Somaclonal Variation and Clonal Fidelity in Commercial Micropropagation: Challenges and Perspectives
Abstract
1. Introduction
2. Clonal Fidelity and Commercial Micropropagation
3. Somaclonal Variation in Micropropagated Plants
3.1. Mechanisms of Somaclonal Variation
3.1.1. Genetic Mechanisms
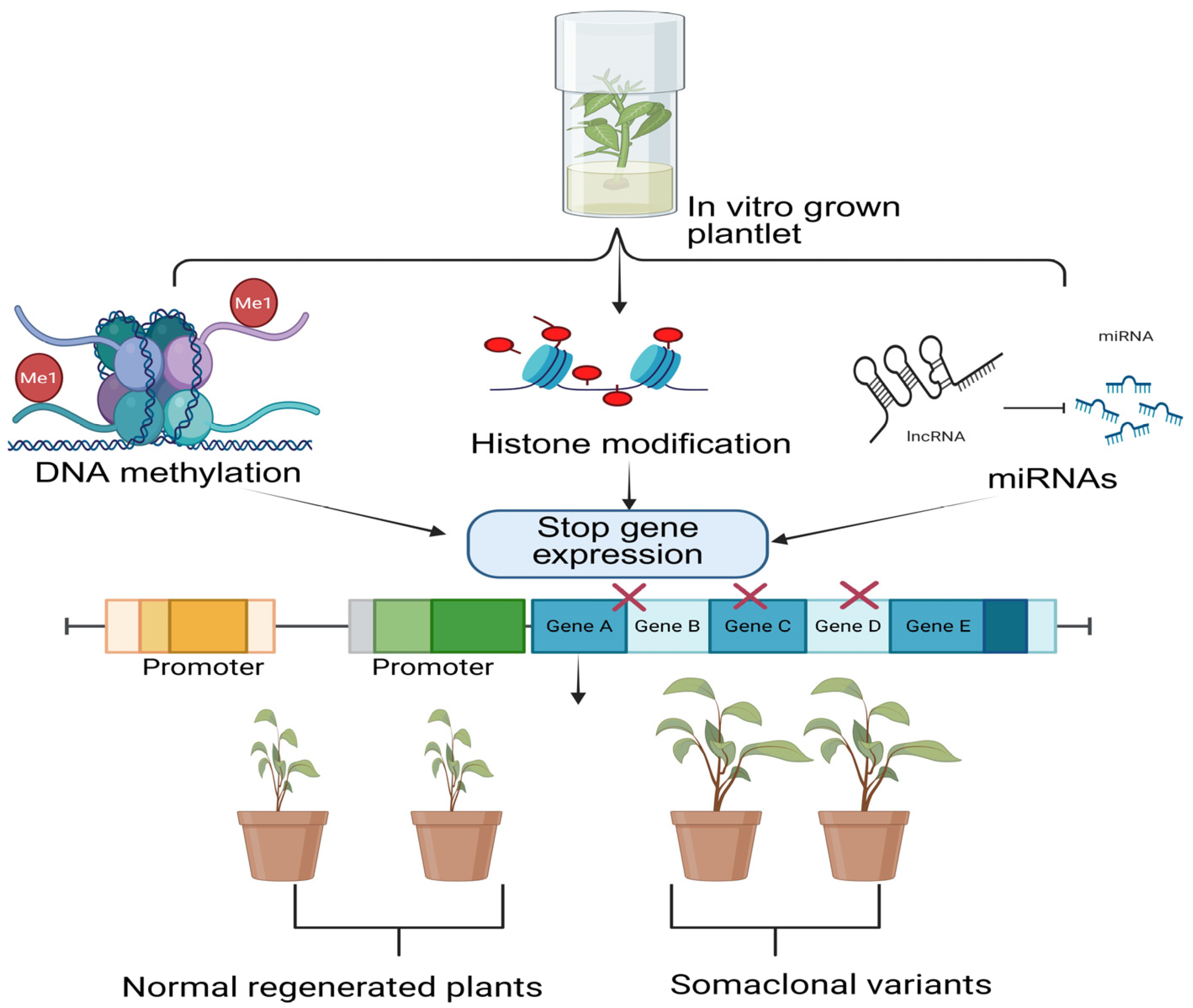
3.1.2. Epigenetic Mechanisms
4. Factors Affecting Somaclonal Variation
5. Detection and Assessment of Somaclonal Variation in Micropropagated Plants
5.1. Detection of Genetic Variation in Micropropagated Plants
5.2. Detection of Epigenetic Variation in Micropropagated Plants
6. Methods for Optimizing Somaclonal Variation
7. Application of Somaclonal Variation as a Crop Improvement Tool
8. Future Prospects
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bahar, N.H.A.; Lo, M.; Sanjaya, M.; Van Vianen, J.; Alexander, P.; Ickowitz, A.; Sunderland, T. Meeting the Food Security Challenge for Nine Billion People in 2050: What Impact on Forests? Glob. Environ. Chang. 2020, 62, 102056. [Google Scholar] [CrossRef]
- Hemanthakumar, A.S.; Preetha, T.S. Macro- and Micropropagation of Plants for Income Generation. In Conservation and Sustainable Utilization of Bioresources; Springer Nature: Singapore, 2023; pp. 409–450. [Google Scholar]
- Lasley, B.L.; Loskutoff, N.M.; Anderson, G.B. The limitation of conventional breeding programs and the need and promise of assisted 548 reproduction in nondomestic species. Theriogenology 1994, 41, 119–132. [Google Scholar] [CrossRef]
- Chandran, H.; Meena, M.; Barupal, T.; Sharma, K. Plant Tissue Culture as a Perpetual Source for Production of Industrially Important Bioactive Compounds. Biotechnol. Rep. 2020, 26, e00450. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, N.; El-Ramady, H.; Seliem, M.K.; El-Mahrouk, M.E.; Taha, N.; Bayoumi, Y.; Shalaby, T.A.; Dobránszki, J. An Academic and Technical Overview on Plant Micropropagation Challenges. Horticulturae 2022, 8, 677. [Google Scholar] [CrossRef]
- Bertsouklis, K.; Kartsonas, E.; Carra, A. Seed Germination and Micropropagation of Ornamental Plants. Horticulturae 2024, 10, 541. [Google Scholar] [CrossRef]
- Bettoni, J.C.; Wang, M.-R.; Wang, Q.-C. In Vitro Regeneration, Micropropagation and Germplasm Conservation of Horticultural Plants. Horticulturae 2024, 10, 45. [Google Scholar] [CrossRef]
- George, I.E.F.; Hall, M.A.; De Klerk, G.-J.; (Eds.). Plant Propagation by Tissue Culture. In Plant Cell Tiss Organ Cult, 3rd ed.; Springer: Dordrecht, The Netherlands, 2008; Volume 1, The Background; pp. 353–355. [Google Scholar] [CrossRef]
- Ozyigit, I.I.; Dogan, I.; Hocaoglu-Ozyigit, A.; Yalcin, B.; Erdogan, A.; Yalcin, I.E.; Cabi, E.; Kaya, Y. Production of Secondary Metabolites Using Tissue Culture-Based Biotechnological Applications. Front. Plant Sci. 2023, 14, 1132555. [Google Scholar] [CrossRef]
- Debnath, S.C.; Vyas, P.; Goyali, J.C.; Igamberdiev, A.U. Morphological and Molecular Analyses in Micropropagated Berry Plants Acclimatized under Ex Vitro Condition. Can. J. Plant Sci. 2012, 92, 1065–1073. [Google Scholar] [CrossRef]
- Taji, A.; Kumar, P.; Lakshmanan, P. In Vitro Plant Breeding; CRC Press: Boca Raton, FL, USA, 2024. [Google Scholar]
- Yancheva, S.; Kondakova, V. Plant Tissue Culture Technology: Present and Future Development. In Bioprocessing of Plant In Vitro Systems; Springer: Cham, Switzerland, 2018; pp. 39–63. [Google Scholar]
- Costa, G.F.D.; Cabral, P.D.S.; Silva, F.G.; Rubio Neto, A.; Mendonça, M.A.C. Clonal Fidelity and Genetic Diversity of Micropropagated Hancornia Speciosa Gomes (Apocynaceae) as Evaluated by Molecular Markers. Forests 2022, 13, 1645. [Google Scholar] [CrossRef]
- Debnath, S.C. Molecular Markers for Genetic Fidelity During In Vitro Propagation of Horticultural Crops. Acta Hortic. 2012, 929, 459–465. [Google Scholar] [CrossRef]
- Venkatachalam, L.; Sreedhar, R.V.; Bhagyalakshmi, N. Micropropagation in Banana Using High Levels of Cytokinins Does Not Involve Any Genetic Changes as Revealed by RAPD and ISSR Markers. Plant Growth Regul. 2007, 51, 193–205. [Google Scholar] [CrossRef]
- Debnath, S. Bioreactors and Molecular Analysis in Berry Crop Micropropagation—A Review. Can. J. Plant Sci. 2011, 91, 147–157. [Google Scholar] [CrossRef]
- Kundu, S.; Barua, R.; Marshall, H.D.; Siow, Y.L.; Igamberdiev, A.U.; Debnath, S.C. Somatic Embryogenesis in Vaccinium vitis-Idaea L.: A High-Fidelity Micropropagation Approach Producing True-to-Type Plants with Enhanced Phytochemical Profiles. Plant Cell Tissue Organ Cult. (PCTOC) 2025, 161, 37. [Google Scholar] [CrossRef]
- Gupta, R.; Modgil, M.; Chakrabarti, S.K. Assessment of Genetic Fidelity of Micropropagated Apple Rootstock Plants, EMLA 111, Using RAPD Markers. Indian J. Exp. Biol. 2009, 47, 925–928. [Google Scholar] [PubMed]
- Amin, A.; Sharma, N.; Sultan, P.; Gandhi, S.G.; Srinivas, K.; Hassan, Q.P.; Ahmed, Z. In Vitro Propagation of Bergenia Stracheyi: An Alternative Approach for Higher Production of Valuable Bioactive Compounds. Vegetos 2024, 1–10. [Google Scholar] [CrossRef]
- Khezri, M.; Asghari-Zakaria, R.; Zare, N. Plant Cell and Tissue Culture: Propagation, Improvement, and Conservation of Medicinal Plants. In Biosynthesis of Natural Products in Plants; Springer Nature: Singapore, 2024; pp. 267–291. [Google Scholar]
- Mohapatra, P.; Ray, A.; Jena, S. Evaluation of Genetic Stability of In Vitro Raised Orchids Using Molecular-Based Markers. In Commercial Scale Tissue Culture for Horticulture and Plantation Crops; Springer Nature: Singapore, 2022; pp. 293–316. [Google Scholar]
- Dorani, E.; Dehghanian, Z.; Gougerdchi, V.; Hamedpour-Darabi, M. Application of Somaclonal Variation in Crop Improvements. In Plant Mutagenesis; Springer: Cham, Switzerland, 2024; pp. 93–109. [Google Scholar]
- Nath, J.; Kumari, A.; Joshi, S.; Gusain, S.; Kumari, K.; Patial, M.; Rawat, M.; Joshi, R. Micropropagation Technology for Improvement of Ornamental Plants. In Ornamental Horticulture: Latest Cultivation Practices and Breeding Technologies; Springer Nature: Singapore, 2024; pp. 121–149. [Google Scholar]
- Reuveni, O.; Golubowicz, S.; Israeli, Y. Factors Influencing the Occurrence of Somaclonal Variations in Micropropagated Bananas. Acta Hortic. 1993, 336, 357–364. [Google Scholar] [CrossRef]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal Variation—A Novel Source of Variability from Cell Cultures for Plant Improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef]
- Bairu, M.W.; Aremu, A.O.; Van Staden, J. Somaclonal Variation in Plants: Causes and Detection Methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Leva, A.R.; Petruccelli, R.; Rinaldi, L.M.R. Somaclonal Variation in Tissue Culture: A Case Study with Olive. In Recent Advances in Plant In Vitro Culture; InTech: London, UK, 2012. [Google Scholar]
