Enhancing Registration Offices’ Communication Through Interpretable Machine-Learning Techniques
Abstract
1. Introduction
- (i).
- Present a communication-oriented protocol for applying IML in variety registration workflows;
- (ii).
- Illustrate how Random Forests and AMBARTI can be integrated to support this protocol by capturing variable importance and G×E interactions;
- (iii).
- Evaluate wheat genotype performance across environments in terms of yield and protein content;
- (iv).
- Provide interpretable visual outputs, such as heatmaps and interaction networks, to facilitate stakeholder engagement and internal communication;
- (v).
- Explore temporal stability and environmental performance, highlighting consistently favorable sites such as Tolentino (protein) and Torino (yield);
- (vi).
- Demonstrate how the protocol can improve regulatory transparency, support agricultural innovation, and contribute to food security.
2. Materials and Methods
2.1. Experimental Design
2.2. Random Forests, Interpretable Machine Learning, Variable Importance, and Interactions
- ●
- MDA evaluates how permuting a variable affects prediction accuracy:
- ●
- Heatmaps (viviHeatmap): These show variable importance on the diagonal and interactions on the off-diagonal;
- ●
- Network graphs (viviNetwork): These represent variables as nodes, with node size and color indicating importance, and edge thickness reflecting interaction strength.
2.3. AMBARTI Method
3. Results
3.1. Illustrating a Communication Protocol with IML Tools
3.2. Visualizing Trial Data with Random Forests
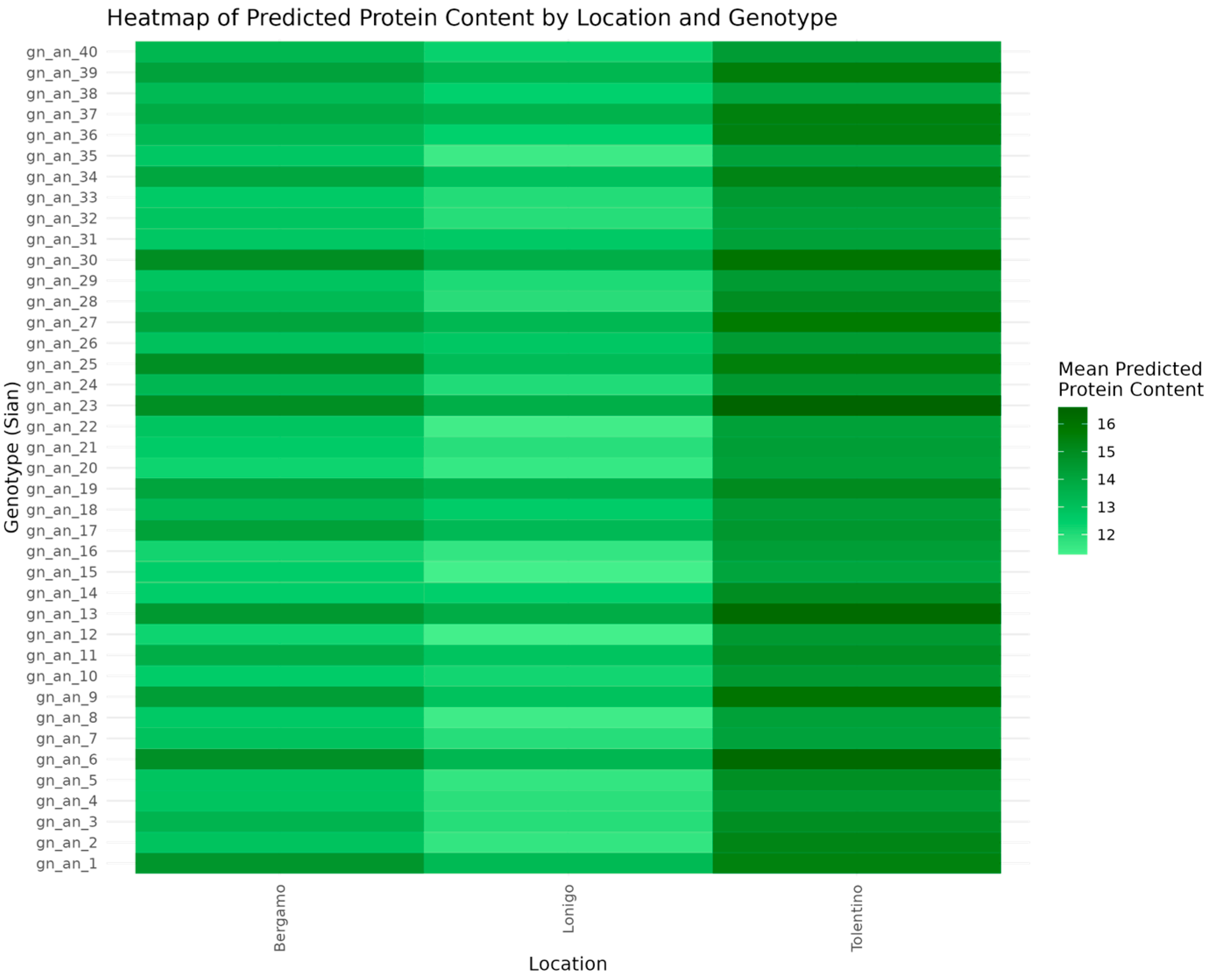
3.2.1. Protein Content Predictions for Communication (2021 and 2022)
