Comparison of Rhizosphere Microbial Diversity in Soybean and Red Kidney Bean Under Continuous Monoculture and Intercropping Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Soil Sampling and Rhizosphere Soil Collection
2.3. DNA Extraction
2.4. Microbial Amplicon Sequencing Library Preparation
2.5. Bioinformatics and Statistical Analysis
3. Results
3.1. Quality Metrics of Amplicon Sequencing
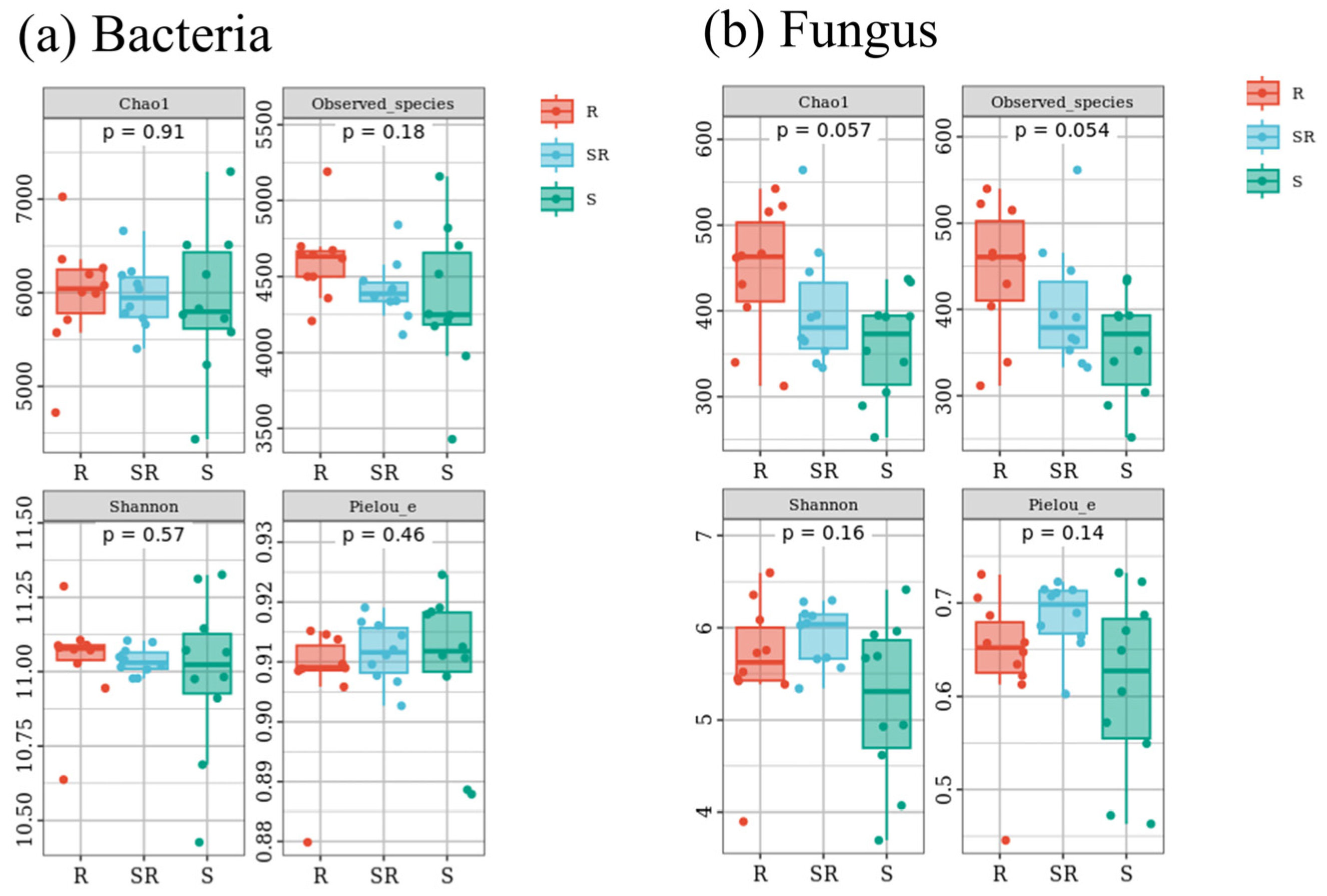
3.2. Alpha Diversity of Bacterial and Fungal Communities
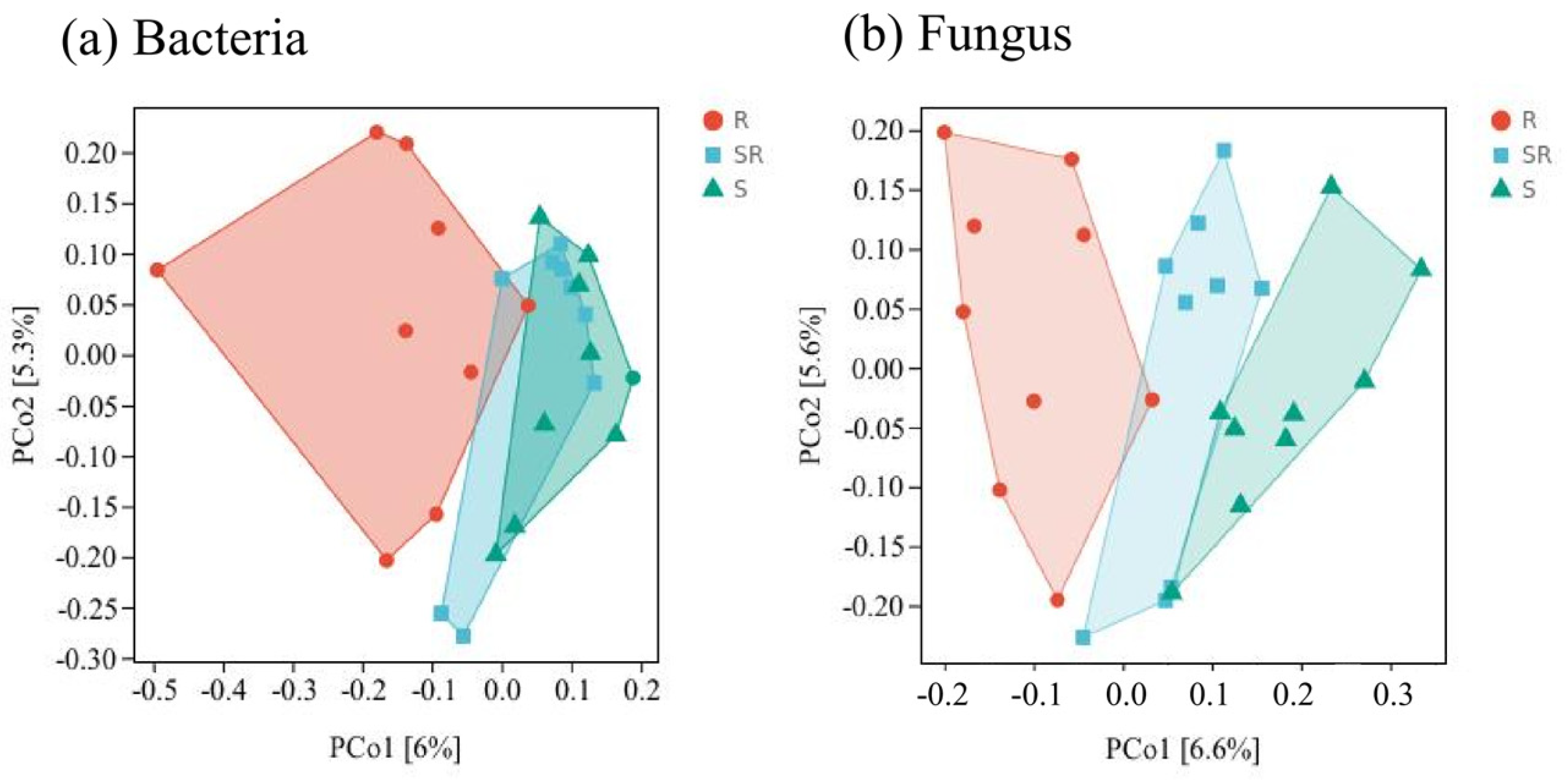
3.3. Beta Diversity Analysis of Bacterial and Fungal Communities Based on Principal Coordinate Analysis (PCoA)
3.4. Rhizosphere Microbial Community Assembly Dynamics Across Cropping Systems and Growth Stages
3.5. Effects of Intercropping Infection on the Bacterial Microbiome
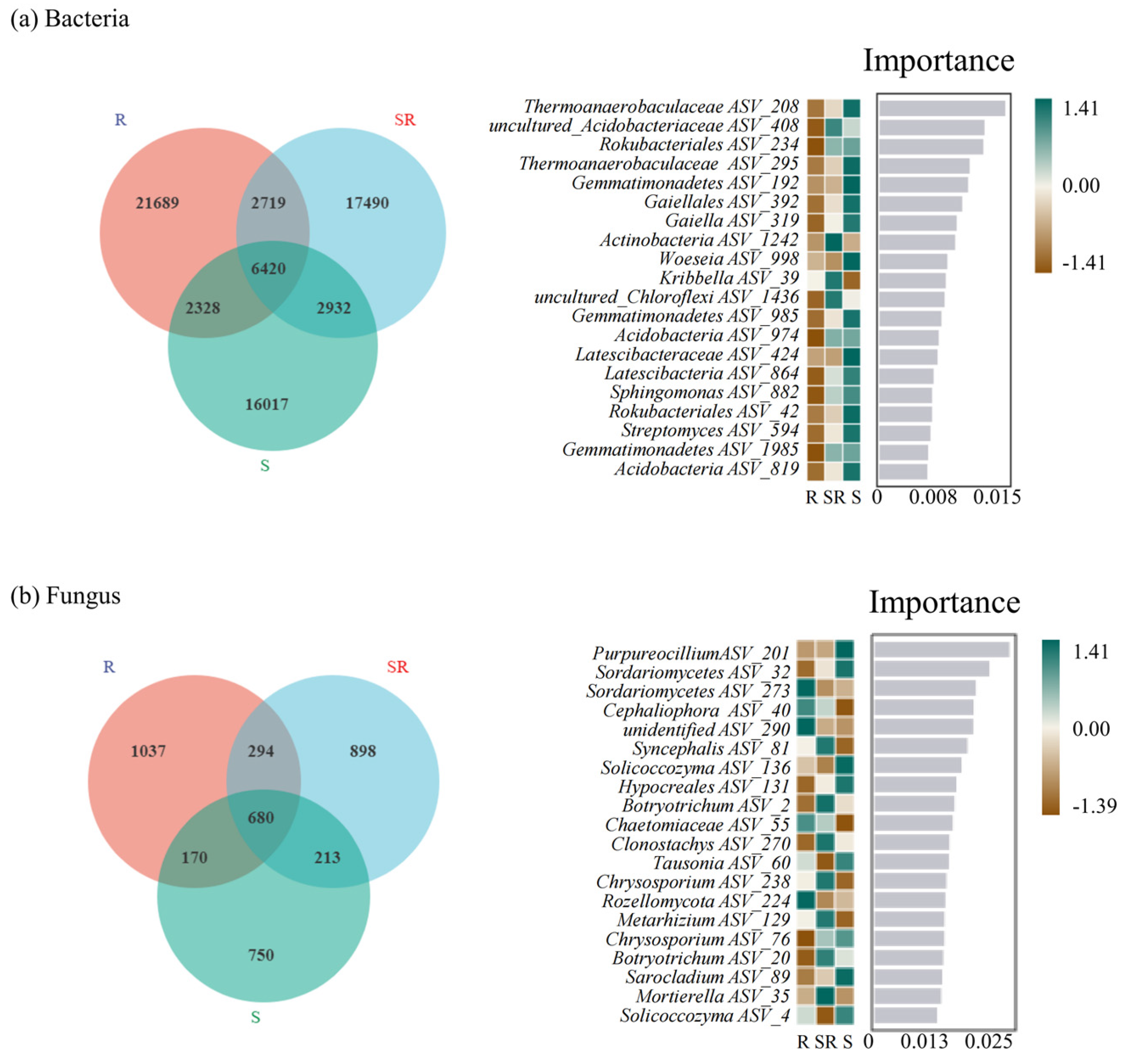
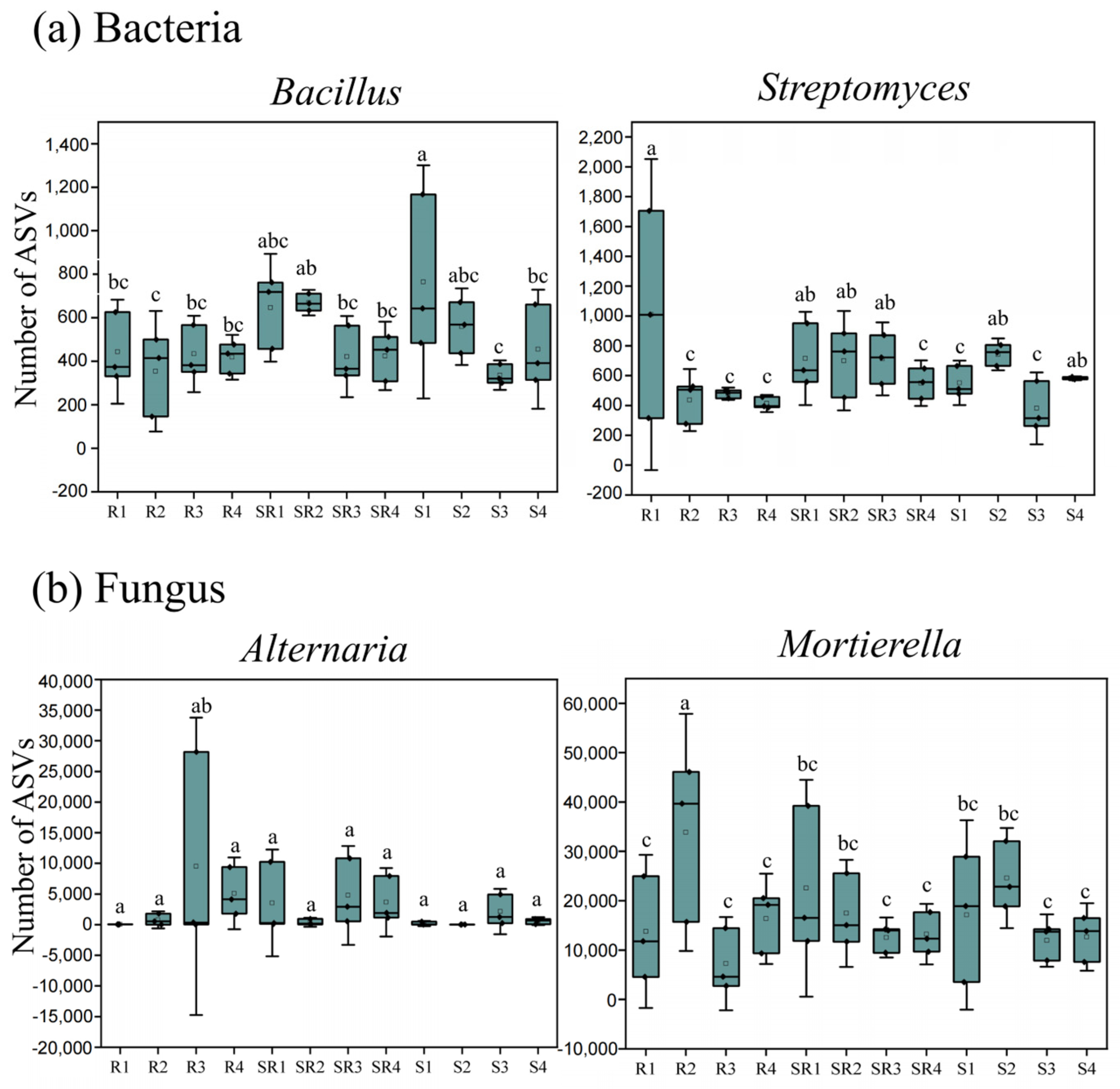
3.6. Distribution of Specific Microorganisms Across Three Cultivation Systems
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xiao, X.; Chen, W.; Zong, L.; Yang, J.; Jiao, S.; Lin, Y.; Wang, E.; Wei, G. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol. Ecol. 2017, 26, 1641–1651. [Google Scholar] [CrossRef]
- Ma, L.; Ma, S.Y.; Chen, G.P.; Lu, X.; Chai, Q.; Li, S. Mechanisms and Mitigation Strategies for the Occurrence of Continuous Cropping Obstacles of Legumes in China. Agronomy 2024, 14, 104. [Google Scholar] [CrossRef]