- Duta-Cornescu, G.; Constantin, N.; Pojoga, D.-M.; Nicuta, D.; Simon-Gruita, A. Somaclonal Variation—Advantage or Disadvantage in Micropropagation of the Medicinal Plants. Int. J. Mol. Sci. 2023, 24, 838. [Google Scholar] [CrossRef]
- Lindner, M.; Verhagen, I.; Mateman, A.C.; van Oers, K.; Laine, V.N.; Visser, M.E. Genetic and Epigenetic Differentiation in Response to Genomic Selection for Avian Lay Date. Evol. Appl. 2024, 17, e13703. [Google Scholar] [CrossRef]
- Hasnain, A.; Naqvi, S.A.H.; Ayesha, S.I.; Khalid, F.; Ellahi, M.; Iqbal, S.; Hassan, M.Z.; Abbas, A.; Adamski, R.; Markowska, D.; et al. Plants In Vitro Propagation with Its Applications in Food, Pharmaceuticals and Cosmetic Industries; Current Scenario and Future Approaches. Front. Plant Sci. 2022, 13, 1009395. [Google Scholar] [CrossRef] [PubMed]
- Savita; Pati, P.K.; Virk, G.S.; Nagpal, A. An Efficient Somatic Embryogenesis Protocol for Citrus jambhiri and Assessment of Clonal Fidelity of Plantlets Using RAPD Markers. J. Plant Growth Regul. 2015, 34, 309–319. [Google Scholar] [CrossRef]
- Hussien, E.T.; Ahmed, M.F.; Ahmed, E.Z. Biometric Measurements and Genetic Instability Assessment of In Vitro Micro-Shoots Culture of Populus alba. Plant Physiol. Rep. 2022, 27, 398–406. [Google Scholar] [CrossRef]
- Shukla, S.; Bhutani, R.; Gupta, N.; Shukla, S.K.; Raman, T.; El-Sheikh, M.A.; Elansary, H.O.; Moussa, I.M. Studies on banana for propagation, conservation and genome analysis. Cogent Food Agric. 2025, 11, 2447898. [Google Scholar] [CrossRef]
- Gimenez, M.D.; Yañez-Santos, A.M.; Paz, R.C.; Quiroga, M.P.; Marfil, C.F.; Conci, V.C.; García-Lampasona, S.C. Assessment of Genetic and Epigenetic Changes in Virus-Free Garlic (Allium sativum L.) Plants Obtained by Meristem Culture Followed by In Vitro Propagation. Plant Cell Rep. 2016, 35, 129–141. [Google Scholar] [CrossRef]
- Verma, D.; Ansari, M.W.; Agrawal, G.K.; Rakwal, R.; Shukla, A.; Tuteja, N. In Vitro Selection and Field Responses of Somaclonal Variant Plants of Rice Cv PR113 for Drought Tolerance. Plant Signal. Behav. 2013, 8, e23519. [Google Scholar] [CrossRef]
- Ali, A.; El-Denary, M.; El-Gendy, A.; Galal, O.; Mohamed, M.; El Sayed, T. Morphological Evaluation of Some Tomato Somaclones Variation under Field Conditions. J. Plant Prod. 2018, 9, 833–838. [Google Scholar] [CrossRef]
- Hannachi, S.; Werbrouck, S.; Bahrini, I.; Abdelgadir, A.; Affan Siddiqui, H. Agronomical, Physiological and Biochemical Characterization of In Vitro Selected Eggplant Somaclonal Variants under NaCl Stress. Plants 2021, 10, 2544. [Google Scholar] [CrossRef]
- Biernacik, B.; Słomnicka, R.; Kaźmińska, K.; Mużacz, S.; Bartoszewski, G. A Single Nucleotide Substitution Introducing Premature Stop Codon within CsTFL1 Explains the Determinate-2 Phenotype in Cucumber (Cucumis sativus L.). Sci. Rep. 2024, 14, 25368. [Google Scholar] [CrossRef]
- Geem, K.R.; Lee, Y.-J.; Lee, J.; Hong, D.; Kim, G.-E.; Sung, J. Role of Carrot (Daucus carota L.) Storage Roots in Drought Stress Adaptation: Hormonal Regulation and Metabolite Accumulation. Metabolites 2025, 15, 56. [Google Scholar] [CrossRef]
- Ferreira, M.d.S.; Rebouças, T.A.; Rocha, A.d.J.; Oliveira, W.D.d.S.; Santos, A.C.L.S.d.; Jesus, J.P.F.L.d.; Ramos, A.P.d.S.; Ferreira, C.F.; Santos-Serejo, J.A.d.; Haddad, F.; et al. Selection and Characterization of Somaclonal Variants of Prata Banana (AAB) Resistant to Fusarium Wilt. Agronomy 2024, 14, 1740. [Google Scholar] [CrossRef]
- Cui, X.; Liu, Q.; Luo, Y.; Zhu, P.; Guan, P.; Zhang, J. Study on Influencing Factors of Embryo Rescue and Germplasm Innovation in Seedless Grape. Plant Cell Tissue Organ Cult. (PCTOC) 2024, 157, 24. [Google Scholar] [CrossRef]
- Joshi, A.; Tripathi, A.; Sharma, N.; Tailor, A. Somaclonal Variation for the Improvement of Tree Species. In Biotechnological Approaches for Sustaining Forest Trees and Their Products; Springer Nature: Singapore, 2024; pp. 77–102. [Google Scholar]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Mirani, A.A.; Teo, C.H.; Markhand, G.S.; Abul-Soad, A.A.; Harikrishna, J.A. Detection of Somaclonal Variations in Tissue Cultured Date Palm (Phoenix dactylifera L.) Using Transposable Element-Based Markers. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 141, 119–130. [Google Scholar] [CrossRef]
- Ferreira, M.d.S.; Rocha, A.d.J.; Nascimento, F.d.S.; Oliveira, W.D.d.S.; Soares, J.M.d.S.; Rebouças, T.A.; Morais Lino, L.S.; Haddad, F.; Ferreira, C.F.; Santos-Serejo, J.A.d.; et al. The Role of Somaclonal Variation in Plant Genetic Improvement: A Systematic Review. Agronomy 2023, 13, 730. [Google Scholar] [CrossRef]
- Purohit, S.D.; Sharma, P.; Nagori, R. Assessment of Molecular Diversity and In Vitro Clonal Fidelity in Some Indian Medicinal Plants. Acta Hortic. 2015, 1098, 47–60. [Google Scholar] [CrossRef]
- Karp, A.; Wu, Q.S.; Steele, S.H.; Jones, M.G.K. Chromosome Variation in Dividing Protoplasts and Cell Suspensions of Wheat. Theor. Appl. Genet. 1987, 74, 140–146. [Google Scholar] [CrossRef]
- Sangthong, R.; Mii, M.; Soonthornchainaksaeng, P.; Supaibulwatana, K. Characteristics of The Tetraploid Plant Derived as A Somaclonal Variation in Lilium longiflorum. Acta Hortic. 2005, 673, 167–174. [Google Scholar] [CrossRef]
- Arnholdt-Schmitt, B. Rapid Changes in Amplification and Methylation Pattern of Genomic DNA in Cultured Carrot Root Explants (Daucus carota L.). Theor. Appl. Genet. 1993, 85, 793–800. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Xu, J.; Liu, X.; Chi, Y.; Li, R.; Mo, L.; Shi, L.; Liang, S.; Yu, W.; et al. Recent Progress of Molecular Mechanisms of DNA Methylation in Plant Response to Abiotic Stress. Environ. Exp. Bot. 2024, 218, 105599. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, C.; Wang, C.; Pan, X.; Zhang, H.; Yao, Y.; Huang, D.; Liao, W. The Involvement of DNA Methylation in Plant Growth Regulators-Mediated Growth in Tomato (Solanum lycopersicum) Seedlings. J. Plant Growth Regul. 2024, 43, 1287–1303. [Google Scholar] [CrossRef]
- Fang, J.; Jiang, J.; Leichter, S.M.; Liu, J.; Biswal, M.; Khudaverdyan, N.; Zhong, X.; Song, J. Mechanistic Basis for Maintenance of CHG DNA Methylation in Plants. Nat. Commun. 2022, 13, 3877. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Q.-M.; An, Q.; Cui, J.; Zhou, Y.; Qi, X.; Zhang, L.; Li, L. A Novel Micropropagation of Lycium ruthenicum and Epigenetic Fidelity Assessment of Three Types of Micropropagated Plants In Vitro and Ex Vitro. PLoS ONE 2021, 16, e0247666. [Google Scholar] [CrossRef] [PubMed]
- Abdulraheem, M.I.; Xiong, Y.; Moshood, A.Y.; Cadenas-Pliego, G.; Zhang, H.; Hu, J. Mechanisms of Plant Epigenetic Regulation in Response to Plant Stress: Recent Discoveries and Implications. Plants 2024, 13, 163. [Google Scholar] [CrossRef]