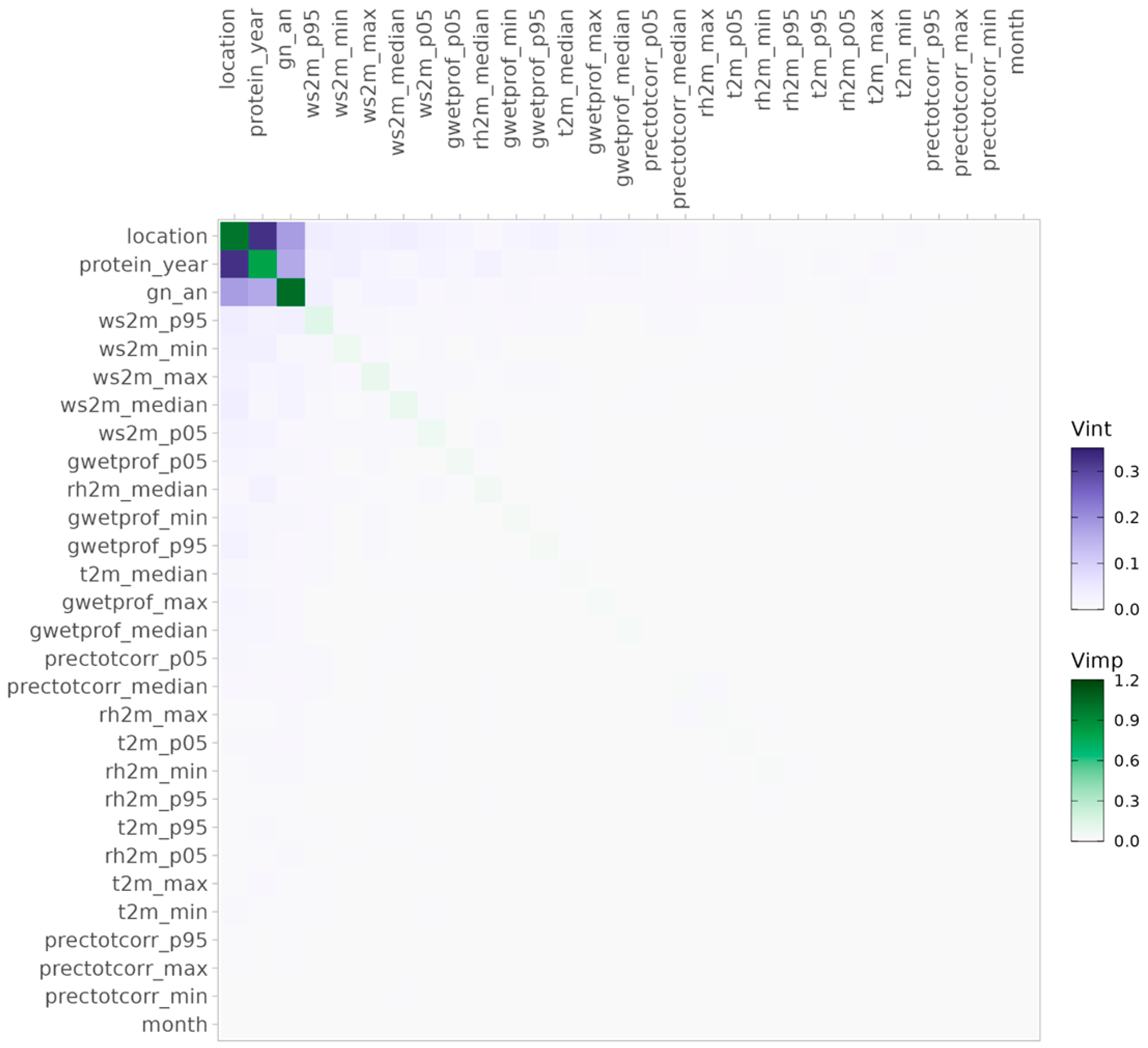
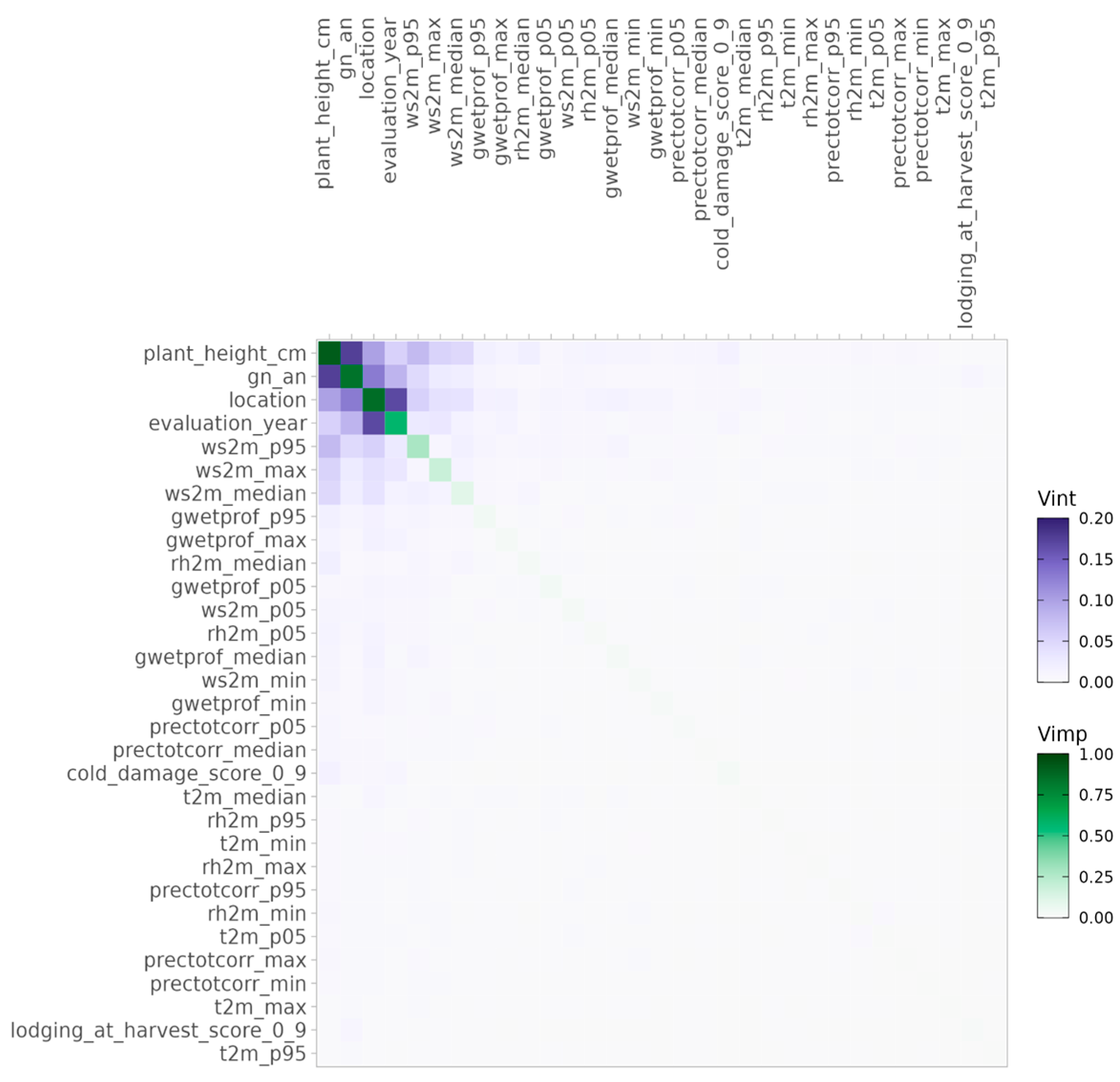
3.2.2. Identifying Key Variables and Interactions for Protein
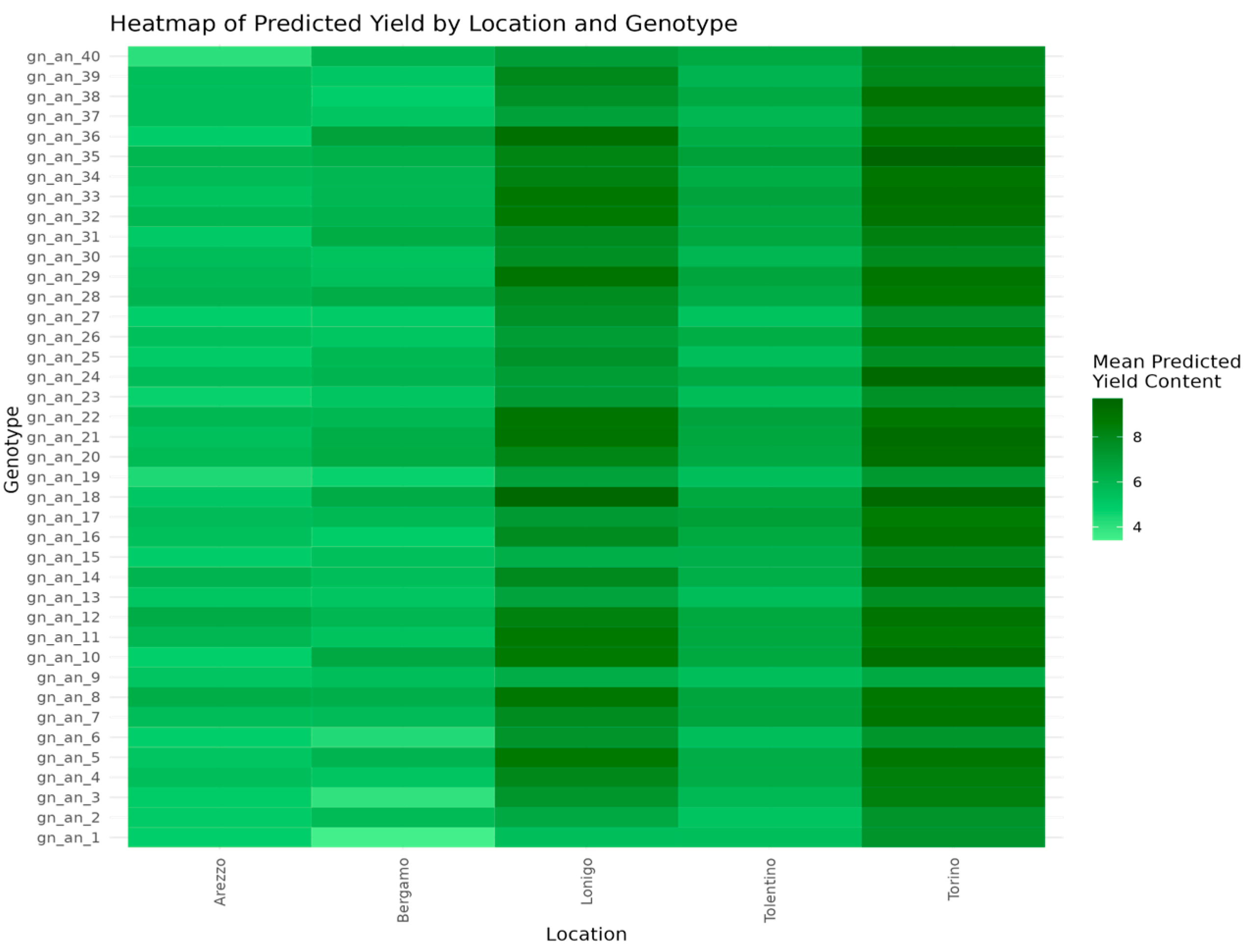
3.2.3. Yield Predictions and Genotypic Stability
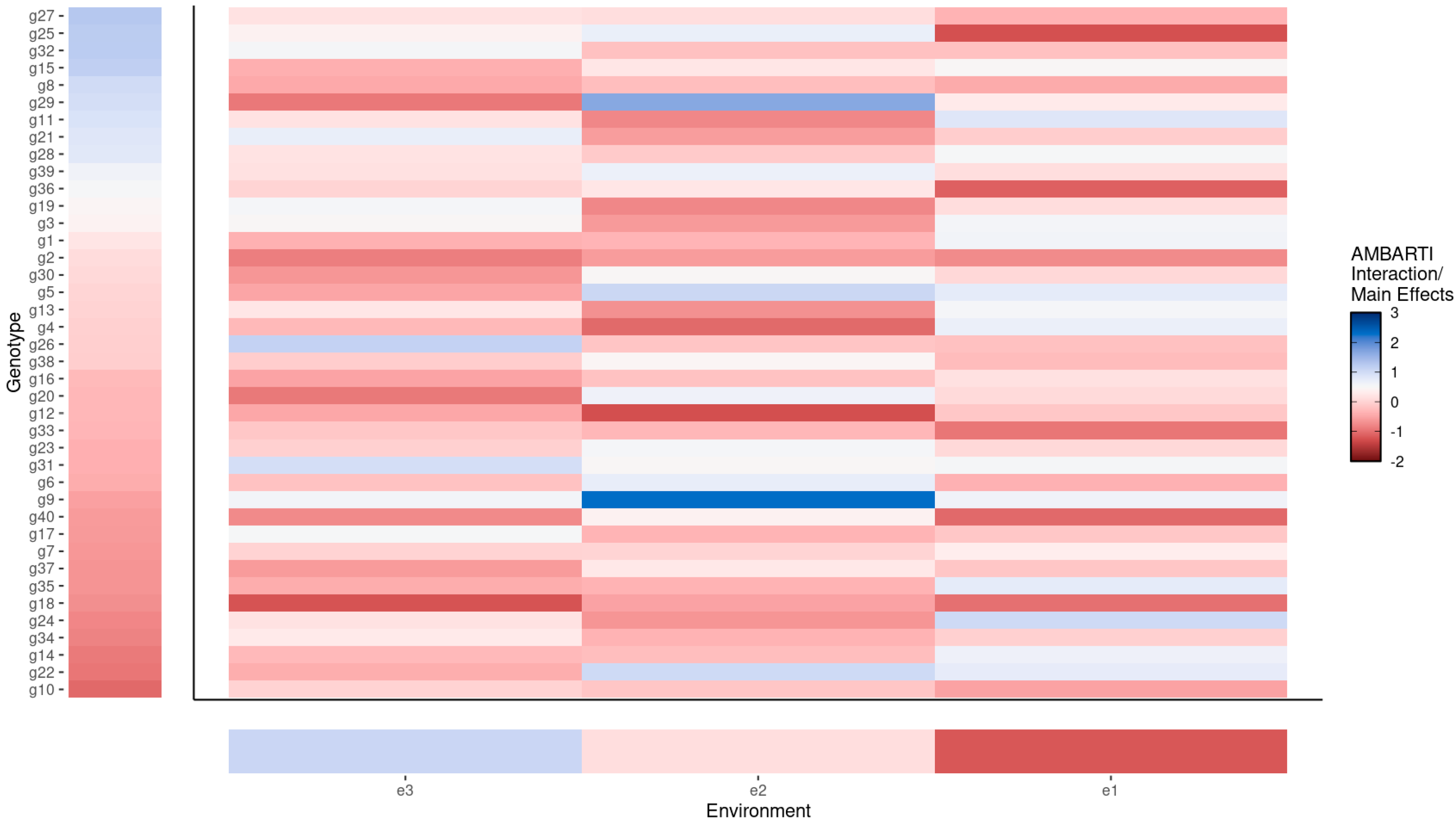
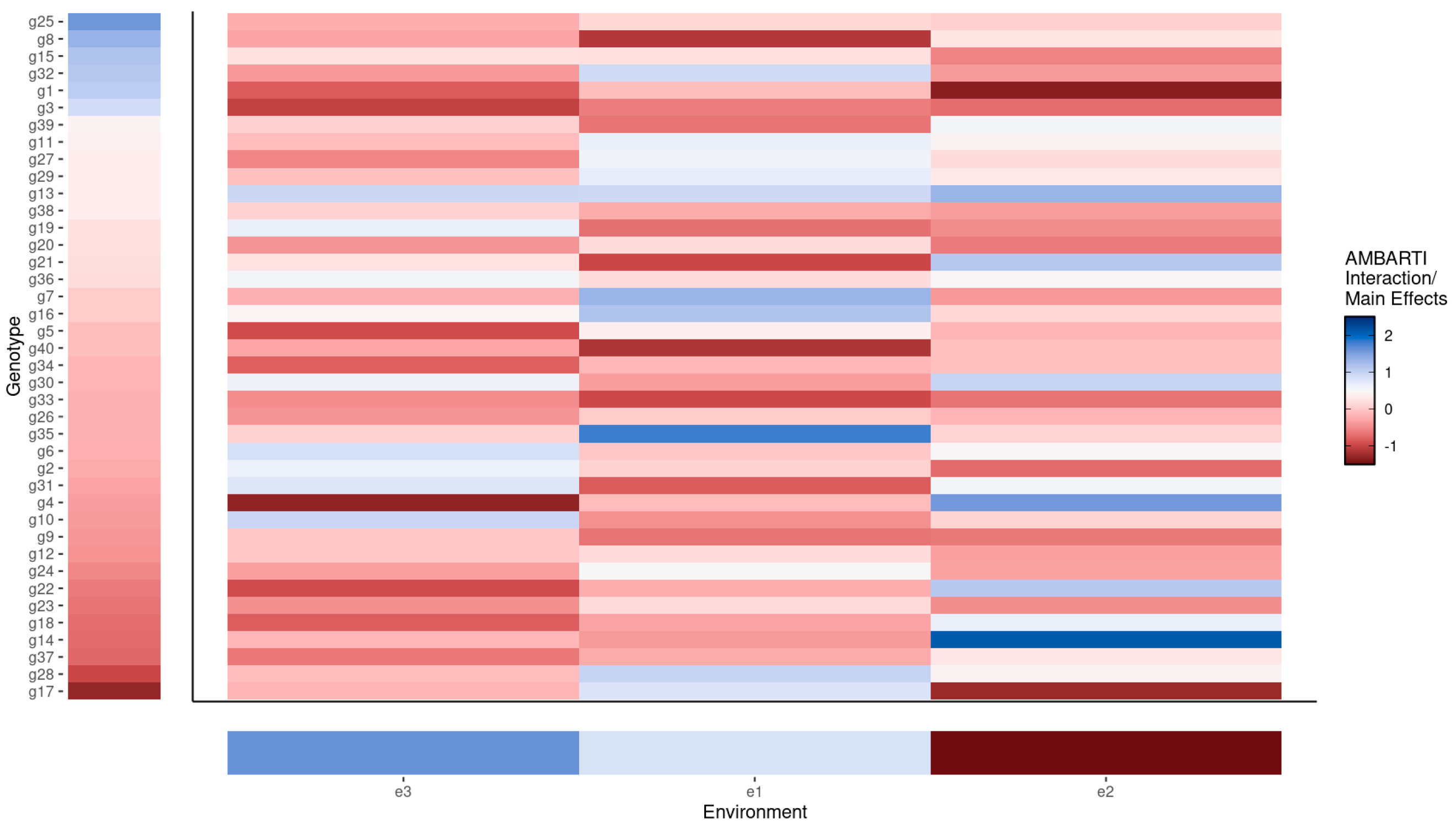
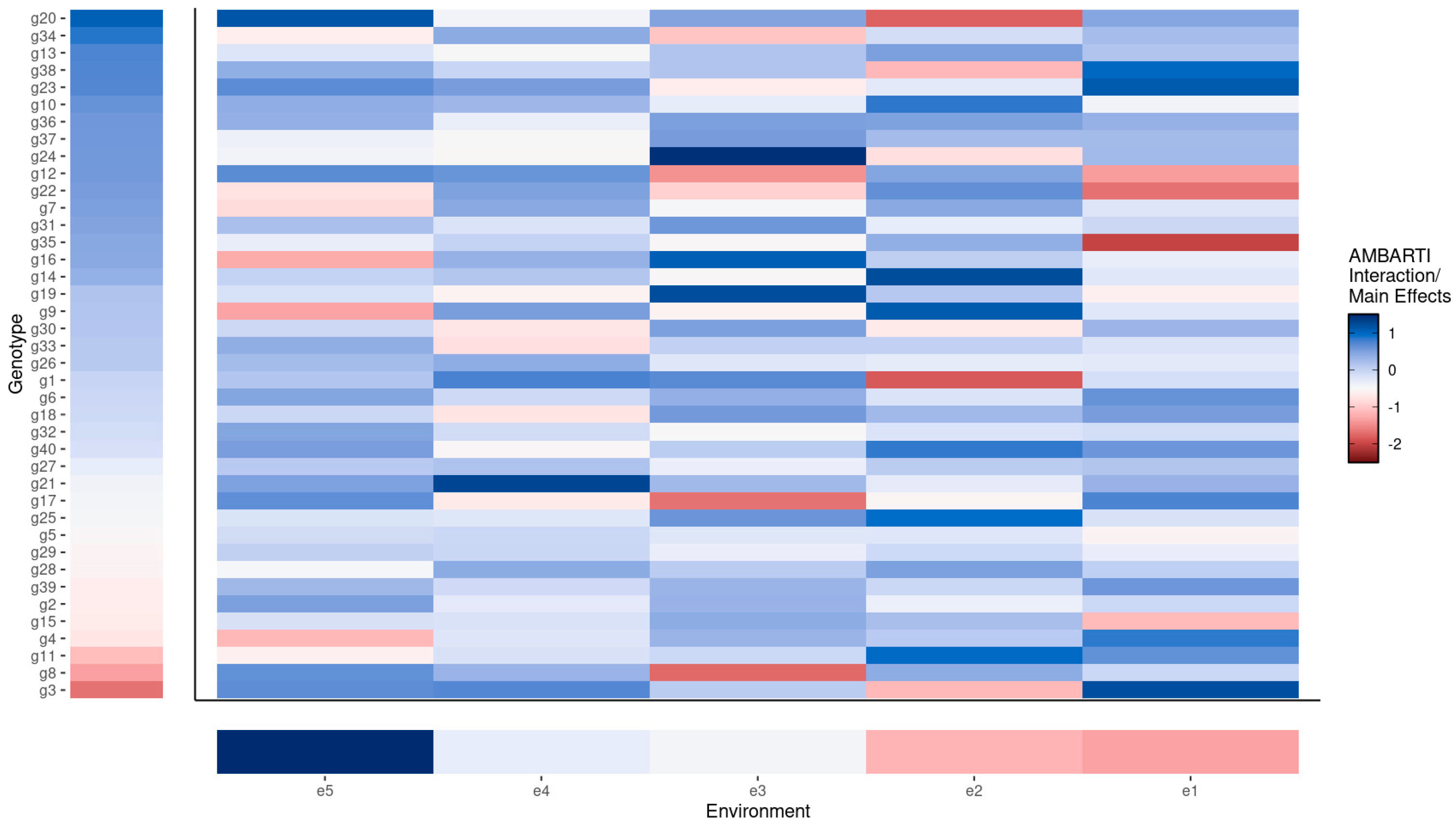
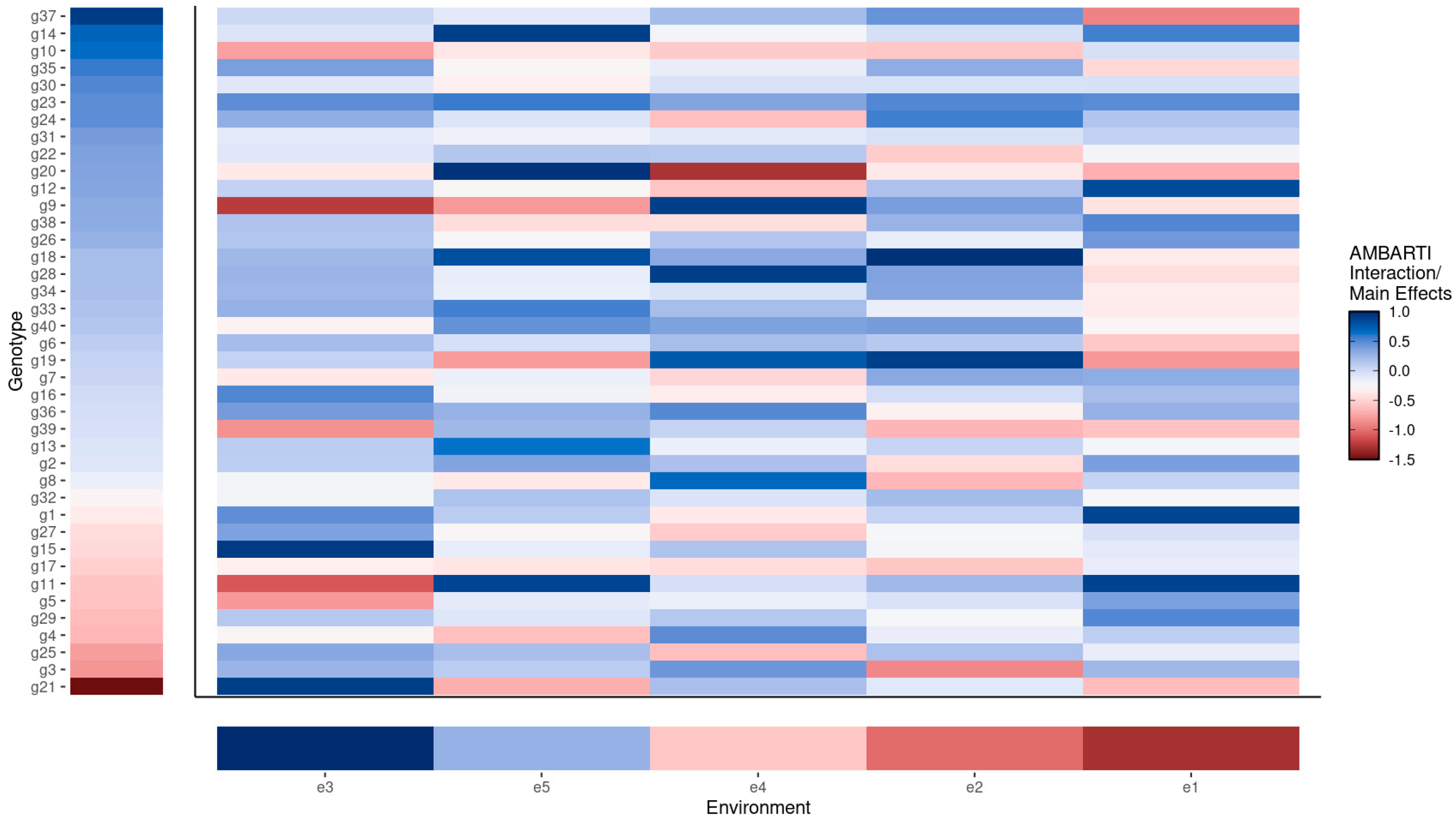
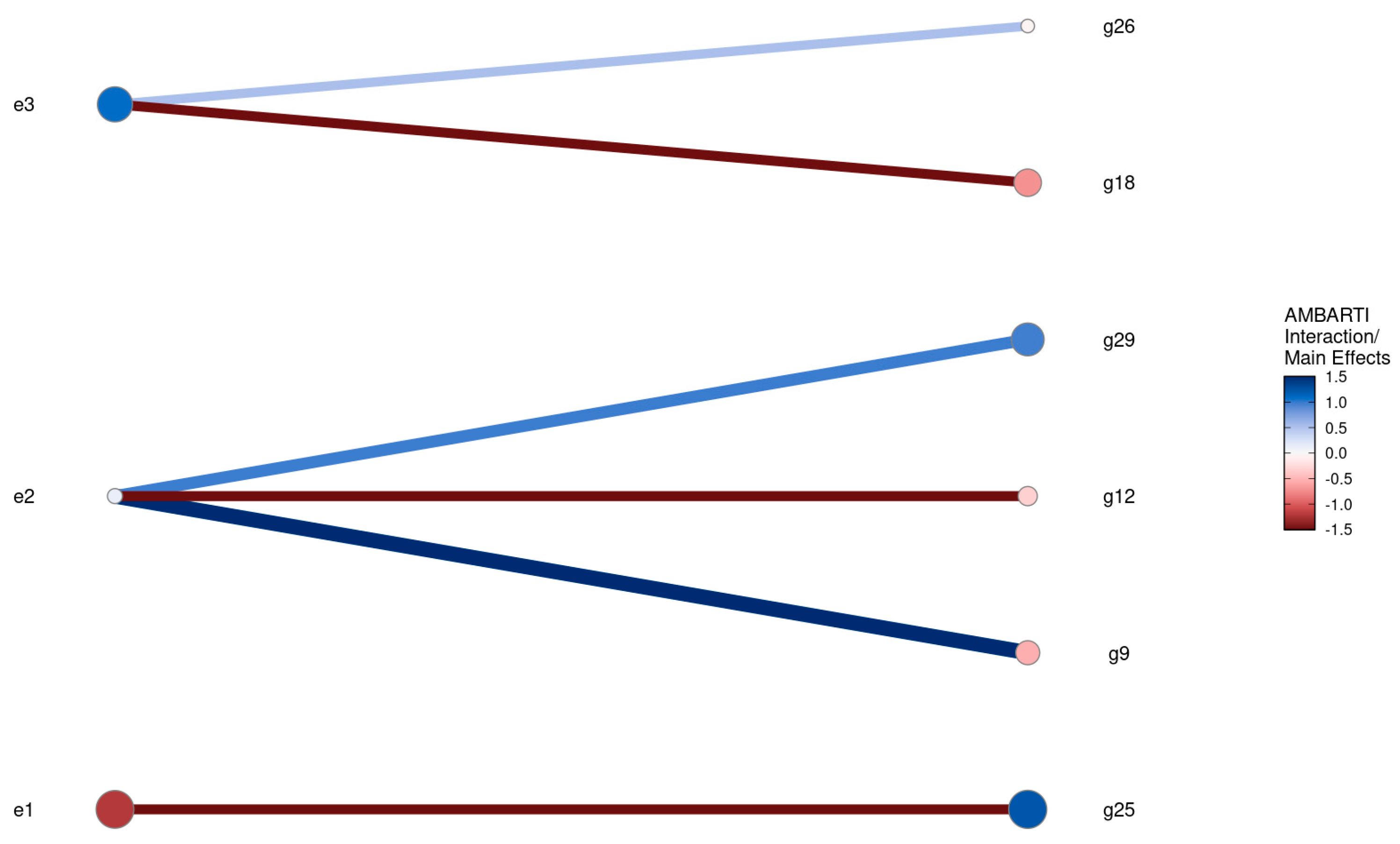
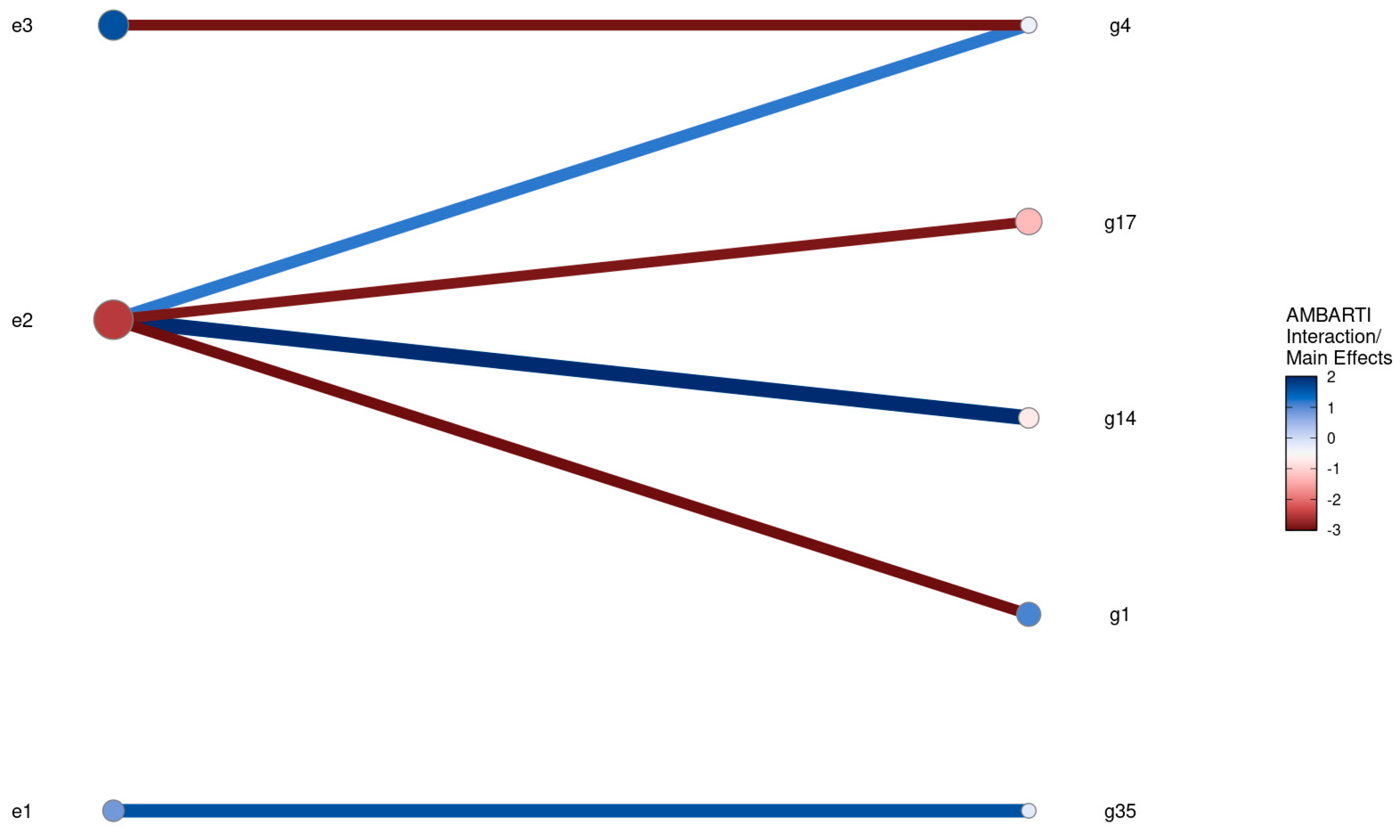
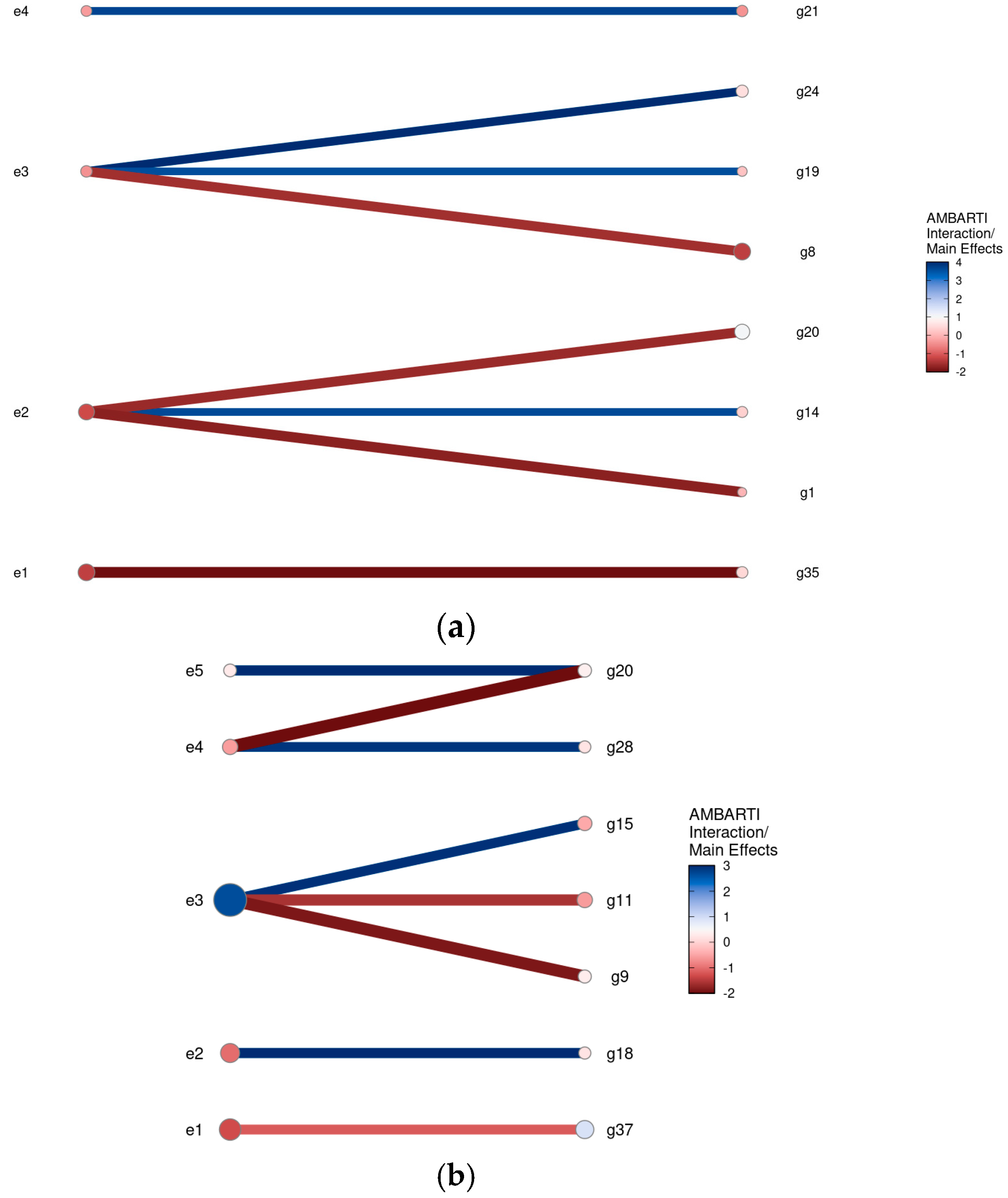
3.3. Visual Summaries via AMBARTI for G×E Understanding
3.3.1. Mapping Protein and Yield Interactions
3.3.2. Highlighting Extreme Interactions via Networks
3.3.3. Synthesizing Genotype and Environment Rankings
3.4. Proposed Four-Step Protocol for IML-Based Communication in Variety Registration
4. Discussion
4.1. Enhancing Communication with IML Visual Tools
4.2. From Predictions to Practice: Translating Model Outputs
4.3. Stability vs. Responsiveness: A Visual Dialogue
4.4. Proposed Protocol for IML-Based Communication
4.5. Final Remarks: Moving Beyond Predictive Accuracy
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz, R. Role of plant breeding to sustain food security under climate change. In Food Security and Climate Change; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; pp. 145–158. [Google Scholar] [CrossRef]
- Wardofa, G.A.; Mohammed, H.; Asnake, D.; Alemu, T. Genotype x environment interaction and yield stability of bread wheat genotypes in Central Ethiopia. J. Plant Breed. Genet. 2019, 7, 87–94. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Khalili, M.; Poczai, P.; Olivoto, T. Stability indices to deciphering the genotype-by-environment interaction (GEI) effect: An applicable review for use in plant breeding programs. Plants 2022, 11, 414. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, A.; Ramezani Moghaddam, M.R.; Najjar, H.; Arab Salmani, M.; Mohajer Abbasi, A. Value of Cultivation and Use (VCU) Evaluation of Some New Promising Genotypes of Upland Cotton (Gossypium hisutum L.) in Razavi Khorasan Province. Agrotech. Ind. Crops 2022, 2, 166–176. [Google Scholar] [CrossRef]
- Bozzoli, M. GWAS Analysis for the Identification of Molecular Basis Responsible for Yield-Related Traits and Disease Resistance in Durum Wheat. Ph.D. Thesis, University of Bologna, Bologna, Italy, 2023. [Google Scholar]
- Wang, L.; Zheng, Y.; Duan, L.; Wang, M.; Wang, H.; Li, H.; Li, R.; Zhang, H. Artificial selection trend of wheat varieties released in Huang-Huai-Hai region in China evaluated using DUS testing characteristics. Front. Plant Sci. 2022, 13, 898102. [Google Scholar] [CrossRef] [PubMed]
- Banjarey, P.; Rani, A.; Pandey, S.; Shukla, R.S.; Kumawat, S.; Singh, S. Morphological characterization of cms based wheat hybrids (Triticum aestivum L.). Int. J. Chem. Stud. 2022, 11, 172–175. [Google Scholar]
- Ghimire, S.; Verdonk, T. International Conference--IP Protection for Plant Innovation 2021. GRUR Int. 2022, 71, 535–539. [Google Scholar] [CrossRef]
- Sarti, D.; Wagner, J.; Palma, F.; Kalvan, H.; Giachini, M.; Lautenschalaeger, D.; Lupianhez, V.; Pires, J.; Sales, M.; Faria, P.; et al. Interpretable machine learning unveils key predictors and default values in an expanded database of human in vitro dermal absorption studies with pesticides. Regul. Toxicol. Pharmacol. 2025, 159, 105801. [Google Scholar] [CrossRef] [PubMed]
