics at the beginning of the 21st century—A review. J. Informetr. 2008, 2, 1–52. [Google Scholar]
- Masic, I.; Begic, E. Evaluation of Scientific Journal Validity, Its Articles and Their Authors. In Unifying the Applications and Foundations of Biomedical and Health Informatics; IOS Press: Amsterdam, The Netherlands, 2016; pp. 9–14. [Google Scholar]
- Torres-Salinas, D.; Valderrama-Baca, P.; Arroyo-Machado, W. Is there a need for a new journal metric? Correlations between JCR Impact Factor metrics and the Journal Citation Indicator—JCI. J. Informetr. 2022, 16, 101315. [Google Scholar]
- Tomaszewski, R. Visibility, impact, and applications of bibliometric software tools through citation analysis. Scientometrics 2023, 128, 4007–4028. [Google Scholar]
- Koushauihhu, K.; Thelwall, M. Artificial intelligence to support publishing and peer review: A summary and review. Learn. Publ. 2024, 37, 4–12. [Google Scholar]
- de Almeida, C.C.; Grácio, M.C.C. Fator de Impacto e a decisão de publicação de um artigo. Páginas a&b Arq. Bibl. 2020, 172–183. [Google Scholar] [CrossRef]
- Junior, A.A.B.; Faria, W.R.; Montenegro, R.L.G.; Bahia, D.S.; Gonçalves, E. Research and development, productive structure and economic effects: Assessing the role of public financing in Brazil. Econ. Model. 2020, 90, 235–253. [Google Scholar]
- Lewis, J.; Schneegans, S.; Straza, T. UNESCO Science Report: The Race Against Time for Smarter Development; Unesco Publishing: Paris, France, 2021. [Google Scholar]
- Hasan, N.; Singh, M. Library and information science research output: A study based on Web of Science. Collnet J. Scientom. Inf. Manag. 2015, 9, 47–64. [Google Scholar]
- Zhao, Z.; Pan, X.; Hua, W. Comparative analysis of the research productivity, publication quality, and collaboration patterns of top ranked library and information science schools in China and the United States. Scientometrics 2021, 126, 931–950. [Google Scholar]
- Jin, W.; Wu, S.; Zhang, Y.; Zhou, G.; Xu, L.; Xu, Y. Review on Chinese agricultural science and technology research from a low-carbon economy perspective: Hotspots, evolution, and frontiers. Front. Environ. Sci. 2023, 11, 1268432. [Google Scholar]
- Cabral, L. Embrapa and the construction of scientific heritage in Brazilian agriculture: Sowing memory. Dev. Policy Rev. 2021, 39, 789–810. [Google Scholar]
- Uchendu, K.; Njoku, D.N.; Paterne, A.; Rabbi, I.Y.; Dzidzienyo, D.; Tongoona, P.; Offei, S.; Egesi, C. Genome-Wide Association Study of Root Mealiness and Other Texture-Associated Traits in Cassava. Front. Plant Sci. 2021, 12, 770434. [Google Scholar]
- Yuan, Z.; Jia, G. Systematic investigation of keywords selection and processing strategy on search engine forecasting: A case of tourist volume in Beijing. Inf. Technol. Tour. 2022, 24, 547–580. [Google Scholar]
- Dirrigl, F.J., Jr.; White, J. The use of keywords in archaeornithology literature. Int. J. Osteoarchaeol. 2023, 33, 787–797. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar]
- Adjebeng-Danquah, J.; Manu-Aduening, J.; Asante, I.K.; Agyare, R.Y.; Gracen, V.; Offei, S.K. Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon 2020, 6, e03154. [Google Scholar]
- Dissanayake, L.; Perera, P.; Attanayaka, T.; Heberle, E.; Jayawardhana, M. Early development of direct embryos in the cultured anthers of Manihot esculenta Crantz. Plants 2020, 9, 1315. [Google Scholar]