- Berr, A.; Shafiq, S.; Shen, W.-H. Histone Modifications in Transcriptional Activation during Plant Development. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2011, 1809, 567–576. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic Regulation in Plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-Genome Analysis of Histone H3 Lysine 27 Trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef]
- Carlsbecker, A.; Lee, J.-Y.; Roberts, C.J.; Dettmer, J.; Lehesranta, S.; Zhou, J.; Lindgren, O.; Moreno-Risueno, M.A.; Vatén, A.; Thitamadee, S.; et al. Cell Signalling by MicroRNA165/6 Directs Gene Dose-Dependent Root Cell Fate. Nature 2010, 465, 316–321. [Google Scholar] [CrossRef]
- Ivey, K.N.; Srivastava, D. MicroRNAs as Regulators of Differentiation and Cell Fate Decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef]
- Li, H.; Zhao, X.; Dai, H.; Wu, W.; Mao, W.; Zhang, Z. Tissue Culture Responsive MicroRNAs in Strawberry. Plant Mol. Biol. Rep. 2012, 30, 1047–1054. [Google Scholar] [CrossRef]
- Zhang, Z. Epigenetic and Genetic Variation in Micro-Propagated Strawberry Plants. Acta Hortic. 2014, 1049, 13–18. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; Huang, F.; Chang, L.; Ma, Y. MicroRNA Expression Profiles in Conventional and Micropropagated Strawberry (Fragaria × Ananassa Duch.) Plants. Plant Cell Rep. 2009, 28, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Tang, Y. Noncoding RNA-Mediated Regulation of DNA Methylation: Insights into Plant Epigenetic Mechanisms. J. Plant Growth Regul. 2025, 44, 373–388. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal Variations and Their Applications in Horticultural Crops Improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef]
- Sianipar, N.F. Somaclonal variations induced by benzylaminopurine to enhance the fruit morphology of horn banana. SABRAO J. Breed. Genet 2024, 56, 2045–2055. [Google Scholar] [CrossRef]
- Miguel, C.; Marum, L. An Epigenetic View of Plant Cells Cultured In Vitro: Somaclonal Variation and Beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar] [CrossRef]
- Bayati, E.; Gomarian, M.; Mirzaie-Nodousha, H.; Changizi, M.; Khaghani, S. Producing a Superior Genotype from Agria Potato Cultivar Using Somaclonal Variation. Nexo Rev. Científica 2021, 34, 671–681. [Google Scholar] [CrossRef]
- Vidyagina, E.O.; Kharchenko, N.N.; Shestibratov, K.A. Efficient Cryopreservation of Populus tremula by In Vitro-Grown Axillary Buds and Genetic Stability of Recovered Plants. Plants 2021, 10, 77. [Google Scholar] [CrossRef]
- Siragusa, M.; Carra, A.; Salvia, L.; Puglia, A.M.; De Pasquale, F.; Carimi, F. Genetic Instability in Calamondin (Citrus madurensis Lour.) Plants Derived from Somatic Embryogenesis Induced by Diphenylurea Derivatives. Plant Cell Rep. 2007, 26, 1289–1296. [Google Scholar] [CrossRef]
- Bychappa, M.; Mishra, M.K.; Jingade, P.; Huded, A.K.C. Genomic Alterations in Coding Region of Tissue Culture Plants of Coffea arabica Obtained through Somatic Embryogenesis Revealed by Molecular Markers. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 139, 91–103. [Google Scholar] [CrossRef]
- Hesami, M.; Adamek, K.; Pepe, M.; Jones, A.M.P. Effect of Explant Source on Phenotypic Changes of In Vitro Grown Cannabis Plantlets over Multiple Subcultures. Biology 2023, 12, 443. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Armitage, D.W. Epigenetic Responses of Trees to Environmental Stress in the Context of Climate Change. Biol. Rev. 2025, 100, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C.; Uğurlar, F.; Adamakis, I.-D.S. Epigenetic Modifications of Hormonal Signaling Pathways in Plant Drought Response and Tolerance for Sustainable Food Security. Int. J. Mol. Sci. 2024, 25, 8229. [Google Scholar] [CrossRef]
- Rudolf, J.; Tomovicova, L.; Panzarova, K.; Fajkus, J.; Hejatko, J.; Skalak, J. Epigenetics and Plant Hormone Dynamics: A Functional and Methodological Perspective. J. Exp. Bot. 2024, 75, 5267–5294. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.; Brown, P.T.H.; Hartke, S.; Lörz, H. DNA Variation in Tissue-Culture-Derived Rice Plants. Theor. Appl. Genet. 1990, 80, 673–679. [Google Scholar] [CrossRef]
- Thorat, A.S.; Sonone, N.A.; Choudhari, V.V.; Devarumath, R.M.; Babu, K.H. Plant Regeneration from Cell Suspension Culture in Saccharum officinarum L. and Ascertaining of Genetic Fidelity through RAPD and ISSR Markers. 3 Biotech 2017, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.S.; Sayed, A.E.; MM Abd Allah, M.M.; EL-shaer, H.F. Molecular Genetic Studies on (Ficus palmata & Ficus carica Subsp Rupestris.) Under the Effect of Gamma Radiation by Tissue Culture Techniques. Al-Azhar J. Agric. Res. 2021, 46, 128–138. [Google Scholar] [CrossRef]
- Pachota, K.A.; Orłowska, R.; Bednarek, P.T. Medium Composition Affects the Tissue Culture-Induced Variation in Triticale Regenerants. Plant Cell Tissue Organ Cult. (PCTOC) 2022, 151, 35–46. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Z.; Wang, N.; Gao, Y.; Liu, Y.; Wu, Y.; Bai, Y.; Zhang, Z.; Lin, X.; Dong, Y.; et al. Tissue Culture-Induced Heritable Genomic Variation in Rice, and Their Phenotypic Implications. PLoS ONE 2014, 9, e96879. [Google Scholar] [CrossRef]
- Tikendra, L.; Potshangbam, A.M.; Dey, A.; Devi, T.R.; Sahoo, M.R.; Nongdam, P. RAPD, ISSR, and SCoT Markers Based Genetic Stability Assessment of Micropropagated Dendrobium Fimbriatum Lindl. Var. Oculatum Hk. f.- an Important Endangered Orchid. Physiol. Mol. Biol. Plants 2021, 27, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Debnath, S.C. Bioreactor-Induced Adventitious Shoot Regeneration Affects Genotype-Dependent Morphology but Maintains Clonal Fidelity in Red Raspberry. Vitr. Cell. Dev. Biol.-Plant 2014, 50, 777–788. [Google Scholar] [CrossRef]
- Bisht, V.; Rawat, J.M.; Gaira, K.S.; Purohit, S.; Anand, J.; Sinha, S.; Mitra, D.; Ataya, F.S.; Elgazzar, A.M.; El-Saber Batiha, G.; et al. Assessment of Genetic Homogeneity of In-Vitro Propagated Apple Root Stock MM 104 Using ISSR and SCoT Primers. BMC Plant Biol. 2024, 24, 240. [Google Scholar] [CrossRef]
- Rathore, M.S.; Chikara, J.; Mastan, S.G.; Rahman, H.; Anand, K.G.V.; Shekhawat, N.S. Assessment of Genetic Stability and Instability of Tissue Culture-Propagated Plantlets of Aloe vera L. by RAPD and ISSR Markers. Appl. Biochem. Biotechnol. 2011, 165, 1356–1365. [Google Scholar] [CrossRef]
- Vishal; Sidhu, G.S.; Gaikwad, P.N.; Mann, S.S.; Gill, M.S.; Manchanda, P. Optimized Protocol for High-Frequency Papaya Propagation: Morpho-Stereomicroscopic Analysis and Genetic Fidelity Assessment. Plant Cell Tissue Organ Cult. (PCTOC) 2024, 156, 81. [Google Scholar] [CrossRef]
- Dyduch-Siemińska, M.; Wawerska, K.; Gawroński, J. The Potential of Plant Tissue Cultures to Improve the Steviol Glycoside Profile of Stevia (Stevia rebaudiana Bertoni) Regenerants. Int. J. Mol. Sci. 2024, 25, 13584. [Google Scholar] [CrossRef]
- Chavez-Cortazar, A.; Mata-Rosas, M.; Oyama, K.; Samain, M.S.; Quesada, M. Induction of Somatic Embryogenesis and Evaluation of Genetic Stability in Regenerated Plants of Magnolia Dealbata. Biol. Plant 2020, 64, 224–233. [Google Scholar] [CrossRef]
