The Genome Era of Forage Selection: Current Status and Future Directions for Perennial Ryegrass Breeding and Evaluation
Abstract
1. Perennial Ryegrass
2. Regional Evaluation Systems
3. Beeding Frameworks
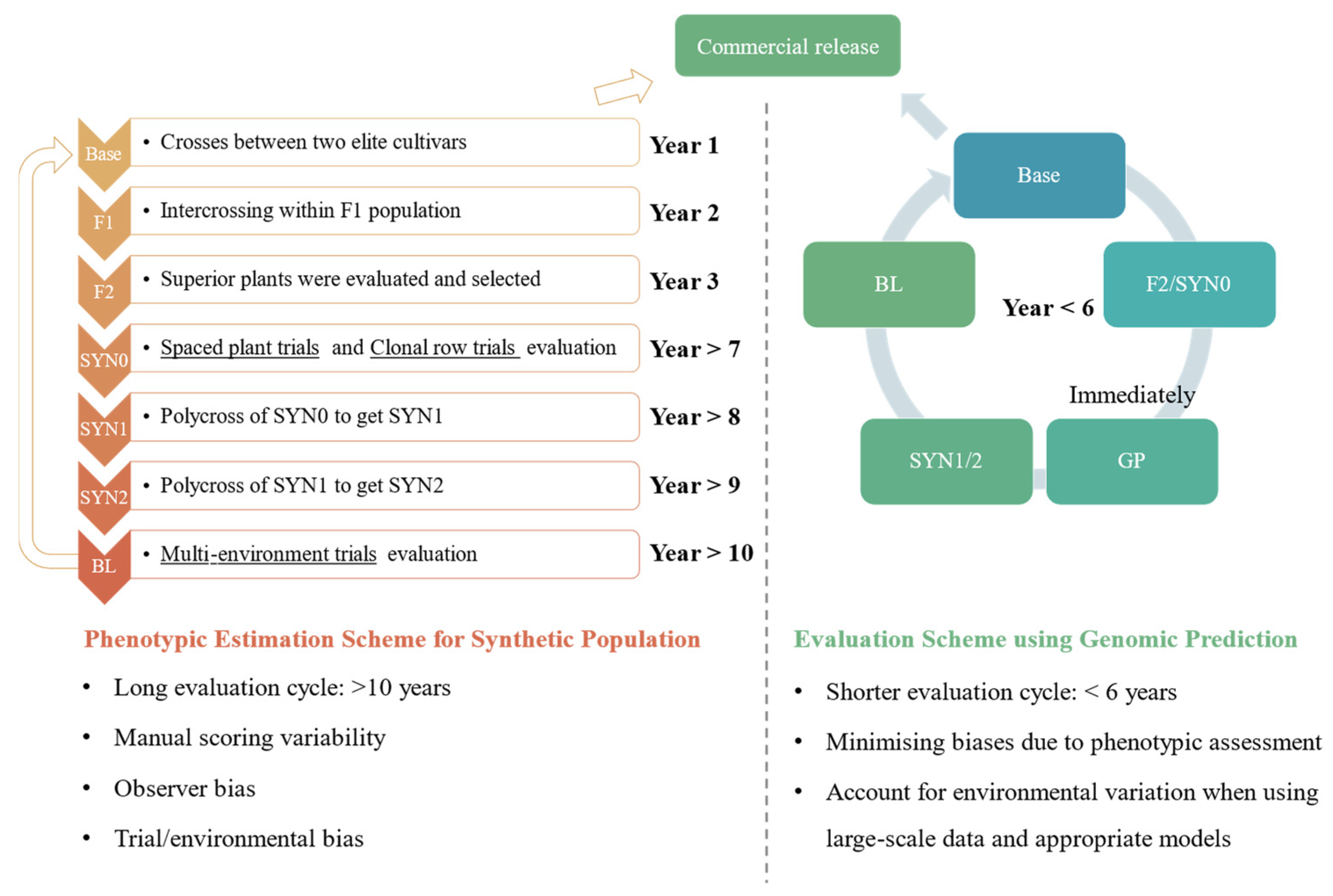
3.1. F2 Family Framework
3.2. Half-Sib Family Framework
3.3. Synthetic Population Framework
3.4. Limitation of Phenotypic Evaluation
4. Technological Advancements in Genetic-Phenotypic Association Studies
4.1. Marker-Assisted Selection
4.2. Advancements in High-Throughput Genotyping Technologies
4.3. Genomic Prediction
5. Statistical Models for Genomic Prediction
5.1. Marker-Based Models
5.2. Kernel-Based Models
5.3. Non-Parametric Models
5.4. Model Selection and Optimization Strategies
5.5. Refinement of Genomic Prediction Models
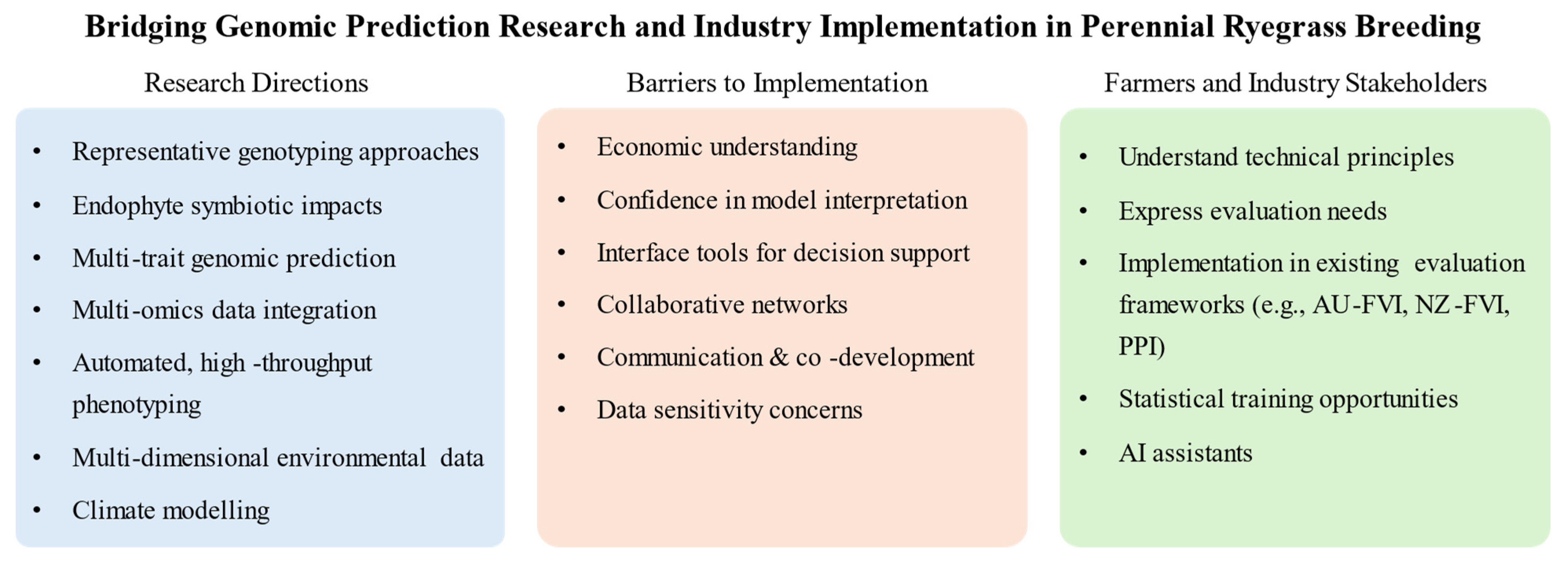
6. Research Gaps and Future Research Directions
6.1. Requirements for Representative Genotyping Approaches
6.2. Endophyte Symbiotic Impacts
6.3. Extension to Broader Agronomic Trait Profiling
6.4. Integration of Multi-Omics Data to Improve Performance Estimation
6.5. Advancement in Phenotypic Data Collection
6.6. Integrating Environmental Data and Modeling Climate Adaptation
6.7. Effective Communication and Implementation
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hannaway, D.; Fransen, S.; Cropper, J.; Teel, M.; Chaney, M.; Griggs, T.; Halse, R.; Hart, J.; Cheeke, P.; Klinger, R.; et al. Perennial Ryegrass (Lolium perenne L.). 1999. Available online: https://ir.library.oregonstate.edu/downloads/6108vb53s (accessed on 1 March 2025).