- van Etten, J.; de Sousa, K.; Aguilar, A.; Barrios, M.; Coto, A.; Dell’Acqua, M.; Fadda, C.; Vicente, J.; Mina, D.; Steinke, J.; et al. Crop variety management for climate adaptation supported by citizen science. Proc. Natl. Acad. Sci. USA 2019, 116, 4194–4199. [Google Scholar] [CrossRef]
- Nehe, A.; Akin, B.; Sanal, T.; Evlice, A.K.; Ünsal, R.; Dinçer, N.; Demir, L.; Geren, H.; Sevim, I.; Orhan, Ş. Genotype x environment interaction and genetic gain for grain yield and grain quality traits in Turkish spring wheat released between 1964 and 2010. PLoS ONE 2019, 14, e0219432. [Google Scholar] [CrossRef]
- Ahakpaz, F.; Abdi, H.; Neyestani, E.; Hesami, A.; Mohammadi, B.; Mahmoudi, K.N.; Abedi-Asl, G.; Noshabadi, M.R.J.; Ahakpaz, F.; Alipour, H. Genotype-by-environment interaction analysis for grain yield of barley genotypes under dryland conditions and the role of monthly rainfall. Agric. Water Manag. 2021, 245, 106665. [Google Scholar] [CrossRef]
- Gupta, V.; Kumar, M.; Singh, V.; Chaudhary, L.; Yashveer, S.; Sheoran, R.; Dalal, M.S.; Nain, A.; Lamba, K.; Gangadharaiah, N. Genotype by environment interaction analysis for grain yield of wheat (Triticum aestivum (L.) em. Thell) genotypes. Agriculture 2022, 12, 1002. [Google Scholar] [CrossRef]
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine learning in agriculture: A comprehensive updated review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.; Boogaard, H.; de Wit, A.; Janssen, S.; Osinga, S.; Pylianidis, C.; Athanasiadis, I.N. Machine learning for large-scale crop yield forecasting. Agric. Syst. 2021, 187, 103016. [Google Scholar] [CrossRef]