- Tongtape, K.; Te-chato, S.; Yenchon, S. Somatic Embryo (SE) Formation from Culturing Floral Explants of Rubber Tree (Hevea brasiliensis Muell. Arg.) and Assessment of Genetic Stability by RAPD and SSR Markers. Trends Sci. 2023, 20, 6728. [Google Scholar] [CrossRef]
- Skała, E.; Olszewska, M.A.; Makowczyńska, J.; Kicel, A. Effect of Sucrose Concentration on Rhaponticum carthamoides (Willd.) Iljin Transformed Root Biomass, Caffeoylquinic Acid Derivative, and Flavonoid Production. Int. J. Mol. Sci. 2022, 23, 13848. [Google Scholar] [CrossRef]
- Safarpour, M.; Sinniah, U.R.; Subramaniam, S.; Swamy, M.K. A Novel Technique for Musa Acuminata Colla ‘Grand Naine’ (AAA) Micropropagation through Transverse Sectioning of the Shoot Apex. Vitr. Cell. Dev. Biol.-Plant 2017, 53, 226–238. [Google Scholar] [CrossRef]
- Ramírez-Mosqueda, M.A.; Iglesias-Andreu, L.G. Indirect Organogenesis and Assessment of Somaclonal Variation in Plantlets of Vanilla planifolia Jacks. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 123, 657–664. [Google Scholar] [CrossRef]
- Han, Z.; Crisp, P.A.; Stelpflug, S.; Kaeppler, S.M.; Li, Q.; Springer, N.M. Heritable Epigenomic Changes to the Maize Methylome Resulting from Tissue Culture. Genetics 2018, 209, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Sikdar, A.; Sharma, U.; Barua, R.; Igamberdiev, A.U.; Debnath, S.C. Epigenomic Insight of Lingonberry and Health-Promoting Traits during Micropropagation. Sci. Rep. 2022, 12, 12487. [Google Scholar] [CrossRef] [PubMed]
- Bobadilla Landey, R.; Cenci, A.; Georget, F.; Bertrand, B.; Camayo, G.; Dechamp, E.; Herrera, J.C.; Santoni, S.; Lashermes, P.; Simpson, J.; et al. High Genetic and Epigenetic Stability in Coffea arabica Plants Derived from Embryogenic Suspensions and Secondary Embryogenesis as Revealed by AFLP, MSAP and the Phenotypic Variation Rate. PLoS ONE 2013, 8, e56372. [Google Scholar] [CrossRef]
- Goyali, J.C.; Igamberdiev, A.U.; Debnath, S.C. DNA Methylation in Lowbush Blueberry (Vaccinium angustifolium Ait.) Propagated by Softwood Cutting and Tissue Culture. Can. J. Plant Sci. 2018, 98, 1035–1044. [Google Scholar] [CrossRef]
- Cao, Q.; Feng, Y.; Dai, X.; Huang, L.; Li, J.; Tao, P.; Crabbe, M.J.C.; Zhang, T.; Qiao, Q. Dynamic Changes of DNA Methylation During Wild Strawberry (Fragaria nilgerrensis) Tissue Culture. Front. Plant Sci. 2021, 12, 765383. [Google Scholar] [CrossRef]
- Lin, W.; Xiao, X.; Zhang, H.; Li, Y.; Liu, S.; Sun, W.; Zhang, X.; Wu, Q. Whole-Genome Bisulfite Sequencing Reveals a Role for DNA Methylation in Variants from Callus Culture of Pineapple (Ananas comosus L.). Genes 2019, 10, 877. [Google Scholar] [CrossRef]
- Orłowska, R. Barley Somatic Embryogenesis-an Attempt to Modify Variation Induced in Tissue Culture. J. Biol. Res.-Thessalon. 2021, 28, 9. [Google Scholar] [CrossRef]
- Himmen, F.D.A.; de Souza, F.A.; de Araújo Silva-Cardoso, I.M.; de Souza, A.L.X.; Scherwinski-Pereira, J.E. Micropropagation, Estimation of DNA Methylation during Multiplication Cycles and Mycorrhization of Seed-Derived Dendrocalamus asper (Schultes f.) Backer Ex Heyne. Plant Cell Tissue Organ Cult. (PCTOC) 2023, 155, 41–56. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Phillips, R.L. Tissue Culture-Induced DNA Methylation Variation in Maize. Proc. Natl. Acad. Sci. USA 1993, 90, 8773–8776. [Google Scholar] [CrossRef]
- Cao, S.; Sawettalake, N.; Li, P.; Fan, S.; Shen, L. DNA Methylation Variations Underlie Lettuce Domestication and Divergence. Genome Biol. 2024, 25, 158. [Google Scholar] [CrossRef] [PubMed]
- Nic-Can, G.I.; López-Torres, A.; Barredo-Pool, F.; Wrobel, K.; Loyola-Vargas, V.M.; Rojas-Herrera, R.; De-la-Peña, C. New Insights into Somatic Embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 Are Epigenetically Regulated in Coffea canephora. PLoS ONE 2013, 8, e72160. [Google Scholar] [CrossRef]
- Linacero, R.; Ballesteros, I. Genetic Basis of Somaclonal Variation. In Somaclonal Variation: Basic and Practical Aspects; Springer International Publishing: Cham, Switzerland, 2024; pp. 1–20. [Google Scholar]
- Noack, F.; Engist, D.; Gantois, J.; Gaur, V.; Hyjazie, B.F.; Larsen, A.; M’Gonigle, L.K.; Missirian, A.; Qaim, M.; Sargent, R.D.; et al. Environmental Impacts of Genetically Modified Crops. Science 2024, 385, eado9340. [Google Scholar] [CrossRef]
- Pandey, S.; Patel, P.; Prasad, A.; Sawant, S.V.; Misra, P. Assessment of Direct Shoot Organogenesis and Genetic Fidelity in Solanum viarum Dunal—A Commercially Important Medicinal Plant. Vitr. Cell. Dev. Biol.-Plant 2020, 56, 538–547. [Google Scholar] [CrossRef]
- Singer, S.D.; Laurie, J.D.; Bilichak, A.; Kumar, S.; Singh, J. Genetic Variation and Unintended Risk in the Context of Old and New Breeding Techniques. CRC Crit. Rev. Plant Sci. 2021, 40, 68–108. [Google Scholar] [CrossRef]
- Rani, V.; Raina, S.N. Genetic Fidelity of Organized Meristem-Derived Micropropagated Plants: A Critical Reappraisal. Vitr. Cell. Dev. Biol.-Plant 2000, 36, 319–330. [Google Scholar] [CrossRef]
- Ding, J.; Deng, X.-X.; Zhang, H.; Cheng, Y.; Xu, Q.; Liu, Q. Somaclonal Variation with Early Juice-Sac Granulation Obtained by Inter-Specific Protoplast Fusion in Citrus. J. Hortic. Sci. Biotechnol. 2009, 84, 567–573. [Google Scholar] [CrossRef]
- Pellicer, J.; Powell, R.F.; Leitch, I.J. The Application of Flow Cytometry for Estimating Genome Size, Ploidy Level Endopolyploidy, and Reproductive Modes in Plants. In Molecular Plant Taxonomy; Humana: New York, NY, USA, 2021; pp. 325–361. [Google Scholar]
- Naing, A.H.; Kim, S.H.; Chung, M.Y.; Park, S.K.; Kim, C.K. In Vitro Propagation Method for Production of Morphologically and Genetically Stable Plants of Different Strawberry Cultivars. Plant Methods 2019, 15, 36. [Google Scholar] [CrossRef]
- Jena, S.; Ray, A.; Sahoo, A.; Sahoo, S.; Kar, B.; Panda, P.C.; Nayak, S. High-Frequency Clonal Propagation of Curcuma Angustifolia Ensuring Genetic Fidelity of Micropropagated Plants. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 135, 473–486. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Sandoval, J.; Kerbellec, F.; Cote, F.; Doumas, P. Distribution of Endogenous Gibberellins in Dwarf and Giant Off-Types Banana (Musa AAA, Cv.Grand Nain) Plants from In Vitro Propagation. Plant Growth Regul. 1995, 17, 219–224. [Google Scholar] [CrossRef]
- Amberger, L.A.; Shoemaker, R.C.; Palmer, R.G. Inheritance of Two Independent Isozyme Variants in Soybean Plants Derived from Tissue Culture. Theor. Appl. Genet. 1992, 84, 600–607. [Google Scholar] [CrossRef] [PubMed]
- González, A.M.; De la Fuente, M.; De Ron, A.M.; Santalla, M. Protein Markers and Seed Size Variation in Common Bean Segregating Populations. Mol. Breed. 2010, 25, 723–740. [Google Scholar] [CrossRef]