- Hunt, W.; Field, T. Growth characteristics of perennial ryegrass. J. N. Z. Grassl. 1978, 104–113. [Google Scholar] [CrossRef]
- Gilliland, T.B.T.; Hennessy, D. Opportunities and challenges for breeding perennial ryegrass cultivars with improved livestock production potential. Ir. J. Agric. Food Res. 2021, 59, 233–245. [Google Scholar] [CrossRef]
- Schubiger, F.X.; Baert, J.; Bayle, B.; Bourdon, P.; Cagas, B.; Cernoch, V.; Czembor, E.; Eickmeyer, F.; Feuerstein, U.; Hartmann, S.; et al. Susceptibility of European cultivars of Italian and perennial ryegrass to crown and stem rust. Euphytica 2010, 176, 167–181. [Google Scholar] [CrossRef]
- Leddin, C.; Giri, K.; Smith, K. Application and Analysis of a Composite Sampling Strategy to Cost-Effectively Compare Nutritive Characteristics of Perennial Ryegrass Cultivars in Field Trials. Agronomy 2020, 10, 1152. [Google Scholar] [CrossRef]
- Wilkins, P.W.; Rognli, O.A. Dry matter yield, herbage quality and persistency of equivalent populations of perennial ryegrass with and without reduced flowering. Plant Breed. 2002, 121, 425–428. [Google Scholar] [CrossRef]
- Waller, R.; Sale, P. Persistence and productivity of perennial ryegrass in sheep pastures in South-Western victoria: A review. Anim. Prod. Sci. 2001, 41, 117–144. [Google Scholar] [CrossRef]
- O’Donovan, M.; McHugh, N.; McEvoy, M.; Grogan, D.; Shalloo, L. Combining seasonal yield, silage dry matter yield, quality and persistency in an economic index to assist perennial ryegrass variety selection. J. Agric. Sci. 2016, 155, 556–568. [Google Scholar] [CrossRef]
- McEvoy, M.; O’Donovan, M.; Shalloo, L. Development and application of an economic ranking index for perennial ryegrass cultivars. J. Dairy Sci. 2011, 94, 1627–1639. [Google Scholar] [CrossRef]
- Leddin, C.; Jacobs, J.; Smith, K.; Giri, K.; Malcolm, B.; Ho, C. Development of a system to rank perennial ryegrass cultivars according to their economic value to dairy farm businesses in south-eastern Australia. Anim. Prod. Sci. 2018, 58, 1552–1558. [Google Scholar] [CrossRef]
- Chapman, D.F.; Bryant, J.R.; Olayemi, M.E.; Edwards, G.R.; Thorrold, B.S.; McMillan, W.H.; Kerr, G.A.; Judson, G.; Cookson, T.; Moorhead, A.; et al. An economically based evaluation index for perennial and short-term ryegrasses in New Zealand dairy farm systems. Grass Forage Sci. 2017, 72, 1–21. [Google Scholar] [CrossRef]
- Lewis, C.D.; Smith, K.F.; Jacobs, J.L.; Ho, C.K.M.; Leddin, C.M.; Moate, P.J.; Malcolm, B. Using a two-price market value framework to value differences in metabolizable energy concentration of pasture across seasons. Agric. Syst. 2024, 217, 103939. [Google Scholar] [CrossRef]
- Leddin, C.; Giri, K.; Smith, K. Variation in the Nutritive Characteristics of Modern Perennial Ryegrass Cultivars in South-Eastern Australian Dairy Environments and Prospects for Inclusion in the Australian Forage Value Index (FVI). Agronomy 2022, 12, 136. [Google Scholar] [CrossRef]
- Malmberg, M.; Smith, C.; Thakur, P.; Drayton, M.; Wilson, J.; Shinozuka, M.; Clayton, W.; Inch, C.; Spangenberg, G.; Smith, K.; et al. Developing an integrated genomic selection approach beyond biomass for varietal protection and nutritive traits in perennial ryegrass (Lolium perenne L.). Theor. Appl. Genet. 2023, 136, 44. [Google Scholar] [CrossRef] [PubMed]
- Arojju, S.K.; Cao, M.; Zulfi Jahufer, M.Z.; Barrett, B.A.; Faville, M.J. Genomic Predictive Ability for Foliar Nutritive Traits in Perennial Ryegrass. G3 Genes|Genomes|Genet. 2020, 10, 695–708. [Google Scholar] [CrossRef]
- Arojju, S.K.; Cao, M.; Trolove, M.; Barrett, B.A.; Inch, C.; Eady, C.; Stewart, A.; Faville, M.J. Multi-Trait Genomic Prediction Improves Predictive Ability for Dry Matter Yield and Water-Soluble Carbohydrates in Perennial Ryegrass. Front. Plant Sci. 2020, 11, 1197. [Google Scholar] [CrossRef]
- Rivero, M.; Oscar, B.; Fabián, L.N.; Siebald, J.A. Grazing Preference of Dairy Cows and Pasture Productivity for Different Cultivars of Perennial Ryegrass under Contrasting Managements. Animals 2019, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Rouquettes, F., Jr.; Bransby, D.; Riewe, M.E. Grazing management and use of ryegrass. Ecol. Prod. Manag. Lolium Forage USA 1997, 24, 79–99. [Google Scholar]
- Bornhofen, E.; Fè, D.; Lenk, I.; Greve, M.; Didion, T.; Jensen, C.S.; Asp, T.; Janss, L. Leveraging spatiotemporal genomic breeding value estimates of dry matter yield and herbage quality in ryegrass via random regression models. Plant Genome 2022, 15, e20255. [Google Scholar] [CrossRef]
- Grinberg, N.F.; Lovatt, A.; Hegarty, M.; Lovatt, A.; Skøt, K.P.; Kelly, R.; Blackmore, T.; Thorogood, D.; King, R.D.; Armstead, I.; et al. Implementation of Genomic Prediction in Lolium perenne (L.) Breeding Populations. Front. Plant Sci. 2016, 7, 133. [Google Scholar] [CrossRef]
- DairyAustralia. Perennial Ryegrass|Dairy Australia. 2025. Available online: https://www.dairyaustralia.com.au/ (accessed on 1 March 2025).