- Duchene, O.; Vian, J.F.; Celette, F. Intercropping with legume for agroecological cropping systems: Complementarity and facilitation processes and the importance of soil microorganisms. A review. AGR Ecosyst. Environ. 2017, 240, 148–161. [Google Scholar] [CrossRef]
- Solanki, M.K.; Wang, Z.; Wang, F.Y.; Li, C.N.; Gupta, C.L.; Singh, R.K.; Malviya, M.K.; Singh, P.; Yang, L.T.; Li, Y.R. Assessment of Diazotrophic Proteobacteria in Sugarcane Rhizosphere When Intercropped with Legumes (Peanut and Soybean) in the Field. Front. Microbiol. 2020, 11, 1814. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Fan, M.; Wang, E.; Chen, W.; Wei, G. Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 2017, 101, 8485–8497. [Google Scholar] [CrossRef]
- Yin, C.; Vargas, J.C.; Schlatter, D.C.; Hagerty, C.; Hulbert, S.H.; Paulitz, T.C. Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 2021, 9, 86. [Google Scholar] [CrossRef]
- Yu, P.; He, X.M.; Baer, M.; Beirinckx, S.; Tian, T.; Moya, Y.A.T.; Zhang, X.C.; Deichmann, M.; Frey, F.P.; Bresgen, V.; et al. Plant flavones enrich rhizosphere Oxalo bacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 2021, 7, 481–499. [Google Scholar] [CrossRef]
- Silva, F.D.; Vieira, V.D.; da Silva, R.C.; Pinheiro, D.G.; Soares, M.A. Introduction of Trichoderma spp. biocontrol strains against Sclerotinia sclerotiorum (Lib.) de Bary change soil microbial community composition in common bean (Phaseolus vulgaris L.) cultivation. Biol. Control 2021, 163, 104755. [Google Scholar] [CrossRef]