- Zednik, C. Solving the black box problem: A normative framework for explainable artificial intelligence. Philos. Technol. 2021, 34, 265–288. [Google Scholar] [CrossRef]
- Molnar, C.; Casalicchio, G.; Bischl, B. Interpretable machine learning—A brief history, state-of-the-art and challenges. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases; Springer: Berlin/Heidelberg, Germany, 2020; pp. 417–431. [Google Scholar]
- Rudin, C.; Chen, C.; Chen, Z.; Huang, H.; Semenova, L.; Zhong, C. Interpretable machine learning: Fundamental principles and 10 grand challenges. Stat. Surv. 2022, 16, 1–85. [Google Scholar] [CrossRef]
- Carter, A.; Imtiaz, S.; Naterer, G.F. Review of interpretable machine learning for process industries. Process Saf. Environ. Prot. 2023, 170, 647–659. [Google Scholar] [CrossRef]
- Chen, Y.; Calabrese, R.; Martin-Barragan, B. Interpretable machine learning for imbalanced credit scoring datasets. Eur. J. Oper. Res. 2024, 312, 357–372. [Google Scholar] [CrossRef]
- Liu, B.; Lu, W.; Olofsson, T.; Zhuang, X.; Rabczuk, T. Stochastic interpretable machine learning-based multiscale modeling in thermal conductivity of Polymeric graphene-enhanced composites. Compos. Struct. 2024, 327, 117601. [Google Scholar] [CrossRef]
- Lee, M.-H. Predicting and analyzing the fill factor of non-fullerene organic solar cells based on material properties and interpretable machine-learning strategies. Sol. Energy 2024, 267, 112191. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y. Spatial distribution and source identification of potentially toxic elements in Yellow River Delta soils, China: An interpretable machine-learning approach. Sci. Total Environ. 2024, 912, 169092. [Google Scholar] [CrossRef]