- Mandal, A.B.; Maiti, A.; Chowdhury, B.; Elanchezhian, R. Isoenzyme Markers in Varietal Identification of Banana. Vitr. Cell. Dev. Biol.-Plant 2001, 37, 599–604. [Google Scholar] [CrossRef]
- Biswas, P.; Kumar, N. Application of Molecular Markers for the Assessment of Genetic Fidelity of In Vitro Raised Plants: Current Status and Future Prospects. In Molecular Marker Techniques; Springer Nature: Singapore, 2023; pp. 233–256. [Google Scholar]
- Roth, E.J.; Frazier, B.L.; Apuya, N.R.; Lark, K.G. Genetic Variation in an Inbred Plant: Variation in Tissue Cultures of Soybean (Glycine max (L.) Merrill). Genetics 1989, 121, 359–368. [Google Scholar] [CrossRef]
- Mohammad, N.; Dahayat, A.; Agrahari, H. Molecular Markers in the Management and Improvement of Forest Genetic Resources. In Biotechnological Approaches for Sustaining Forest Trees and Their Products; Springer Nature: Singapore, 2024; pp. 181–198. [Google Scholar]
- Orbović, V.; Ćalović, M.; Viloria, Z.; Nielsen, B.; Gmitter, F.G.; Castle, W.S.; Grosser, J.W. Analysis of Genetic Variability in Various Tissue Culture-Derived Lemon Plant Populations Using RAPD and Flow Cytometry. Euphytica 2008, 161, 329–335. [Google Scholar] [CrossRef]
- Hashmi, G.; Huettel, R.; Meyer, R.; Krusberg, L.; Hammerschlag, F. RAPD Analysis of Somaclonal Variants Derived from Embryo Callus Cultures of Peach. Plant Cell Rep. 1997, 16, 624–627. [Google Scholar] [CrossRef]
- Kamle, M.; Kumar, P.; Bajpai, A.; Kalim, S.; Chandra, R. Assessment of Genetic Fidelity of Somatic Embryogenesis Regenerated Guava (Psidium guajava L.) Plants Using DNA-Based Markers. N. Zeal. J. Crop Hortic. Sci. 2014, 42, 1–9. [Google Scholar] [CrossRef]
- Martín, C.; González-Benito, M.E. Molecular Markers for the Detection and Analysis of Somaclonal Variation. In Somaclonal Variation: Basic and Practical Aspects; Springer International Publishing: Cham, Switzerland, 2024; pp. 57–82. [Google Scholar]
- Sahoo, A.; Behura, S.; Singh, S.; Jena, S.; Ray, A.; Dash, B.; Kar, B.; Panda, P.C.; Nayak, S. EST-SSR Marker-Based Genetic Diversity and Population Structure Analysis of Indian Curcuma Species: Significance for Conservation. Braz. J. Bot. 2021, 44, 411–428. [Google Scholar] [CrossRef]
- Sadhu, S.; Jogam, P.; Thampu, R.K.; Abbagani, S.; Penna, S.; Peddaboina, V. High Efficiency Plant Regeneration and Genetic Fidelity of Regenerants by SCoT and ISSR Markers in Chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. (PCTOC) 2020, 141, 465–477. [Google Scholar] [CrossRef]
- Al-Aizari, A.A.; Dewir, Y.H.; Ghazy, A.H.; Al-Doss, A.; Al-Obeed, R.S. Micropropagation and Genetic Fidelity of Fegra Fig (Ficus palmata Forssk.) and Grafting Compatibility of the Regenerated Plants with Ficus Carica. Plants 2024, 13, 1278. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, Y.; Yu, S.; Li, S.; Cai, X. Genetic Stability Evaluation of Caladium Somaclonal Variants by Morphological, Cytological, and SSR Analysis in Three Successive Generations. Not. Bot. Horti Agrobot. Cluj-Napoca 2024, 52, 13746. [Google Scholar] [CrossRef]
- Ioannidis, K.; Tomprou, I.; Mitsis, V.; Koropouli, P. Genetic Evaluation of In Vitro Micropropagated and Regenerated Plants of Cannabis sativa L. Using SSR Molecular Markers. Plants 2022, 11, 2569. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Sikdar, A.; Igamberdiev, A.U.; Debnath, S.C. Exploring Genetic and Epigenetic Changes in Lingonberry Using Molecular Markers: Implications for Clonal Propagation. Curr. Issues Mol. Biol. 2023, 45, 6296–6310. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Mamgain, J.; Mujib, A.; Ejaz, B.; Gulzar, B.; Malik, M.Q.; Syeed, R. Flow Cytometry and Start Codon Targeted (SCoT) Genetic Fidelity Assessment of Regenerated Plantlets in Tylophora indica (Burm.f.) Merrill. Plant Cell Tissue Organ Cult. (PCTOC) 2022, 150, 129–140. [Google Scholar] [CrossRef]
- Pandey, N.; Meena, R.P.; Rai, S.K.; Pandey-Rai, S. In vitro generation of high artemisinin yielding salt tolerant somaclonal variant and development of SCAR marker in Artemisia annua L. Plant Cell Tiss. Organ Cult. (PCTOC) 2016, 127, 301–314. [Google Scholar] [CrossRef]
- Adly, W.M.R.M.; Niedbała, G.; EL-Denary, M.E.; Mohamed, M.A.; Piekutowska, M.; Wojciechowski, T.; Abd El-Salam, E.-S.T.; Fouad, A.S. Somaclonal Variation for Genetic Improvement of Starch Accumulation in Potato (Solanum tuberosum) Tubers. Plants 2023, 12, 232. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, S.; Zhang, Z.; Zhang, J.; Li, H. Comparative Characterization of Fruit Volatiles and Volatile-Related Genes Expression of ‘Benihoppe’ Strawberry and Its Somaclonal Mutant. Plants 2023, 12, 1109. [Google Scholar] [CrossRef]
- Ting, N.-C.; Jansen, J.; Nagappan, J.; Ishak, Z.; Chin, C.-W.; Tan, S.-G.; Cheah, S.-C.; Singh, R. Identification of QTLs Associated with Callogenesis and Embryogenesis in Oil Palm Using Genetic Linkage Maps Improved with SSR Markers. PLoS ONE 2013, 8, e53076. [Google Scholar] [CrossRef]
- Siril, E.A.; Joseph, N. Micropropagation of Annatto (Bixa orellana L.) from Mature Tree and Assessment of Genetic Fidelity of Micropropagated Plants with RAPD Markers. Physiol. Mol. Biol. Plants 2013, 19, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Silva, A.P.; Santos, A.A.; Carnide, V. Diversity in Hazelnut Using RAPD and ISSR Markers. Acta Hortic. 2009, 845, 145–150. [Google Scholar] [CrossRef]
- Yu, S.; Zhao, X.; Wang, Y.; Jiang, D.; Zhang, Y.; Hu, L.; Liu, Y.; Cai, X. Morphological, Cytological, and Molecular-Based Genetic Stability Analysis of In Vitro-Propagated Plants from Newly Induced Aneuploids in Caladium. Agriculture 2022, 12, 1708. [Google Scholar] [CrossRef]
- Ploetz, R.C. Management of Fusarium Wilt of Banana: A Review with Special Reference to Tropical Race 4. Crop Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Swain, D.; Lenka, S.; Hota, T.; Rout, G.R. Micro-Propagation of Hypericum gaitii Haines, an Endangered Medicinal Plants: Assessment of Genetic Fidelity. Nucleus 2016, 59, 7–13. [Google Scholar] [CrossRef]
- Sathish, D.; Vasudevan, V.; Theboral, J.; Elayaraja, D.; Appunu, C.; Siva, R.; Manickavasagam, M. Efficient Direct Plant Regeneration from Immature Leaf Roll Explants of Sugarcane (Saccharum officinarum L.) Using Polyamines and Assessment of Genetic Fidelity by SCoT Markers. Vitr. Cell. Dev. Biol.-Plant 2018, 54, 399–412. [Google Scholar] [CrossRef]
- Jogam, P.; Sandhya, D.; Shekhawat, M.S.; Alok, A.; Abbagani, S.; Allini, V.R. Genetic Stability Analysis Using DNA Barcoding and Molecular Markers and Foliar Micro-Morphological Analysis of In Vitro Regenerated and In Vivo Grown Plants of Artemisia Vulgaris L. Ind. Crops Prod. 2020, 151, 112476. [Google Scholar] [CrossRef]
- Monica, H.; Kumaria, S. Exogenous Application of Chitosan, a Potent Biotic Elicitor Enhances Micropropagation Efficiency of Cymbidium aloifolium (L.) Sw., an Orchid of Medicinal and Horticultural Importance. Vegetos 2024, 38, 58–69. [Google Scholar] [CrossRef]