- Lai, H.; Gao, F.; Su, H.; Zheng, P.; Li, Y.; Yao, H. Nitrogen Distribution and Soil Microbial Community Characteristics in a Legume–Cereal Intercropping System: A Review. Agronomy 2022, 12, 1900. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, C.; Bei, S.; Wang, G.; Geisen, S.; Bedoussac, L.; Christie, P.; Zhang, J. High bacterial diversity and siderophore-producing bacteria collectively suppress Fusarium oxysporum in maize/faba bean intercropping. Front. Microbiol. 2022, 13, 972587. [Google Scholar] [CrossRef]
- Zhao, Y.; Yan, C.; Hu, F.; Luo, Z.; Zhang, S.; Xiao, M.; Chen, Z.; Fan, H. Intercropping Pinto Peanut in Litchi Orchard Effectively Improved Soil Available Potassium Content, Optimized Soil Bacterial Community Structure, and Advanced Bacterial Community Diversity. Front. Microbiol. 2022, 13, 868312. [Google Scholar] [CrossRef]
- Punyalue, A.; Jamjod, S.; Rerkasem, B. Intercropping Maize With Legumes for Sustainable Highland Maize Production. Mt. Res. Dev. 2018, 38, 35–44. [Google Scholar] [CrossRef]
- Klichowska, E.; Nobis, M.; Piszczek, P.; Blaszkowski, J.; Zubek, S. Soil properties rather than topography, climatic conditions, and vegetation type shape AMF-feathergrass relationship in semi-natural European grasslands. Appl. Soil. Ecol. 2019, 144, 22–30. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellin, L.E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Van Acker, R.; Van Montagu Boerjan, W.; Vangronsveld, J. Lignin engineering in field-grown poplar trees affects the endospherebacterial microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 2312–2317. [Google Scholar] [CrossRef]