- Giri, K.; Chia, K.; Chandra, S.; Smith, K.F.; Leddin, C.M.; Ho, C.K.M.; Jacobs, J.L. Modelling and prediction of dry matter yield of perennial ryegrass cultivars sown in multi-environment multi-harvest trials in south-eastern Australia. Field Crops Res. 2019, 243, 107614. [Google Scholar] [CrossRef]
- Lewis, C.D.; Smith, K.F.; Jacobs, J.L.; Ho, C.K.M.; Leddin, C.M.; Malcolm, B. Using a two-price market value method to value extra pasture DM in different seasons. Agric. Syst. 2020, 178, 102729. [Google Scholar] [CrossRef]
- Chapman, D.F.; Edwards, G.R.; Stewart, A.V.; McEvoy, M.; O’Donovan, M.; Waghorn, G.C. Valuing forages for genetic selection: What traits should we focus on? Anim. Prod. Sci. 2015, 55, 869–882. [Google Scholar] [CrossRef]
- Crush, J.R.; Woodward, S.L.; Eerens, J.P.J.; MacDonald, K.A. Growth and milk solids production in pastures of older and more recent ryegrass and white clover cultivars under dairy grazing. N. Z. J. Agric. Res. 2006, 49, 119–135. [Google Scholar] [CrossRef]
- Fè, D.; Cericola, F.; Byrne, S.; Lenk, I.; Ashraf, B.; Pedersen, M.G.; Roulund, N.; Asp, T.; Janss, L.; Jensen, C.S.; et al. Genomic Dissection and Prediction of Heading Date in Perennial Ryegrass. BMC Genom. 2015, 16, 921. [Google Scholar] [CrossRef]
- Fè, D.; Ashraf, B.H.; Pedersen, M.G.; Janss, L.; Byrne, S.; Roulund, N.; Lenk, I.; Didion, T.; Asp, T.; Jensen, C.S.; et al. Accuracy of Genomic Prediction in a Commercial Perennial Ryegrass Breeding Program. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cericola, F.; Lenk, I.; Fè, D.; Byrne, S.; Jensen, C.S.; Pedersen, M.G.; Asp, T.; Jensen, J.; Janss, L. Optimized Use of Low-Depth Genotyping-by-Sequencing for Genomic Prediction Among Multi-Parental Family Pools and Single Plants in Perennial Ryegrass (Lolium perenne L.). Front. Plant Sci. 2018, 9, 369. [Google Scholar] [CrossRef]
- Faville, M.J.; Ganesh, S.; Cao, M.; Jahufer, M.Z.Z.; Bilton, T.P.; Easton, H.S.; Ryan, D.L.; Trethewey, J.A.K.; Rolston, M.P.; Griffiths, A.G.; et al. Predictive ability of genomic selection models in a multi-population perennial ryegrass training set using genotyping-by-sequencing. Theor. Appl. Genet. 2018, 131, 703–720. [Google Scholar] [CrossRef]
- Jahufer, M.Z.Z.; Arojju, S.K.; Faville, M.J.; Ghamkhar, K.; Luo, D.; Arief, V.; Yang, W.-H.; Sun, M.; DeLacy, I.H.; Griffiths, A.G.; et al. Deterministic and stochastic modelling of impacts from genomic selection and phenomics on genetic gain for perennial ryegrass dry matter yield. Sci. Rep. 2021, 11, 13265. [Google Scholar] [CrossRef]
- Faville, M.J.; Ganesh, S.; Moraga, R.; Easton, H.S.; Jahufer, M.Z.Z.; Elshire, R.E.; Asp, T.; Barrett, B.A. Development of Genomic Selection for Perennial Ryegrass. In Breeding in a World of Scarcity; Springer International Publishing: Cham, Switzerland, 2016; pp. 139–143. [Google Scholar]
- Lin, Z.; Cogan, N.O.I.; Pembleton, L.W.; Spangenberg, G.C.; Forster, J.W.; Hayes, B.J.; Daetwyler, H.D. Genetic Gain and Inbreeding from Genomic Selection in a Simulated Commercial Breeding Program for Perennial Ryegrass. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Hayes, B.J.; Cogan, N.O.I.; Pembleton, L.W.; Goddard, M.E.; Wang, J.; Spangenberg, G.C.; Forster, J.W. Prospects for genomic selection in forage plant species. Plant Breed. 2013, 132, 133–143. [Google Scholar] [CrossRef]
- Conaghan, P.; Casler, M.D. A theoretical and practical analysis of the optimum breeding system for perennial ryegrass. Ir. J. Agric. Food Res. 2011, 50, 47–63. [Google Scholar]
- Casler, M.D.; Brummer, E.C. Theoretical Expected Genetic Gains for Among-and-Within-Family Selection Methods in Perennial Forage Crops. Crop Sci. 2008, 48, 890–902. [Google Scholar] [CrossRef]
- Barre, P.; Asp, T.; Byrne, S.; Casler, M.; Faville, M.; Rognli, O.A.; Roldan-Ruiz, I.; Skøt, L.; Ghesquière, M. Genomic Prediction of Complex TraitsComplex traits in Forage Plants Species: Perennial Grasses Case. In Genomic Prediction of Complex Traits: Methods and Protocols; Ahmadi, N., Bartholomé, J., Eds.; Springer: New York, NY, USA, 2022; pp. 521–541. [Google Scholar] [CrossRef]
- Zhu, J.; Giri, K.; Cogan, N.O.; Smith, K.F.; Jacobs, J.L. Genotype-by-environment interaction analysis of dry matter yield of perennial ryegrass cultivars across south-eastern Australia using factor analytic models. Field Crops Res. 2023, 303, 109143. [Google Scholar] [CrossRef]
- Zhu, J.; Giri, K.; Lin, Z.; Cogan, N.O.; Jacobs, J.L.; Smith, K.F. Estimation of ryegrass (Lolium) dry matter yield using genomic prediction considering genotype by environment interaction across south-eastern Australia. Front. Plant Sci. 2025, 16, 1579376. [Google Scholar] [CrossRef]
- Zhu, J. Utilising Genomic Relationships During the Estimation of Perennial Ryegrass Performance; The University of Melbourne: Parkville, VIC, Australia, 2025. [Google Scholar]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Unêda-Trevisoli, S.H.; da Silva, F.M.; Di Mauro, A.O. Marker-assisted selection and genomic selection. In Soybean Breeding, 1st ed.; da Silva, F.L., Borém, A., Sediyama, T., Ludke, W.H., Eds.; Springer: Cham, Switzerland, 2017; pp. 275–291. [Google Scholar] [CrossRef]
- Sim, S.; Diesburg, K.; Casler, M.; Jung, G. Mapping and Comparative Analysis of QTL for Crown Rust Resistance in an Italian × Perennial Ryegrass Population. Phytopathology 2007, 97, 767–776. [Google Scholar] [CrossRef]
- Schejbel, B.; Jensen, L.B.; Xing, Y.; Lübberstedt, T. QTL analysis of crown rust resistance in perennial ryegrass under conditions of natural and artificial infection. Plant Breed. 2007, 126, 347–352. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Paina, C.; Byrne, S.; Studer, B.; Rognli, O.A.; Asp, T. Using a Candidate Gene-Based Genetic Linkage Map to Identify QTL for Winter Survival in Perennial Ryegrass. PLoS ONE 2016, 11, e0152004. [Google Scholar] [CrossRef]
- Emebiri, L.C.; Moody, D.B. Heritable basis for some genotype–environment stability statistics: Inferences from QTL analysis of heading date in two-rowed barley. Field Crops Res. 2006, 96, 243–251. [Google Scholar] [CrossRef]
- Keep, T.; Sampoux, J.-P.; Blanco-Pastor, J.L.; Dehmer, K.J.; Hegarty, M.J.; Ledauphin, T.; Litrico, I.; Muylle, H.; Roldán-Ruiz, I.; Roschanski, A.M.; et al. High-Throughput Genome-Wide Genotyping To Optimize the Use of Natural Genetic Resources in the Grassland Species Perennial Ryegrass (Lolium perenne L.). G3 Genes|Genomes|Genet. 2020, 10, 3347–3364. [Google Scholar] [CrossRef] [PubMed]