- Wani, N.A.; Kumar, R.; Bedi, J. DeepXplainer: An interpretable deep learning-based approach for lung cancer detection using explainable artificial intelligence. Comput. Methods Programs Biomed. 2024, 243, 107879. [Google Scholar] [CrossRef] [PubMed]
- Inglis, A.; Parnell, A. Vivid: An R package for Variable Importance and Variable Interactions Displays for Machine Learning Models. arXiv 2022, arXiv:2210.11391. [Google Scholar] [CrossRef]
- Inglis, A.; Parnell, A.; Hurley, C.B. Visualizing variable importance and variable interaction effects in machine learning models. J. Comput. Graph. Stat. 2022, 31, 766–778. [Google Scholar] [CrossRef]
- Sarti, D.A.; Prado, E.B.; Inglis, A.N.; Dos Santos, A.A.L.; Hurley, C.B.; Moral, R.A.; Parnell, A.C. Bayesian Additive Regression Trees for Genotype by Environment Interaction Models. Ann. Appl. Stat. 2023, 7, 1936–1957. [Google Scholar] [CrossRef]
- Sarti, D.A. The Statistical Paradigm: Probabilistic and Multivariate Analysis Applied Through Computational Stimulation in the Interaction Between Genotype x Environment. Ph.D. Thesis, Universidade de Sao Paulo, Sao Paulo, Brazil, 2019. [Google Scholar]
- Sarti, D.A.; Dias, C.T.S. Comparison between AMMI, W-AMMI and GGE methodology in the context of simulated data. Rev. Bras. de Biom. 2020, 38, 290–393. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.M. Random Forests; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Chipman, H.A.; George, E.I.; McCulloch, R.E. BART: Bayesian additive regression trees. Ann. Appl. Stat. 2010, 4, 266–298. [Google Scholar] [CrossRef]