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterialmicrobiome of feld-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef]
- Mori, H.; Maruyama, F.; Kato, H.; Toyoda, A.; Dozono, A.; Ohtsubo, Y.; Nagata, Y.; Fujiyama, A.; Tsuda, M.; Kurokawa, K. Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 2014, 21, 217–227. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet GAAl-Ghalith, J.; Caporaso, G. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v2. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. Dada2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. NAR 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaudoise Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Koljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Sun, C.; Qiu, M.; Lu, X.; Wang, Y.W. Core microbiota play important roles in maintaining soil multi-nutrient cycling in lakeshore wetland of plateau lake Caohai. Land Degrad. Dev. 2024, 35, 1308–1319. [Google Scholar]
- Lim, S.K.; Ten, L.N.; Avalos-Ruiz, D.; Ryu, J.J.; Kang, I.K.; Lee, S.Y.; Jung, H.Y. Isolation and Identification of Two Unreported Sordariomycetes Fungi in Korea: Pestalotiopsis clavata and Botryotrichum iranicum. Kor. J. Mycol. 2022, 50, 183–194. [Google Scholar]
- Abbasi, S.; Sadeghi, A.; Safaie, N. Streptomyces alleviate drought stress in tomato plants and modulate the expression of transcription factors ERF1 and WRKY70 genes. Sci. Hortic. 2020, 265, 109206. [Google Scholar] [CrossRef]