- Jaškūnė, K.; Aleliūnas, A.; Statkevičiūtė, G.; Kemešytė, V.; Studer, B.; Yates, S. Genome-Wide Association Study to Identify Candidate Loci for Biomass Formation Under Water Deficit in Perennial Ryegrass. Front. Plant Sci. 2020, 11, 570204. [Google Scholar] [CrossRef] [PubMed]
- Fois, M.; Bellucci, A.; Malinowska, M.; Greve, M.; Ruud, A.K.; Asp, T. Genome-wide association mapping of crown and brown rust resistance in perennial Ryegrass. Genes 2021, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- de Souza, N. Get out the map. Nat. Rev. Genet. 2007, 8, S10. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Niklas, A.; Olszewska, D. Application of the RAPD technique to identify genetic diversity in cultivated forms of Capsicum annuum L. Biotechnologia 2021, 102, 209–223. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Stewart, C.N., Jr. Functional Markers for Precision Plant Breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef]
- Gonçalves-Vidigal, M.C.; Rubiano, L.B. Development and application of microsatellites in plant breeding. Crop Breed. Appl. Biotechnol. 2011, 11, 66–72. [Google Scholar] [CrossRef]
- Birrer, M.; Kölliker, R.; Manzanares, C.; Asp, T.; Studer, B. A DNA Marker Assay Based on High-Resolution Melting Curve Analysis for Distinguishing Species of the Festuca–Lolium Complex. Mol. Breed. 2014, 34, 421–429. [Google Scholar] [CrossRef]
- Cogan, N.O.I.; Ponting, R.C.; Vecchies, A.C.; Drayton, M.C.; George, J.; Dracatos, P.M.; Dobrowolski, M.P.; Sawbridge, T.I.; Smith, K.F.; Spangenberg, G.C.; et al. Gene-associated single nucleotide polymorphism discovery in perennial ryegrass (Lolium perenne L.). Mol. Genet. Genom. 2006, 276, 101–112. [Google Scholar] [CrossRef]
- Byrne, S.L.; Conaghan, P.; Barth, S.; Arojju, S.K.; Casler, M.; Michel, T.; Velmurugan, J.; Milbourne, D. Using variable importance measures to identify a small set of SNPs to predict heading date in perennial ryegrass. Sci. Rep. 2017, 7, 3566. [Google Scholar] [CrossRef]
- Degen, B.; Müller, N.A. A simulation study comparing advanced marker-assisted selection with genomic selection in tree breeding programs. G3 Genes|Genomes|Genet. 2023, 13, jkad164. [Google Scholar] [CrossRef] [PubMed]
- Kushanov, F.N.; Turaev, O.S.; Ernazarova, D.K.; Gapparov, B.M.; Oripova, B.B.; Kudratova, M.K.; Rafieva, F.U.; Khalikov, K.K.; Erjigitov, D.S.; Khidirov, M.T.; et al. Genetic Diversity, QTL Mapping, and Marker-Assisted Selection Technology in Cotton (Gossypium spp.). Front. Plant Sci. 2021, 12, 779386. [Google Scholar] [CrossRef]
- Ebdon, J.S.; Gauch, H. Additive Main Effect and Multiplicative Interaction Analysis of National Turfgrass Performance Trials: I. Interpretation of Genotype × Environment Interaction. Crop Sci. 2002, 42, 489–496. [Google Scholar] [CrossRef]
- Fois, M.; Malinowska, M.; Schubiger, F.X.; Asp, T. Genomic Prediction and Genotype-by-Environment Interaction Analysis of Crown and Stem Rust in Ryegrasses in European Multi-Site Trials. Agronomy 2021, 11, 1119. [Google Scholar] [CrossRef]
- Conaghan, P.; Casler, M.D.; McGilloway, D.A.; O’Kiely, P.; Dowley, L.J. Genotype × environment interactions for herbage yield of perennial ryegrass sward plots in Ireland. Grass Forage Sci. 2008, 63, 107–120. [Google Scholar] [CrossRef]
- Isik, F.; Holland, J.; Maltecca, C. Genomic Relationships and GBLUP. In Genetic Data Analysis for Plant and Animal Breeding; Isik, F., Holland, J., Maltecca, C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 311–354. [Google Scholar] [CrossRef]
- Wang, J.; Pembleton, L.W.; Baillie, R.C.; Drayton, M.C.; Hand, M.L.; Bain, M.; Sawbridge, T.I.; Spangenberg, G.C.; Forster, J.W.; Cogan, N.O.I. Development and implementation of a multiplexed single nucleotide polymorphism genotyping tool for differentiation of ryegrass species and cultivars. Mol. Breed. 2014, 33, 435–451. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Drayton, M.C.; Bain, M.; Baillie, R.C.; Inch, C.; Spangenberg, G.C.; Wang, J.; Forster, J.W.; Cogan, N.O. Targeted genotyping-by-sequencing permits cost-effective identification and discrimination of pasture grass species and cultivars. Theor. Appl. Genet. 2016, 129, 991–1005. [Google Scholar] [CrossRef]
- Malmberg, M.M.; Pembleton, L.W.; Baillie, R.C.; Drayton, M.C.; Sudheesh, S.; Kaur, S.; Shinozuka, H.; Verma, P.; Spangenberg, G.C.; Daetwyler, H.D.; et al. Genotyping-by-sequencing through transcriptomics: Implementation in a range of crop species with varying reproductive habits and ploidy levels. Plant Biotechnol. J. 2018, 16, 877–889. [Google Scholar] [CrossRef]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef]
- Arojju, S.K.; Conaghan, P.; Barth, S.; Milbourne, D.; Casler, M.D.; Hodkinson, T.R.; Michel, T.; Byrne, S.L. Genomic prediction of crown rust resistance in Lolium perenne. BMC Genet. 2018, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Genomic selection: A paradigm shift in animal breeding. Anim. Front. 2016, 6, 6–14. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D.C.; et al. Genomic Selection: A Tool for Accelerating the Efficiency of Molecular Breeding for Development of Climate-Resilient Crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.; Åstrand, J.; Montesinos-López, O.; Sánchez, J.; Fernández-Gónzalez, J.; Tadesse, W.; Vetukuri, R.; Carlsson, A.; Ceplitis, A.; Crossa, J.; et al. Genomic selection in plant breeding: Key factors shaping two decades of progress. Mol. Plant 2024, 17, 552–578. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.R.; Carrillo, J.A. Genomic selection in United States dairy cattle. Front Genet 2022, 13, 994466. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef]
- Chen, Y.; Xiong, Y.; Hong, H.; Li, G.; Gao, J.; Guo, Q.; Sun, R.; Ren, H.; Zhang, F.; Wang, J.; et al. Genetic dissection of and genomic selection for seed weight, pod length, and pod width in soybean. Crop J. 2023, 11, 832–841. [Google Scholar] [CrossRef]
- Shahi, D.; Guo, J.; Pradhan, S.; Khan, J.; Avci, M.; Khan, N.; McBreen, J.; Bai, G.; Reynolds, M.; Foulkes, J.; et al. Multi-trait genomic prediction using in-season physiological parameters increases prediction accuracy of complex traits in US wheat. BMC Genom. 2022, 23, 298. [Google Scholar] [CrossRef]
- Lu, F.; Lipka, A.E.; Glaubitz, J.; Elshire, R.; Cherney, J.H.; Casler, M.D.; Buckler, E.S.; Costich, D.E. Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PLoS Genet. 2013, 9, e1003215. [Google Scholar] [CrossRef]