- Josse, J.; van Eeuwijk, F.; Piepho, H.P.; Denis, J.B. Another look at Bayesian analysis of AMMI models for genotype-environment data. J. Agric. Biol. Environ. Stat. 2014, 19, 240–257. [Google Scholar] [CrossRef]
- Brown, D.; van den Bergh, I.; de Bruin, S.; Machida, L.; van Etten, J. Data synthesis for crop variety evaluation. A review. Agron. Sustain. Dev. 2020, 40, 25. [Google Scholar] [CrossRef]


| Location | Code |
|---|---|
| Bergamo | e1 |
| Lonigo | e2 |
| Tolentino | e3 |
| Location | GPS Coordinates of Field Trials | Code |
|---|---|---|
| Arezzo | 43.184500–11.494800 | e1 |
| Bergamo | 45.661050–9.659516 | e2 |
| Lonigo | 45.392700–11.380719 | e3 |
| Tolentino | 13.346147–43.233995 | e4 |
| Torino | 44.894023–7.878138 | e5 |
| Rank | Rank_Protein_Year1 | Rank_Protein_Year2 | Rank_Yield_Year1 | Rank_Yield_Year2 |
|---|---|---|---|---|
| 1 | Tolentino | Lonigo | Torino | Torino |
| 2 | Lonigo | Tolentino | Toentino | Tolentino |
| 3 | Bergamo | Bergamo | Lonigo | Arezzo |
| 4 | Bergamo | Lonigo | ||
| 5 | Arezzo | Bergamo |
| Genotype | Rank Prot Year 1 | Rank Prot Year 2 | Rank Yld Year 1 | Rank Yld Year 2 |
|---|---|---|---|---|
| g1 | 14 | 5 | 22 | 30 |
| g10 | 40 | 30 | 6 | 3 |
| g11 | 7 | 8 | 38 | 34 |
| g12 | 24 | 32 | 10 | 11 |
| g13 | 18 | 11 | 3 | 26 |
| g14 | 38 | 37 | 16 | 2 |
| g15 | 4 | 3 | 36 | 32 |
| g16 | 22 | 18 | 15 | 23 |
| g17 | 31 | 40 | 29 | 33 |
| g18 | 35 | 36 | 24 | 15 |
| g19 | 12 | 13 | 17 | 21 |
| g2 | 15 | 27 | 35 | 27 |
| g20 | 23 | 14 | 1 | 10 |
| g21 | 8 | 15 | 28 | 40 |
| g22 | 39 | 34 | 11 | 9 |
| g23 | 26 | 35 | 5 | 6 |