- Meteyer, C.U.; Boyles, J.G. Fungal chimera: A lethal mammalian fungus with invasion strategies of plant pathogens. Virulence 2025, 16, 2439497. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.J.; Ahn, D.H.; Lee, J.K.; Park, H. Draft genome sequence of Sphingomonas echinoides ATCC 14820. J. Bacteriol. 2012, 194, 1843. [Google Scholar] [CrossRef]
- Rafii, F.; Crawford, D.L.; Bleakley, B.H.; Wang, Z. Assessing the risks of releasing recombinant Streptomyces in soil. Microbiol. Sci. 1988, 5, 358–362. [Google Scholar]
- Marcheva, M.; Petkova, M.; Slavova, V.; Popov, V. Positive Effect of Camelina Intercropping with Legumes on Soil Microbial Diversity by Applying NGS Analysis and Mobile Fluorescence Spectroscopy. Appl. Sci. 2024, 14, 9046. [Google Scholar] [CrossRef]
- Vora, S.M.; Joshi, P.; Belwalkar, M.; Archana, G. Root exudates influence chemotaxis and colonization of diverse plant growth promoting rhizobacteria in the pigeon pea-maize intercropping system. Rhizophere 2021, 18, 100331. [Google Scholar] [CrossRef]
- Lanzavecchia, G.; Frascarelli, G.; Rocchetti, L.; Bellucci, E.; Bitocchi, E.; Di Vittori, V.; Sillo, F.; Ferraris, I.; Carta, G.; Delledonne, M.; et al. Genotype Combinations Drive Variability in the Microbiome Configuration of the Rhizosphere of Maize/Bean Intercropping System. Int. J. Mol. Sci. 2024, 25, 1288. [Google Scholar] [CrossRef]
- Chamkhi, I.; Cheto, S.; Geistlinger, J.; Zeroual, Y.; Kouisni, L.; Bargaz, A.; Ghoulam, C. Legume-based intercropping systems promote beneficial rhizobacterial community and crop yield under stressing conditions. Ind. Crop. Prod. 2022, 183, 114958. [Google Scholar] [CrossRef]