- Oliya, B.K.; Chand, K.; Thakuri, L.S.; Baniya, M.K.; Sah, A.K.; Pant, B. Assessment of Genetic Stability of Micropropagated Plants of Rhynchostylis retusa (L.) Using RAPD Markers. Sci. Hortic. 2021, 281, 110008. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, Z.; Khan, M.A.; Zhou, Y. In Vitro Selection of Resistant Mutant Garlic Lines by Using Crude Pathogen Culture Filtrate of Sclerotium cepivorum. Australas. Plant Pathol. 2012, 41, 211–217. [Google Scholar] [CrossRef]
- Martins, A.A.; da Silva, M.F.; Pinto, L.R. Epigenetic Diversity of Saccharum Spp. Accessions Assessed by Methylation-Sensitive Amplification Polymorphism (MSAP). 3 Biotech 2020, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Rodríguez, R.; Cañal, M.J. Rapid quantification of DNA methylation by high performance capillary electrophoresis. Electrophoresis 2000, 21, 2990–2994. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hao, J.-L.; Wang, Z.; Song, K.-J.; Ye, J.-H.; Zheng, X.-Q.; Liang, Y.-R.; Lu, J.-L. DNA Methylation Levels in Different Tissues in Tea Plant via an Optimized HPLC Method. Hortic. Environ. Biotechnol. 2019, 60, 967–974. [Google Scholar] [CrossRef]
- Zhao, L.; Xie, L.; Zhang, Q.; Ouyang, W.; Deng, L.; Guan, P.; Ma, M.; Li, Y.; Zhang, Y.; Xiao, Q.; et al. Integrative Analysis of Reference Epigenomes in 20 Rice Varieties. Nat. Commun. 2020, 11, 2658. [Google Scholar] [CrossRef]
- Saleh, A.; Alvarez-Venegas, R.; Avramova, Z. An Efficient Chromatin Immunoprecipitation (ChIP) Protocol for Studying Histone Modifications in Arabidopsis Plants. Nat. Protoc. 2008, 3, 1018–1025. [Google Scholar] [CrossRef]
- Gong, Z.; Zheng, J.; Yang, N.; Li, X.; Qian, S.; Sun, F.; Geng, S.; Liang, Y.; Wang, J. Whole-Genome Bisulfite Sequencing (WGBS) Analysis of Gossypium hirsutum under High-Temperature Stress Conditions. Genes 2024, 15, 1241. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Eichten, S.R.; Hermanson, P.J.; Springer, N.M.; Kaeppler, S.M. Consistent and Heritable Alterations of DNA Methylation Are Induced by Tissue Culture in Maize. Genetics 2014, 198, 209–218. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Wu, D.; Tian, Q.; Su, S.; Cheng, C.; Nie, J.; Yuan, Y.; Wang, Y.; Xu, X. DNA Methylation Variation Is Crucial to Restore Adventitious Rooting Ability during In Vitro Shoot Culture-induced Rejuvenation in Apple Rootstock. Plant J. 2023, 114, 554–569. [Google Scholar] [CrossRef]
- Mueller, S.A.; Merondun, J.; Lečić, S.; Wolf, J.B.W. Epigenetic Variation in Light of Population Genetic Practice. Nat. Commun. 2025, 16, 1028. [Google Scholar] [CrossRef]
- Liu, D.; Mu, Q.; Li, X.; Xu, S.; Li, Y.; Gu, T. The Callus Formation Capacity of Strawberry Leaf Explant Is Modulated by DNA Methylation. Hortic. Res. 2022, 9, uhab073. [Google Scholar] [CrossRef]
- Machczyńska, J.; Zimny, J.; Bednarek, P.T. Tissue Culture-Induced Genetic and Epigenetic Variation in Triticale (× Triticosecale Spp. Wittmack Ex A. Camus 1927) Regenerants. Plant Mol. Biol. 2015, 89, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Peredo, E.L.; Revilla, M.A.; Arroyo-García, R. Assessment of Genetic and Epigenetic Variation in Hop Plants Regenerated from Sequential Subcultures of Organogenic Calli. J. Plant Physiol. 2006, 163, 1071–1079. [Google Scholar] [CrossRef]
- Kitimu, S.R.; Taylor, J.; March, T.J.; Tairo, F.; Wilkinson, M.J.; Rodríguez López, C.M. Meristem Micropropagation of Cassava (Manihot esculenta) Evokes Genome-Wide Changes in DNA Methylation. Front. Plant Sci. 2015, 6, 590. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, R.; Machczyńska, J.; Oleszczuk, S.; Zimny, J. DNA Methylation Changes and TE Activity Induced in Tissue Cultures of Barley (Hordeum vulgare L.). J. Biol. Res.-Thessaloniki 2016, 23, 19. [Google Scholar] [CrossRef]
- Kastelec, D.; Troha, S.; Svetik, S.; Trafela, T.; Murovec, J. Optimization of Micropropagation Protocols and Assessment of Epigenetic Changes in Tissue Cultures of Diverse Medical Cannabis (Cannabis sativa L.) Genotypes. Ind. Crops Prod. 2025, 229, 121015. [Google Scholar] [CrossRef]
- Khodaeiaminjan, M.; Gomes, C.; Pagano, A.; Kruszka, D.; Sulima, P.; Przyborowski, J.A.; Krajewski, P.; Paiva, J.A.P. Impacts of In-Vitro Zebularine Treatment on Genome-Wide DNA Methylation and Transcriptomic Profiles in Salix purpurea L. Physiol. Plant 2024, 176, e14403. [Google Scholar] [CrossRef]
- Rai, M.K. Somaclonal Variation in Improvement of Agricultural Crops: Recent Progress. In Agricultural Biotechnology: Latest Research and Trends; Springer Nature: Singapore, 2021; pp. 129–146. [Google Scholar]
- Rodrigues, P.H.V.; Tulmann Neto, A.; Cassieri Neto, P.; Mendes, B.M.J. Influence of the Number of Subcultures on Somaclonal Variation in Micropropagated Nanicão (Musa spp., aaa group). Acta Hortic. 1998, 490, 469–474. [Google Scholar] [CrossRef]
- Gomes, H.T.; Monteiro, T.R.; Camillo, J.; Nogueira, G.F.; da Silva Costa, F.H.; de Araújo Silva-Cardoso, I.M.; Scherwinski-Pereira, J.E. Integrated Seed Cryopreservation, Bioreactor and Conventional Micropropagation, and Genetic Stability Assessment by Flow Cytometry of Uncaria guianensis (Aubl.) J.F.Gmel, a High-Value Amazonian Medicinal Plant. Plant Cell Tissue Organ Cult. (PCTOC) 2025, 161, 45. [Google Scholar] [CrossRef]
- Rajeswari, S.; Thirugnanakumar, S.; Anandan, A.; Krishnamurthi, M. Somaclonal Variation in Sugarcane through Tissue Culture and Evaluation for Quantitative and Quality Traits. Euphytica 2009, 168, 71. [Google Scholar] [CrossRef]
- Kessel-Domini, A.; Pérez-Brito, D.; Guzmán-Antonio, A.; Barredo-Pool, F.A.; Mijangos-Cortés, J.O.; Iglesias-Andreu, L.G.; Cortés-Velázquez, A.; Canto-Flick, A.; Avilés-Viñas, S.A.; Rodríguez-Llanes, Y.; et al. Indirect Somatic Embryogenesis: An Efficient and Genetically Reliable Clonal Propagation System for Ananas comosus L. Merr. Hybrid “MD2.”. Agriculture 2022, 12, 713. [Google Scholar] [CrossRef]
- Kukreja, A.K.; Dhawan, O.P.; Mathur, A.K.; Ahuja, P.S.; Mandal, S. Screening and Evaluation of Agronomically Useful Somaclonal Variations in Japanese Mint (Mentha arvensis L.). Euphytica 1991, 53, 183–191. [Google Scholar] [CrossRef]
- Cioloca, M.; Tican, A.; Popa, M. Evaluation of Virus-Free Potato Minitubers Derived from “in Vitro” Meristem Cultures for High-Quality Seed Potato Production. Rom. J. Hortic. 2024, 5, 15–22. [Google Scholar] [CrossRef]
- Kritskaya, T.A.; Kashin, A.S.; Kasatkin, M.Y. Micropropagation and Somaclonal Variation of Tulipa Suaveolens (Liliaceae) in Vitro. Russ. J. Dev. Biol. 2019, 50, 209–215. [Google Scholar] [CrossRef]
- Sridhar, R.; Harish Babu, B.N. A Novel Pomegranate (Punica granatum L.) Variant with High Anthocyanin Content. Acta Hortic. 2019, 1255, 149–152. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.S.; Ganapathi, T.R. Characterization of Fusarium Wilt Resistant Somaclonal Variants of Banana Cv. Rasthali by CDNA-RAPD. Mol. Biol. Rep. 2014, 41, 7929–7935. [Google Scholar] [CrossRef]