- Guo, X.; Cericola, F.; Fè, D.; Pedersen, M.G.; Lenk, I.; Jensen, C.S.; Jensen, J.; Janss, L.L. Genomic Prediction in Tetraploid Ryegrass Using Allele Frequencies Based on Genotyping by Sequencing. Front. Plant Sci. 2018, 9, 1165. [Google Scholar] [CrossRef] [PubMed]
- Konkolewska, A.; Phang, S.; Conaghan, P.; Milbourne, D.; Lawlor, A.; Byrne, S. Genomic prediction of seasonal forage yield in perennial ryegrass. Grassl. Res. 2023, 2, 167–181. [Google Scholar] [CrossRef]
- Henderson, C.R. Best linear unbiased estimation and prediction under a selection model. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Faville, M.; Schmidt, J.; Trolove, M.; Moran, P.; Hong, W.; Cao, M.; Ganesh, S.; George, R.; Barrett, B. Empirical assessment of a genomic breeding strategy in perennial ryegrass. J. N. Z. Grassl. 2021, 83, 115–122. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, J.; Cogan, N.O.; Pembleton, L.W.; Badenhorst, P.; Forster, J.W.; Spangenberg, G.C.; Hayes, B.J.; Daetwyler, H.D. Optimizing resource allocation in a genomic breeding program for perennial ryegrass to balance genetic gain, cost, and inbreeding. Crop Sci. 2017, 57, 243–252. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Inch, C.; Baillie, R.C.; Drayton, M.C.; Thakur, P.; Ogaji, Y.O.; Spangenberg, G.C.; Forster, J.W.; Daetwyler, H.D.; Cogan, N.O.I. Exploitation of data from breeding programs supports rapid implementation of genomic selection for key agronomic traits in perennial ryegrass. Theor. Appl. Genet. 2018, 131, 1891–1902. [Google Scholar] [CrossRef]
- Lourenço, V.M.; Ogutu, J.O.; Rodrigues, R.A.P.; Posekany, A.; Piepho, H.P. Genomic prediction using machine learning: A comparison of the performance of regularized regression, ensemble, instance-based and deep learning methods on synthetic and empirical data. BMC Genom. 2024, 25, 152. [Google Scholar] [CrossRef]
- Esfandyari, H.; Fè, D.; Tessema, B.B.; Janss, L.L.G.; Jensen, J. Effects of Different Strategies for Exploiting Genomic Selection in Perennial Ryegrass Breeding Programs. G3 Genes|Genome|Genet. 2020, 10, 3783–3795. [Google Scholar] [CrossRef]
- Robinson, G.K. That BLUP is a Good Thing: The Estimation of Random Effects. Stat. Sci. 1991, 6, 15–32. [Google Scholar] [CrossRef]
- Gianola, D.; van Kaam, J.B. Reproducing kernel hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 2008, 178, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Runcie, D.; Cheng, H. Pitfalls and Remedies for Cross Validation with Multi-trait Genomic Prediction Methods. G3 (Bethesda Md.) 2019, 9, 3727–3741. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Li, X.; Fang, L.; Liu, A.; Su, G.; Zhang, Y.; Luobu, B.; Ding, X.; Zhang, S. Genomic Prediction Using Bayesian Regression Models With Global–Local Prior. Front. Genet. 2021, 12, 628205. [Google Scholar] [CrossRef]
- Zhu, S.; Guo, T.; Yuan, C.; Liu, J.; Li, J.; Han, M.; Zhao, H.; Wu, Y.; Sun, W.; Wang, X.; et al. Evaluation of Bayesian alphabet and GBLUP based on different marker density for genomic prediction in Alpine Merino sheep. G3 Genes|Genomes|Genet. 2021, 11, jkab206. [Google Scholar] [CrossRef] [PubMed]
- Schrauf, M.F.; de Los Campos, G.; Munilla, S. Comparing Genomic Prediction Models by Means of Cross Validation. Front. Plant Sci. 2021, 12, 734512. [Google Scholar] [CrossRef]
- Resende, M.F., Jr.; Muñoz, P.; Resende, M.D.; Garrick, D.J.; Fernando, R.L.; Davis, J.M.; Jokela, E.J.; Martin, T.A.; Peter, G.F.; Kirst, M. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 2012, 190, 1503–1510. [Google Scholar] [CrossRef]
- Werner, C.R.; Gaynor, R.C.; Gorjanc, G.; Hickey, J.M.; Kox, T.; Abbadi, A.; Leckband, G.; Snowdon, R.J.; Stahl, A. How Population Structure Impacts Genomic Selection Accuracy in Cross-Validation: Implications for Practical Breeding. Front. Plant Sci. 2020, 11, 592977. [Google Scholar] [CrossRef]
- Guan, X.; Yuyama, N.; Stewart, A.; Ding, C.; Xu, N.; Kiyoshi, T.; Cai, H. Genetic Diversity and Structure of Lolium Species Surveyed on Nuclear Simple Sequence Repeat and Cytoplasmic Markers. Front. Plant Sci. 2017, 8, 584. [Google Scholar] [CrossRef]
- Blackmore, T.; Thorogood, D.; Skøt, L.; McMahon, R.; Powell, W.; Hegarty, M. Germplasm dynamics: The role of ecotypic diversity in shaping the patterns of genetic variation in Lolium perenne. Sci. Rep. 2016, 6, 22603. [Google Scholar] [CrossRef]
- Zhu, J.; Malmberg, M.M.; Shinozuka, M.; Retegan, R.M.; Cogan, N.O.; Jacobs, J.L.; Giri, K.; Smith, K.F. Machine learning solutions for integrating partially overlapping genetic datasets and modelling host–endophyte effects in ryegrass (Lolium) dry matter yield estimation. Front. Plant Sci. 2025, 16, 1543956. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Kemper, K.E.; van der Werf, J.H.J.; Hayes, B.J. Components of the accuracy of genomic prediction in a multi-breed sheep population. J. Anim. Sci. 2012, 90, 3375–3384. [Google Scholar] [CrossRef] [PubMed]
- Daetwyler, H.D.; Villanueva, B.; Woolliams, J.A. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 2008, 3, e3395. [Google Scholar] [CrossRef] [PubMed]
- Daetwyler, H.D.; Pong-Wong, R.; Villanueva, B.; Woolliams, J.A. The impact of genetic architecture on genome-wide evaluation methods. Genetics 2010, 185, 1021–1031. [Google Scholar] [CrossRef]
- Wientjes, Y.C.J.; Veerkamp, R.F.; Bijma, P.; Bovenhuis, H.; Schrooten, C.; Calus, M.P.L. Empirical and deterministic accuracies of across-population genomic prediction. Genet. Sel. Evol. 2015, 47, 5. [Google Scholar] [CrossRef]
- Brard, S.; Ricard, A. Is the use of formulae a reliable way to predict the accuracy of genomic selection? J. Anim. Breed. Genet. 2015, 132, 207–217. [Google Scholar] [CrossRef]
- Newell, M.A.; Jannink, J.L. Genomic Selection in Plant Breeding. In Crop Breeding; Methods in Molecular Biology; Fleury, D., Whitford, R., Eds.; Humana Press: New York, NY, USA, 2014; Volume 1145, pp. 117–130. [Google Scholar] [CrossRef]
- Barrett, B.; Jahufer, Z.; Arojju, S.; Sise, J.; Faville, M. Forecasting the genetic and economic impacts of genomic selection in perennial ryegrass. J. N. Z. Grassl. 2022, 83, 92–98. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Dekkers, J.C.M. The Impact of Genetic Relationship Information on Genome-Assisted Breeding Values. Genetics 2007, 177, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M. Genomic selection: Prediction of accuracy and maximisation of long term response. Genetica 2008, 136, 245–257. [Google Scholar] [CrossRef]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef]