- Blessing, D.J.; Gu, Y.; Cao, M.; Cui, Y.; Wang, X.; Asante-Badu, B. Overview of the advantages and limitations of maize-soybean intercropping in sustainable agriculture and future prospects: A review. Chil. J. Agric. Res. 2022, 82, 177–188. [Google Scholar] [CrossRef]
- Yang, Q.; Niu, A.; Li, S.; Liu, J.; Zhou, G. Unveiling Metabolic Crosstalk: Bacillus-Mediated Defense Priming in Pine Needles Against Pathogen Infection. Metabolites 2024, 14, 646. [Google Scholar] [CrossRef]
- Muok, A.R.; Claessen, D.; Briegel, A. Microbial hitchhiking: How Streptomyces spores are transported by motile soil bacteria. ISME J. 2021, 15, 2591–2600. [Google Scholar] [CrossRef]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [PubMed]
- Worsley, S.F.; Newitt, J.; Rassbach, J.; Batey, S.F.D.; Holmes, N.A.; Murrell, J.C.; Wilkinson, B.; Hutchings, M. Streptomyces Endophytes Promote Host Health and Enhance Growth across Plant Species. Appl. Environ. Microb. 2020, 86, e01053-20. [Google Scholar] [CrossRef] [PubMed]
- Witte, T.E.; Villenueve, N.; Shields, S.W.; Sproule, A.; Eggertson, Q.; Kim, N.E.; Boddy, C.N.; Dettman, J.R.; Overy, D.P. Untargeted metabolomics screening reveals unique secondary metabolite production from Alternaria section Alternaria. Front. Mol. Biosci. 2022, 9, 1038299. [Google Scholar] [CrossRef] [PubMed]
- Vélez, M.L.; Marfetán, J.A.; Salgado Salomón, M.E.; Taccari, L.E.; Santini, A. Mortierella species from declining Araucaria araucana trees in Patagonia, Argentina. For. Pathol. 2020, 50, e12591. [Google Scholar] [CrossRef]