- Khadijat, S.J.; Maimuna, A.M.; Sagir, M.M. Screening for Drought Tolerance in Cotton (Gossypium Hirsutum L.) Using In Vitro Technique. J. Dryland Agric. 2021, 7, 52–59. [Google Scholar] [CrossRef]
- Popoola, A.R.; Durosomo, A.H.; Afolabi, C.G.; Idehen, E.O. Regeneration of Somaclonal Variants of Tomato (Solanum lycopersicum L.) for Resistance to Fusarium Wilt. J. Crop Improv. 2015, 29, 636–649. [Google Scholar] [CrossRef]
- Dhurve, L.; Ajith Kumar, K.; Bhaskar, J.; Sobhana, A.; Francies, R.M.; Mathew, D. Wide Variability among the ‘Mauritius’ Somaclones Demonstrates Somaclonal Variation as a Promising Improvement Strategy in Pineapple (Ananas comosus L.). Plant Cell Tissue Organ Cult. (PCTOC) 2021, 145, 701–705. [Google Scholar] [CrossRef]
- Arun, B.; Singh, B.D.; Sharma, S.; Paliwal, R.; Joshi, A.K. Development of Somaclonal Variants of Wheat (Triticum aestivum L.) for Yield Traits and Disease Resistance Suitable for Heat Stressed and Zero-till Conditions. Field Crops Res. 2007, 103, 62–69. [Google Scholar] [CrossRef]
- Dolcet-Sanjuan, R.; Clavería, E.; Llauradó, L.; Ortigosa, A.; Arús, P. Carnation (Dianthus caryophyllus L.) Dihaploid Lines Resistant to Fusarium oxysporum f.sp. dianthi. Acta Hortic. 2001, 560, 145–148. [Google Scholar] [CrossRef]
- Ramasamy, Y.; Arunachalam, S.; Dasgupta, M.; Nambiar-Veetil, M. Genetic Markers, Genomics and Genetic Modification in Forest Trees: Current Status and Prospects. In Textbook of Forest Science; Mandal, A.K., Nicodemus, A., Eds.; Springer: Singapore, 2025. [Google Scholar] [CrossRef]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-Target Effects in CRISPR/Cas9 Gene Editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, D.; Martínez, M.T.; Sánchez-Romero, C.; Montalbán, I.A.; Sales, E.; Moncaleán, P.; Arrillaga, I.; Corredoira, E. Current Status of the Cryopreservation of Embryogenic Material of Woody Species. Front. Plant Sci. 2024, 14, 1337152. [Google Scholar] [CrossRef] [PubMed]


| Plant Name | Explants | Media Composition and PGRs | Genotype | Subcultures Cycles | Environmental Factors | Key Findings | Reference |
|---|---|---|---|---|---|---|---|
| Populus alba (White Poplar) | Shoot tips, leaf explants, or nodal segments | MS medium with sucrose (PGRs inferred) | Not specified | Not specified | Standard in vitro conditions | Significant variation was observed in morphological, physiological, and molecular traits. | [32] |
| Citrus jambhiri (Orange) | Leaf, nodal, root segments, stigmas, styles, ovaries, nucellar tissues, cotyledons, juice vesicles | MS medium with BAP, KN, ME, 2,4-D for callus induction; BA, 2,4-D, ABA for somatic embryo induction, sucrose for germination | Not specified | Not specified | Controlled Environment | Callus was induced from most explants except styles and juice vesicles; somatic embryos formed from ovary and nucellar tissues with BA and ME, and 20–30% variation in ovary-derived plantlets. | [31] |
| Vaccinium vitis-idaea (Lingonberry) | Juvenile leaf explants from two genotypes | Berry basal medium + 5.5 µM thidiazuron for callus induction; 4.0 µM zeatin for plantlet regeneration | Not specified | Not specified | Standard in vitro conditions | 92% embryogenic callus induction with thidiazuron, enhanced plantlet regeneration with 4.0 µM zeatin, confirmed somatic embryo development, and demonstrated increased flavonoids, anthocyanins, antioxidants, stress resistance. | [17] |
| Musa acuminata (Banana) | Leaf base explants | MS medium with high levels of BAP (up to 53.28 µM) and kinetin (55.80 µM) for shoot proliferation | Nanjanagudu Rasabale | Not specified | Standard in vitro conditions | High cytokinin levels (BAP and kinetin) produced up to 80 shoots per segment and ensuring genetic stability for conserving the endangered NR cultivar. | [33] |
| Saccharum officinarum (Sugarcane) | Callus cultures derived from embryogenic tissue | Callus induction medium supplemented with PVP, casein hydrolysate, MES buffer, and PEG 8000 for somatic embryogenesis | Co 86,032 and Q117 | Not specified | Standard in vitro conditions | Medium VI with PVP, casein hydrolysate, and MES buffer induced 79.66% callus in Co 86,032 and 82.83% in Q117, with PEG 8000 enhancing somatic embryogenesis and minimal somaclonal variation. | [77] |
| Dendrobium fimbriatum (Orchid) | Not specified | Mitra medium supplemented with KN (0.8–4.8 mg/L), IBA, NAA, or BAP for regeneration | Not specified | Not specified | Standard in vitro conditions | Effective shoot formation was achieved with KN, and root development with IBA and BAP/KN combinations, while low genetic polymorphism was observed. | [81] |
| Oryza sativa (Rice) | Callus cultures from various explants | Not specified | Not specified | More prolonged incubation (67 days) and shorter (28 days) | Standard in vitro conditions | Regenerants from more extended callus incubation periods (67 days) exhibited higher genetic instability, with DNA polymorphisms and methylation changes linked to somaclonal variation in both structural and housekeeping genes. | [76] |
| Rubus idaeus (Red Raspberry) | Leaf segments | Liquid medium + 2.3–9.0 μM thidiazuron (TDZ) for regeneration, 4.4 μM BA for shoot elongation, PGRs for rooting | ‘Latham’, ‘Heritage’, ‘Festival’, ‘Nova’ | Not specified | Bioreactor system, controlled field conditions | Tissue culture plants of ‘Latham’ and ‘Festival’ outperformed root cutting (RC) plants in cane and berry yields, with clonal fidelity observed. In contrast, bioreactor culture influenced juvenile branching, promoting growth and berry production. | [82] |
| Malus domestica (Apple Rootstock MM 104) | Axillary buds | MS basal medium, 3.0% sucrose, 0.8% agar, BA (5.0 µM), NAA (1.0 µM) for bud establishment, PGRs BA (1.0 µM) + NAA (1.0 µM) for shoot multiplication, IBA (0.1 µM) for rooting | MM 104 | Not specified | 5-day dark period for rooting, acclimatization in vermiculite: perlite: sand: soil (2:2:1:1) mix | High shoot multiplication (100%) with 9.8 shoots/explant and 76% survival under field conditions. | [83] |
| Citrus mitis (Calamondin) | Style-stigma explants | BAP (6 µM), 4-CPPU, PBU, 2,3-MDPU (12 µM each), hormone-free (HF) conditions | Not specified | Not specified | In vitro culture conditions | Phenylurea derivatives (4-CPPU, PBU, 2,3-MDPU) enhanced embryogenic potential, with 2,3-MDPU and PBU causing 3.7% somaclonal variation, while BAP/HF medium showed no variability | [70] |
| Coffea arabica (Coffee) | Foliar explants | MS + 0.4–0.6 mg/L 2,4-D for callus induction; IAA and Kn also tested | S.4202, S.4932 | Not specified | In vitro culture followed by field testing | Efficient somatic embryogenesis was achieved in both hybrids, with SRAP and SCoT marker analyses showing high genetic similarity to mother plants. | [71] |