- Vassiliadis, S.; Reddy, P.; Hemsworth, J.; Spangenberg, G.C.; Guthridge, K.M.; Rochfort, S.J. Quantitation and Distribution of Epichloë-Derived Alkaloids in Perennial Ryegrass Tissues. Metabolites 2023, 13, 205. [Google Scholar] [CrossRef]
- Popay, A.J.; Hume, D.E. Endophytes for Improving Ryegrass Performance: Current Status and Future Possibilities. In Proceedings of the 22nd International Grassland Congress, Sydney, Australia, 15–19 September 2013; pp. 1625–1626. [Google Scholar]
- Eady, C. The Impact of Alkaloid-Producing Epichloë Endophyte on Forage Ryegrass Breeding: A New Zealand Perspective. Toxins 2021, 13, 158. [Google Scholar] [CrossRef] [PubMed]
- Karpyn Esqueda, M.; Yen, A.L.; Rochfort, S.; Guthridge, K.M.; Powell, K.S.; Edwards, J.; Spangenberg, G.C. A Review of Perennial Ryegrass Endophytes and Their Potential Use in the Management of African Black Beetle in Perennial Grazing Systems in Australia. Front. Plant Sci. 2017, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, S.-m.; Jiang, Y. Genotypic Variations in Plant Growth and Nutritional Elements of Perennial Ryegrass Accessions under Salinity Stress. J. Am. Soc. Hortic. Sci. 2017, 142, 476–483. [Google Scholar] [CrossRef]
- Miao, C.; Zhang, Y.; Bai, X.; Qin, T. Insights into the Response of Perennial Ryegrass to Abiotic Stress: Underlying Survival Strategies and Adaptation Mechanisms. Life 2022, 12, 860. [Google Scholar] [CrossRef] [PubMed]
- Kemesyte, V.; Statkeviciute, G.; Brazauskas, G. Perennial Ryegrass Yield Performance under Abiotic Stress. Crop Sci. 2017, 57, 1935–1940. [Google Scholar] [CrossRef]
- Fuchun, X.; Rahul, D.; Dong, Q. Plant Growth and Morphophysiological Modifications in Perennial Ryegrass under Environmental Stress. In Abiotic Stress in Plants; Shah, F., Shah, S., Yajun, C., Chao, W., Depeng, W., Eds.; IntechOpen: Rijeka, Croatia, 2020; p. Ch. 17. [Google Scholar] [CrossRef]
- Nagy, I.; Veeckman, E.; Liu, C.; Bel, M.V.; Vandepoele, K.; Jensen, C.S.; Ruttink, T.; Asp, T. Chromosome-scale assembly and annotation of the perennial ryegrass genome. BMC Genom. 2022, 23, 505. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Chen, W.; Zhu, J.; Chen, Y.; Li, D. Transcriptomic analysis of the metabolic regulatory mechanism of Schizochytrium limacinum B4D1 using sodium acetate to produce DHA. Biochem. Eng. J. 2023, 197, 108963. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.; Chai, X.; Liu, W.; Wang, Y.; Yang, Q.; Wang, Z.; Liu, Z. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef]
- Jinqiu, Y.; Bing, L.; Tingting, S.; Jinglei, H.; Zelai, K.; Lu, L.; Wenhua, H.; Tao, H.; Xinyu, H.; Zengqing, L.; et al. Integrated Physiological and Transcriptomic Analyses Responses to Altitude Stress in Oat (Avena sativa L.). Front. Genet. 2021, 12, 638683. [Google Scholar] [CrossRef]
- Varoquaux, N.; Cole, B.; Gao, C.; Pierroz, G.; Baker, C.R.; Patel, D.; Madera, M.; Jeffers, T.; Hollingsworth, J.; Sievert, J.; et al. Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. USA 2019, 116, 27124–27132. [Google Scholar] [CrossRef]
- Lu, A.; Zeng, S.; Pi, K.; Long, B.; Mo, Z.; Liu, R. Transcriptome analysis reveals the key role of overdominant expression of photosynthetic and respiration-related genes in the formation of tobacco (Nicotiana tabacum L.) biomass heterosis. BMC Genom. 2024, 25, 598. [Google Scholar] [CrossRef]
- Faizi, M.; Steuer, R. Optimal proteome allocation strategies for phototrophic growth in a light-limited chemostat. Microb. Cell Factories 2019, 18, 165. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Xiao, Y.; Ming, Y.; Peng, R.; Hu, J.; Wang, H.-B.; Jin, H.-L. Quantitative proteomics reveals redox-based functional regulation of photosynthesis under fluctuating light in plants. J. Integr. Plant Biol. 2022, 64, 2168–2186. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, X.; Wu, S.; Zhao, M.; Hu, Z. Transcriptomic and metabolomic analyses revealed regulation mechanism of mixotrophic Cylindrotheca sp. glycerol utilization and biomass promotion. Biotechnol. Biofuels Bioprod. 2023, 16, 84. [Google Scholar] [CrossRef]
- Gao, B.; Wang, F.; Huang, L.; Liu, H.; Zhong, Y.; Zhang, C. Biomass, lipid accumulation kinetics, and the transcriptome of heterotrophic oleaginous microalga Tetradesmus bernardii under different carbon and nitrogen sources. Biotechnol. Biofuels 2021, 14, 4. [Google Scholar] [CrossRef]
- Pranga, J.; Borra-Serrano, I.; Aper, J.; De Swaef, T.; Ghesquiere, A.; Quataert, P.; Roldán-Ruiz, I.; Janssens, I.A.; Ruysschaert, G.; Lootens, P. Improving Accuracy of Herbage Yield Predictions in Perennial Ryegrass with UAV-Based Structural and Spectral Data Fusion and Machine Learning. Remote Sens. 2021, 13, 3459. [Google Scholar] [CrossRef]
- Tanaka, T.S.T.; Wang, S.; Jørgensen, J.R.; Gentili, M.; Vidal, A.Z.; Mortensen, A.K.; Acharya, B.S.; Beck, B.D.; Gislum, R. Review of Crop Phenotyping in Field Plot Experiments Using UAV-Mounted Sensors and Algorithms. Drones 2024, 8, 212. [Google Scholar] [CrossRef]
- Gebremedhin, A.; Badenhorst, P.; Wang, J.; Shi, F.; Breen, E.; Giri, K.; Spangenberg, G.C.; Smith, K. Development and Validation of a Phenotyping Computational Workflow to Predict the Biomass Yield of a Large Perennial Ryegrass Breeding Field Trial. Front. Plant Sci. 2020, 11, 689. [Google Scholar] [CrossRef]
- Wang, J.; Badenhorst, P.; Phelan, A.; Pembleton, L.; Shi, F.; Cogan, N.; Spangenberg, G.; Smith, K. Using Sensors and Unmanned Aircraft Systems for High-Throughput Phenotyping of Biomass in Perennial Ryegrass Breeding Trials. Front. Plant Sci. 2019, 10, 1381. [Google Scholar] [CrossRef]
- Parasurama, S.; Banan, D.; Yun, K.; Doty, S.; Kim, S.-H. Bridging Time-series Image Phenotyping and Functional–Structural Plant Modeling to Predict Adventitious Root System Architecture. Plant Phenomics 2023, 5, 0127. [Google Scholar] [CrossRef]
- Wang, H.; Singh, K.D.; Poudel, H.P.; Natarajan, M.; Ravichandran, P.; Eisenreich, B. Forage Height and Above-Ground Biomass Estimation by Comparing UAV-Based Multispectral and RGB Imagery. Sensors 2024, 24, 5794. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, M.M.; Chen, D.; Gillani, Z.; Klukas, C.; Chen, M. Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front. Plant Sci. 2015, 6, 619. [Google Scholar] [CrossRef] [PubMed]