| g24 | 36 | 33 | 9 | 7 |
| g25 | 2 | 1 | 30 | 38 |
| g26 | 20 | 24 | 21 | 14 |
| g27 | 1 | 9 | 27 | 31 |
| g28 | 9 | 39 | 33 | 16 |
| g29 | 6 | 10 | 32 | 36 |
| g3 | 13 | 6 | 40 | 39 |
| g30 | 16 | 22 | 19 | 5 |
| g31 | 27 | 28 | 13 | 8 |
| g32 | 3 | 4 | 25 | 29 |
| g33 | 25 | 23 | 20 | 18 |
| g34 | 37 | 21 | 2 | 17 |
| g35 | 34 | 25 | 14 | 4 |
| g36 | 11 | 16 | 7 | 24 |
| g37 | 33 | 38 | 8 | 1 |
| g38 | 21 | 12 | 4 | 13 |
| g39 | 10 | 7 | 34 | 25 |
| g4 | 19 | 29 | 37 | 37 |
| g40 | 30 | 20 | 26 | 19 |
| g5 | 17 | 19 | 31 | 35 |
| g6 | 28 | 26 | 23 | 20 |
| g7 | 32 | 17 | 12 | 22 |
| g8 | 5 | 2 | 39 | 28 |
| g9 | 29 | 31 | 18 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarti, D.A.; Bardelli, T.; Bianchi, P.G.; Giulini, A.P.M. Enhancing Registration Offices’ Communication Through Interpretable Machine-Learning Techniques. Agronomy 2025, 15, 1603. https://doi.org/10.3390/agronomy15071603
Sarti DA, Bardelli T, Bianchi PG, Giulini APM. Enhancing Registration Offices’ Communication Through Interpretable Machine-Learning Techniques. Agronomy. 2025; 15(7):1603. https://doi.org/10.3390/agronomy15071603
Chicago/Turabian StyleSarti, Danilo Augusto, Tommaso Bardelli, Pier Giacomo Bianchi, and Anna Pia Maria Giulini. 2025. "Enhancing Registration Offices’ Communication Through Interpretable Machine-Learning Techniques" Agronomy 15, no. 7: 1603. https://doi.org/10.3390/agronomy15071603
APA StyleSarti, D. A., Bardelli, T., Bianchi, P. G., & Giulini, A. P. M. (2025). Enhancing Registration Offices’ Communication Through Interpretable Machine-Learning Techniques. Agronomy, 15(7), 1603. https://doi.org/10.3390/agronomy15071603