- Olsen, K.M. SNPs, SSRs and inferences on cassava’s origin. Plant Mol. Biol. 2004, 56, 517–526. [Google Scholar]
- Lopez, C.E.; Piégu, B.; Cooke, R.; Delseny, M.; Tohme, J.; Verdier, V. Using cDNA and genomic sequences as tools to develop SNP strategies in cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 2005, 110, 425–431. [Google Scholar]
- Xavier, C.A.; Nogueira, A.M.; Bello, V.H.; Watanabe, L.F.M.; Barbosa, T.M.C.; Júnior, M.A.; Zerbini, F.M. Assessing the diversity of whiteflies infesting cassava in Brazil. PeerJ 2021, 9, e11741. [Google Scholar]
- Masumba, E.A.; Kapinga, F.; Mkamilo, G.; Salum, K.; Kulembeka, H.; Rounsley, S.; Bredeson, J.V.; Myburg, A.A.; Ferguson, M.E. QTL associated with resistance to cassava brown streak and cassava mosaic diseases in a bi-parental cross of two Tanzanian farmer varieties, Namikonga and Albert. Theor. Appl. Genet. 2017, 130, 2069–2090. [Google Scholar]
- Nzuki, I.; Katari, M.S.; Bredeson, J.V.; Masumba, E.; Kapinga, F.; Salum, K.; Mkamilo, G.S.; Shah, T.; Lyons, J.B.; Rokhsar, D.S.; et al. QTL mapping for pest and disease resistance in cassava and coincidence of some QTL with introgression regions derived from Manihot glaziovii. Front. Plant Sci. 2017, 8, 1168. [Google Scholar]
- Morales, N.; Bauchet, G.J.; Tantikanjana, T.; Powell, A.F.; Ellerbrock, B.J.; Tecle, I.Y.; Mueller, L.A. High density genotype storage for plant breeding in the Chado schema of Breedbase. PLoS ONE 2020, 15, e0240059. [Google Scholar]
- Rabbi, I.Y.; Hamblin, M.T.; Gedil, M.A.; Esuma, W.; Gore, M.A.; Kulakow, P.A. Genome-Wide Association Study of Root Mealiness and Other Texture-Associated Traits in Cassava. Front. Plant Sci. 2022, 11, 623123. [Google Scholar]
- Srisawad, N.; Sraphet, S.; Suksee, N.; Boontung, R.; Smith, D.R.; Triwitayakorn, K. Use of Diversity Arrays Technology (DArT) for detection of QTL underlying plant architecture and yield-related traits in cassava. J. Crop Improv. 2023, 37, 99–118. [Google Scholar]
- Fu, J.; Hong, Z.M.; Huang, W.Z. Harnessing genomic tools for cassava improvement: Advances and prospects. Biosci. Evid. 2024, 14, 32–38. [Google Scholar]
- Yin, H.; Yan, Y.; Hu, W.; Liu, G.; Zeng, H.; Wei, Y.; Shi, H. Genome-wide association studies reveal genetic basis of ionomic variation in cassava. Plant J. 2022, 112, 1212–1223. [Google Scholar]
- Ige, A.D.; Olasanmi, B.; Mbanj, E.G.N.; Kayondo, I.S.; Parkes, E.Y.; Kulakow, P.; Rabbi, I.Y. Conversion and validation of uniplex SNP markers for selection of resistance to cassava mosaic disease in cassava breeding programs. Agronomy 2021, 11, 420. [Google Scholar]
- Adu, M.O.; Asare, P.A.; Yawson, D.O.; Nyarko, M.A.; Abdul Razak, A.; Kusi, A.K.; Amenorpe, G. The search for yield predictors for mature field-grown plants from juvenile pot-grown cassava (Manihot esculenta Crantz). PLoS ONE 2020, 15, e0232595. [Google Scholar]
- Chiewchankaset, P.; Thaiprasit, J.; Kalapanulak, S.; Wojciechowski, T.; Boonjing, P.; Saithong, T. Effective metabolic carbon utilization and shoot-to-root partitioning modulate distinctive yield in high yielding cassava variety. Front. Plant Sci. 2022, 13, 832304. [Google Scholar]
- Simon, M.F.; Mendoza Flores, J.M.; Liu, H.L.; Martins, M.L.L.; Drovetski, S.V.; Przelomska, N.A.; Kistler, L. Phylogenomic analysis points to a South American origin of Manihot and illuminates the primary gene pool of cassava. New Phytol. 2021, 233, 534–545. [Google Scholar]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).