- Ludovisi, R.; Tauro, F.; Salvati, R.; Khoury, S.; Mugnozza Scarascia, G.; Harfouche, A. UAV-Based Thermal Imaging for High-Throughput Field Phenotyping of Black Poplar Response to Drought. Front. Plant Sci. 2017, 8, 1681. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Shi, F.; Wang, J.; Badenhorst, P.E.; Spangenberg, G.C.; Smith, K.F.; Daetwyler, H.D. Within and combined season prediction models for perennial ryegrass biomass yield using ground- and air-based sensor data. Front. Plant Sci. 2022, 13, 950720. [Google Scholar] [CrossRef]
- Wedderburn, M.E.; Crush, J.R.; Pengelly, W.J.; Walcroft, J.L. Root growth patterns of perennial ryegrasses under well-watered and drought conditions. N. Z. J. Agric. Res. 2010, 53, 377–388. [Google Scholar] [CrossRef]
- Colas, V.; Barre, P.; van Parijs, F.; Wolters, L.; Quitté, Y.; Ruttink, T.; Roldán-Ruiz, I.; Escobar Gutiérrez, A.J.; Muylle, H. Seasonal differences in structural and genetic control of digestibility in perennial ryegrass. Front. Plant Sci. 2022, 12, 801145. [Google Scholar] [CrossRef]
- Piepho, H.-P.; Eckl, T. Analysis of series of variety trials with perennial crops. Grass Forage Sci. 2014, 69, 431–440. [Google Scholar] [CrossRef]
- Förster, L.; Grant, J.; Michel, T.; Ng, C.; Barth, S. Growth under cold conditions in a wide perennial ryegrass panel is under tight physiological control. PeerJ 2018, 6, e5520. [Google Scholar] [CrossRef]
- Waldron, B.; Asay, K.; Jensen, K. Stability and Yield of Cool-Season Pasture Grass Species Grown at Five Irrigation Levels. Crop Sci. 2002, 42, 890–896. [Google Scholar] [CrossRef]
- Olsen, C.; Cain, A.; Gould, M.; Mattox, C.; Kowalewski, A. Optimizing Irrigation Rates and Frequency for Perennial Ryegrass in Western Oregon. Crop Forage Turfgrass Manag. 2019, 5, 180094. [Google Scholar] [CrossRef]
- Basford, K.E.; Cooper, M. Genotype×environment interactions and some considerations of their implications for wheat breeding in Australia. Aust. J. Agric. Res. 1998, 49, 153–174. [Google Scholar] [CrossRef]
- Smith, A.B.; Stringer, J.K.; Wei, X.; Cullis, B.R. Varietal selection for perennial crops where data relate to multiple harvests from a series of field trials. Euphytica 2007, 157, 253–266. [Google Scholar] [CrossRef]
- Smith, A.B.; Ganesalingam, A.; Kuchel, H.; Cullis, B.R. Factor analytic mixed models for the provision of grower information from national crop variety testing programs. Theor. Appl. Genet. 2015, 128, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Gauch, H.G.; Wei, J.; Xie, J.; Chen, S.; Peng, H.; Bu, J.; Jiang, X. Genotype by Environment Interaction Analysis for Grain Yield and Yield Components of Summer Maize Hybrids across the Huanghuaihai Region in China. Agriculture 2022, 12, 602. [Google Scholar] [CrossRef]
- Annicchiarico, P. Additive main effects and multiplicative interaction (AMMI) analysis of genotype-location interaction in variety trials repeated over years. Theor. Appl. Genet. 1997, 94, 1072–1077. [Google Scholar] [CrossRef]
- Jung, M.; Quesada-Traver, C.; Roth, M.; Aranzana, M.J.; Muranty, H.; Rymenants, M.; Guerra, W.; Holzknecht, E.; Pradas, N.; Lozano, L.; et al. Integrative multi-environmental genomic prediction in apple. Hortic. Res. 2024, 12, uhae319. [Google Scholar] [CrossRef]
- Mienye, I.D.; Swart, T.G.; Obaido, G. Recurrent Neural Networks: A Comprehensive Review of Architectures, Variants, and Applications. Information 2024, 15, 517. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L.; Archontoulis, S.V. A CNN-RNN Framework for Crop Yield Prediction. Front. Plant Sci. 2019, 10, 1750. [Google Scholar] [CrossRef]
- Robert, P.; Le Gouis, J.; BreedWheat Consortium; Rincent, R. Combining Crop Growth Modeling With Trait-Assisted Prediction Improved the Prediction of Genotype by Environment Interactions. Front. Plant Sci. 2020, 11, 827. [Google Scholar] [CrossRef]
- Boote, K.J.; Jones, J.W.; Hoogenboom, G. Incorporating realistic trait physiology into crop growth models to support genetic improvement. In Silico Plants 2021, 3, diab002. [Google Scholar] [CrossRef]
- Holzworth, D.P.; Huth, N.I.; deVoil, P.G.; Zurcher, E.J.; Herrmann, N.I.; McLean, G.; Chenu, K.; van Oosterom, E.J.; Snow, V.; Murphy, C.; et al. APSIM—Evolution towards a new generation of agricultural systems simulation. Environ. Model. Softw. 2014, 62, 327–350. [Google Scholar] [CrossRef]
- Holzworth, D.; Huth, N.I.; Fainges, J.; Brown, H.; Zurcher, E.; Cichota, R.; Verrall, S.; Herrmann, N.I.; Zheng, B.; Snow, V. APSIM Next Generation: Overcoming challenges in modernising a farming systems model. Environ. Model. Softw. 2018, 103, 43–51. [Google Scholar] [CrossRef]
- Chapman, S.C.; Cooper, M.; Hammer, G.L. Using crop simulation to generate genotype by environment interaction effects for sorghum in water-limited environments. Aust. J. Agric. Res. 2002, 53, 379–389. [Google Scholar] [CrossRef]
- Onogi, A. Integration of Crop Growth Models and Genomic PredictionGenomic predictions (GP). In Genomic Prediction of Complex Traits: Methods and Protocols; Ahmadi, N., Bartholomé, J., Eds.; Springer: New York, NY, USA, 2022; pp. 359–396. [Google Scholar] [CrossRef]
- Adnan, A.A.; Diels, J.; Jibrin, J.M.; Kamara, A.Y.; Shaibu, A.S.; Craufurd, P.; Menkir, A. CERES-Maize model for simulating genotype-by-environment interaction of maize and its stability in the dry and wet savannas of Nigeria. Field Crops Res. 2020, 253, 107826. [Google Scholar] [CrossRef]
- Li, T.; Angeles, O.; Marcaida, M.; Manalo, E.; Manalili, M.P.; Radanielson, A.; Mohanty, S. From ORYZA2000 to ORYZA (v3): An improved simulation model for rice in drought and nitrogen-deficient environments. Agric. For. Meteorol. 2017, 237–238, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Stöckle, C.O.; Donatelli, M.; Nelson, R. CropSyst, a cropping systems simulation model. Eur. J. Agron. 2003, 18, 289–307. [Google Scholar] [CrossRef]
- Basso, B.; Liu, L.; Ritchie, J.T. A Comprehensive Review of the CERES-Wheat, -Maize and -Rice Models’ Performances. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 136, pp. 27–132. [Google Scholar]
- Technow, F.; Messina, C.D.; Totir, L.R.; Cooper, M. Integrating Crop Growth Models with Whole Genome Prediction through Approximate Bayesian Computation. PLoS ONE 2015, 10, e0130855. [Google Scholar] [CrossRef]
- McGranahan, D.A.; Yurkonis, K.A. Variability in grass forage quality and quantity in response to elevated CO2 and water limitation. Grass Forage Sci. 2018, 73, 517–521. [Google Scholar] [CrossRef]
- Elgersma, A.; Smith, K.F. Editorial—Topics from the XXIV International Grassland Congress held in 2021. Grass Forage Sci. 2022, 77, 107–110. [Google Scholar] [CrossRef]