- Zhang, S.; Li, S.M.; Meng, L.B.; Liu, X.D.; Zhang, Y.H.; Zhao, S.C.; Zhao, H.B. Root exudation under maize/soybean intercropping system mediates the arbuscular mycorrhizal fungi diversity and improves the plant growth. Front. Plant Sci. 2024, 15, 1375194. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, H.; Li, A.; Zhong, S.; Zhang, Y.; Li, C.; Mu, Z.; Zhang, H.; Wu, J. Comparison of Rhizosphere Microbial Diversity in Soybean and Red Kidney Bean Under Continuous Monoculture and Intercropping Systems. Agronomy 2025, 15, 1705. https://doi.org/10.3390/agronomy15071705
Qin H, Li A, Zhong S, Zhang Y, Li C, Mu Z, Zhang H, Wu J. Comparison of Rhizosphere Microbial Diversity in Soybean and Red Kidney Bean Under Continuous Monoculture and Intercropping Systems. Agronomy. 2025; 15(7):1705. https://doi.org/10.3390/agronomy15071705
Chicago/Turabian StyleQin, Huibin, Aohui Li, Shuyu Zhong, Yingying Zhang, Chuhui Li, Zhixin Mu, Haiping Zhang, and Jing Wu. 2025. "Comparison of Rhizosphere Microbial Diversity in Soybean and Red Kidney Bean Under Continuous Monoculture and Intercropping Systems" Agronomy 15, no. 7: 1705. https://doi.org/10.3390/agronomy15071705
APA StyleQin, H., Li, A., Zhong, S., Zhang, Y., Li, C., Mu, Z., Zhang, H., & Wu, J. (2025). Comparison of Rhizosphere Microbial Diversity in Soybean and Red Kidney Bean Under Continuous Monoculture and Intercropping Systems. Agronomy, 15(7), 1705. https://doi.org/10.3390/agronomy15071705