- Marinoni, L.d.R.; Zabala, J.M.; Taleisnik, E.L.; Schrauf, G.E.; Richard, G.A.; Tomas, P.A.; Giavedoni, J.A.; Pensiero, J.F. Wild halophytic species as forage sources: Key aspects for plant breeding. Grass Forage Sci. 2019, 74, 321–344. [Google Scholar] [CrossRef]
- Smith, K.F.; Elgersma, A. Editorial: Grass and Forage Science—75 years of impact and service to the science of grasslands. Grass Forage Sci. 2020, 75, 351–356. [Google Scholar] [CrossRef]
- Fei, Y.F.; Yang, A.L.; Li, W.J.; Yuan, X.Q.; Fenech, A. Forecasting Crop Yield Under Climate Change Using Crop Growth Models in China: A Review. In Proceedings of the 14th International Conference on Environmental Science and Development (ICESD 2023), Xiamen, China, 25–27 May 2024; Springer Nature: Cham, Switzerland, 2024. [Google Scholar]
- Paleari, L.; Tondelli, A.; Cattivelli, L.; Igartua, E.; Casas, A.M.; Visoni, A.; Schulman, A.H.; Rossini, L.; Waugh, R.; Russell, J.; et al. Extending genomic prediction to future climates through crop modelling. A case study on heading time in barley. Agric. For. Meteorol. 2025, 368, 110560. [Google Scholar] [CrossRef]
- Jighly, A.; Weeks, A.; Christy, B.; O’Leary, G.J.; Kant, S.; Aggarwal, R.; Hessel, D.; Forrest, K.L.; Technow, F.; Tibbits, J.F.G.; et al. Integrating biophysical crop growth models and whole genome prediction for their mutual benefit: A case study in wheat phenology. J. Exp. Bot. 2023, 74, 4415–4426. [Google Scholar] [CrossRef] [PubMed]
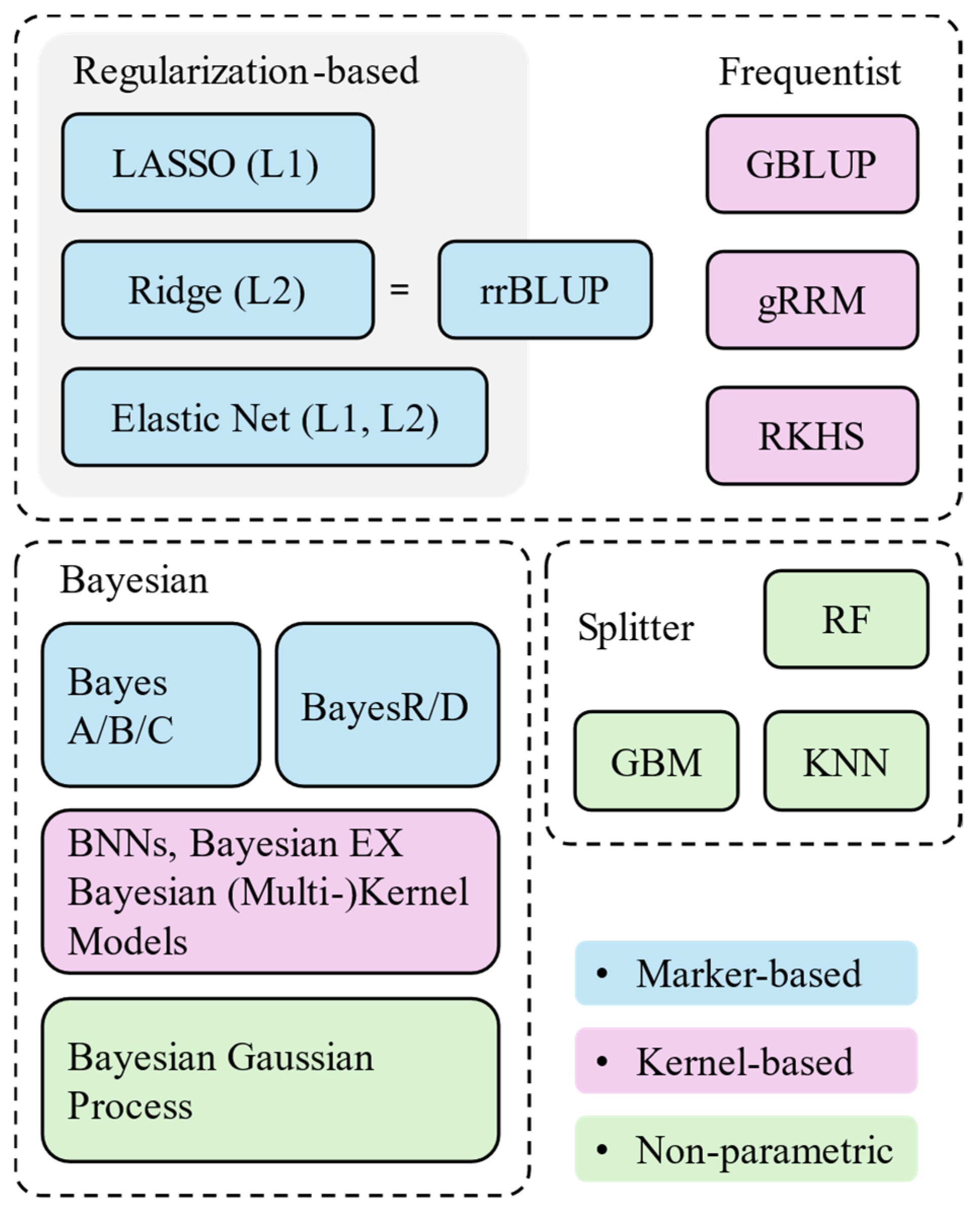
| Model | Structures | Assumptions | Frameworks | Applications |
|---|---|---|---|---|
| LASSO | + L1 penalty: | Linear additive marker effects; unified variances across markers | Sparse Marker-Based; Frequentist | High-dimensional additive genotyping data, feature selection [79] |
| Ridge regression | + L2 penalty: ; this is mathematically equivalent to | Linear additive marker effects; unified variances across markers | Dense Marker-Base; Frequentist | Traits with additive effects per marker, also called rrBLUP [20,29,30,31,47,57,68]; when markers are SNPs, also called snpBLUP |
| Elastic Net | + L1 and L2 penalties | Linear additive marker effects; unified variances across markers | Sparse Marker-Base; Frequentist | Correlated markers with additive effects per marker |
| GBLUP | Variance components are in a genomic relationship matrix (). Coefficients are solved by Henderson mixed model equation [80]. | Linear association between genetic markers and phenotypes | Kernel-Based; Frequentist | Traits with linear additive effects; computational efficiency [15,26,28,29,30,32,38,79,81,82]. The basic GBLUP uses a genomic relationship matrix (GRM) [83]. When the GRM is expanded with Legendre polynomials to model environmental variances, it becomes a genomic random regression model (gRRM) [19]. |
| RKHS | Akin to GBLUP but uses non-linear kernel . | Non-linear association between markers and phenotypes | Kernel-Based; Frequentist | Traits with non-linear effects and epistatic interactions [16,79] |
| BayesA/B/C | Linear additive marker effects; Variances can be marker-specific () or unified | Marker-Based: Dense when , Sparse when ; Bayesian | Traits with flexible genetic architecture [14,15,68,79,84] | |
| BayesR/D | , where | Linear additive marker effects with flexible shrinkage across multiple effect sizes; marker-specific variances | Sparse Marker-Based; Bayesian | Traits with mixed levels of effect sizes |
| Bayesian EX | General-purpose Bayesian framework: , where | Linear additive marker effects; prior choice determines shrinkage or sparsity level | Marker-Based; Bayesian | Flexible prior choices for traits with additive effects per marker. The prior could be Gaussian (Bayesian Ridge) [79], Laplace (Bayesian LASSO) [68,79], or other distributions. |
| Bayesian Neural Networks (BNNs) | Network learns , mapping from markers to phenotypes via weights . | Flexible marker effects; Implicit variance components | Marker-Based; Bayesian | Flexible genetic associations [79] |
| Bayesian Kernel Models | Flexible kernel is derived from the markers to account for genetic variance components, then estimates genetic effects. | Flexible genotype similarities; predefined variance components | Kernel-Based; Bayesian | Traits with non-linear genetic associations. When combining multiple kernels: , it becomes a Bayesian Multikernel Model. |
| Bayesian Gaussian Process | Flexible genotype similarities; stochastic variance components | Non-parametric; Kernel-Based; Bayesian | Traits requiring uncertainty quantification in genetic relationships. | |
| Random Forest (RF) | is trained on bootstrapped samples based on certain splitting criteria. RF parallelly aggregates tree predictions: . | Similarity-based prediction: patterns among similar data points provide higher prediction reliability | Non-parametric; Tree-Based Splitter | Non-linear additive prediction; noisy datasets with missing data; marker interactions [20,28,29,31,57,68] |
| Gradient Boosting Machines (GBM) | GBM sequentially corrects tree predictions: , where is negative gradient of the loss function in each tree, is the learning rate. | Similarity-based prediction | Non-parametric; Tree-Based Splitter | Non-linear prediction [20] |
| K-Nearest Neighbours (KNN) | Predicts based on closest training samples in the marker feature space: where indexes the nearest neighbours. | Similarity-based prediction | Non-parametric; Splitter | Non-linear prediction [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Smith, K.F.; Cogan, N.O.; Giri, K.; Jacobs, J.L. The Genome Era of Forage Selection: Current Status and Future Directions for Perennial Ryegrass Breeding and Evaluation. Agronomy 2025, 15, 1494. https://doi.org/10.3390/agronomy15061494
Zhu J, Smith KF, Cogan NO, Giri K, Jacobs JL. The Genome Era of Forage Selection: Current Status and Future Directions for Perennial Ryegrass Breeding and Evaluation. Agronomy. 2025; 15(6):1494. https://doi.org/10.3390/agronomy15061494
Chicago/Turabian StyleZhu, Jiashuai, Kevin F. Smith, Noel O. Cogan, Khageswor Giri, and Joe L. Jacobs. 2025. "The Genome Era of Forage Selection: Current Status and Future Directions for Perennial Ryegrass Breeding and Evaluation" Agronomy 15, no. 6: 1494. https://doi.org/10.3390/agronomy15061494
APA StyleZhu, J., Smith, K. F., Cogan, N. O., Giri, K., & Jacobs, J. L. (2025). The Genome Era of Forage Selection: Current Status and Future Directions for Perennial Ryegrass Breeding and Evaluation. Agronomy, 15(6), 1494. https://doi.org/10.3390/agronomy15061494