| Aloe vera (Aloe vera) | Axillary buds (direct); Inflorescence base (indirect via callus) | Not specified | Sweet genotype | Not specified | In vitro regeneration with and without callus | Direct regeneration from axillary buds showed 0% polymorphism (true to type), while indirect regeneration via callus from the inflorescence base exhibited 80% polymorphism. | [84] |
| Ficus palmata (Fig) | Leaves and stems | MS basal medium, 2,4-D (2.0 mg/L) + Kin (0.2 mg/L) | Not specifically named | Not mentioned | Gamma radiation at 0, 20, 30, 40 Gy | Highest callus induction at 30–40 Gy. A total of 99.58% polymorphism due to gamma treatment, yet tissue culture plants remained genetically similar to mother plants. GC-MS revealed increased secondary metabolites at 30 and 40 Gy. | [78] |
| Carica papaya (Papaya) | Apical, nodal, petiole, leaf, and root segments | MS basal medium, BAP, Zeatin, NAA (direct); MS + BAP (1.0 mg/L), TDZ (0.3 mg/L), NAA (0.10 mg/L), 30 g/L sucrose (indirect) | Red Lady 786 | Not specified | Standard in vitro conditions | Direct regeneration with 65–88% success; indirect regeneration with 75–85% somatic embryogenesis; efficient medium for somatic embryos with TDZ, BAP, NAA, and sucrose. | [85] |
| Stevia rebaudiana (Candy leaf) | Not specified | MS + BAP (4.0 mg/L), NAA (2.0 mg/L), 2,4-D (2.0 mg/L) | Not specified | 6 (Six) subcultures | Standard in vitro conditions | Indirect organogenesis induced somaclonal variation, improving rebaudioside A content and Rebaudioside A/stevioside ratio, with SCoT analysis showing genetic variation, while subculturing had no effect on genotype or glycoside profile. | [86] |
| Magnolia dealbata (Cloudforest magnolia) | Not specified (embryogenic tissues) | MS medium + 2,4-D (2.26 or 4.52 μM) | Not specified | 2 cycles of secondary somatic embryogenesis | Standard in vitro conditions | Highest embryo yield at 4.52 μM 2,4-D. Genetic similarity between donor and regenerants was 0.90, indicating low somaclonal variation. | [87] |
| Hevea brasiliensis (Rubber tree) | Mixed floral explant | MS + 2.0 mg/L BA + 1.5 mg/L 2,4-D; for germination: MS + 0.25 mg/L GA3 | Early introduced clone (white root disease resistant) | 3 passages (4 weeks each) | Standard in vitro conditions | Highest SE induction (39.84%) and 3.25 cotyledonary embryos per callus. A total of 50% of somatic embryos developed embryonic axes, 25% formed shoots, and confirmed genetic stability. | [88] |
| Alhagi maurorum (Camelthorn) | Leaf (direct), hypocotyl, cotyledon, root (indirect) | MS + BA, IBA or NAA (0.2 or 0.5 mg/L); rooting on ½ MS ± IBA (0.2–2.0 mg/L) | Not specified | Not specified | Standard in vitro conditions | Direct organogenesis produced high chlorogenic acid, while indirect regeneration via callus yielded higher 20-HE content, with genetic similarity ranging from 0.765–0.947. | [89] |
| Musa acuminata (Banana) | Individual shoot apex; transverse sections | MS + 10, 20, or 30 µM BAP + 1.0 µM NAA (proliferation); MS + 6 µM IBA (rooting) | ‘Grand Naine’ | Monthly subcultures for 6 months | Standard in vitro conditions | Transverse sectioning of shoot apex improved shoot production (up to 32.77 shoots/section in 6 months). No genetic variation in plants from transverse sections; 23.46% polymorphism detected in plants from whole shoot apex. | [90] |
| Vanilla planifolia (Vanilla) | Immature seeds | MS + 2.27 µM TDZ (callus induction); MS + 8.88 µM BA (shoot regeneration); MS without PGRs (rooting) | ‘Vanilla planifolia’ | Indirect organogenesis (not specified) | Standard in vitro conditions | Transverse sectioning of the shoot apex improved shoot production (up to 32.77 shoots/section in 6 months), with no genetic variation in plants from transverse sections. | [91] |
| Zea mays (Maize) | Callus culture | MS medium | Multiple independent lines | Multiple cycles | Standard in vitro conditions | DNA methylation changes were observed at specific loci in tissue culture, including gains and losses of methylation, primarily affecting CG and CHG contexts, with some methylation changes being heritable. | [92] |
| Vaccinium vitis-idaea (Lingonberry) | Leaf, Node, and Cutting explants | Zeatin-induced media; Liquid medium (NC1), Semi-solid medium (NC2), Node culture-derived (NC3), Leaf culture-derived (LC1) | Erntedank cultivar | Multiple subcultures, both in vitro and ex vitro | Tissue culture conditions | Highest methylation was observed in culture-derived plants) with 108 methylation bands, NC1, NC2, NC3, and LC1 showed higher methylation than cutting-propagated (ED) plants (79 bands), with higher secondary metabolites than micropropagated shoots and plants. | [93] |
| Coffea arabica (Coffee) | Embryogenic suspensions | Low 2,4-D concentrations, short proliferation periods | Hybrids | 6 months of culture | Field plots for phenotypic assessment | Somaclonal variation was very low (0.74%), with minimal genetic polymorphism, chromosome loss in rare variants, and no phenotype-MSAP pattern correlation. | [94] |
| X Triticosecale spp. (Winter triticale) | Explants from cultured tissues | Cu2+ (0.1, 5, 10 µM), Ag+ (0, 2, 10 µM), MS medium modifications | Not specified | 9 different conditions tested | Incubation time: 35, 42, 49 days | 51% tissue culture-induced variation was observed, and variation was influenced by culture medium composition. | [79] |
| Lowbush Blueberry (Blue berry) | Softwood cutting (SC), Tissue culture (TC) | Standard tissue culture medium | Wild clone QB9C, cultivar Fundy | Not specified | Conventional propagation for SC, in vitro culture for TC | Tissue culture plants showed higher DNA methylation (29% in TC QB9C, 20% in TC Fundy) than SC plants (25% and 19%, respectively), with polymorphism detected only in TC plants. | [95] |
| Fragaria x ananassa (Strawberry) | Shoot tips | Standard tissue culture medium | Fragaria nilgerrensis | 6 stages (explants to acclimation) | Tissue culture conditions | Dedifferentiation/redifferentiation stages affecting genes in hormone metabolism, development, and stress response, are linked to epigenetic variation. | [96] |
| Oryza sativa (Rice) | Somaclonal line (TC-reg-2008) | Standard tissue culture medium | Oryza sativa (cv. Hitomebore) | Extensive selfing of somaclonal line | Normal growing conditions and abiotic stress conditions | Tissue culture extensively induced heritable genomic variation, non-randomly distributed across 12 chromosomes, affecting functional genes with stress-responsive phenotypic effects, linked to transposable element mobilization and DNA methylation changes. | [80] |
| Ananas comosus (Pineapple) | Callus culture-derived plants | Standard tissue culture medium | Ananas comosus | Not specified | Controlled in vitro conditions | DNA methylation changes observed in somaclonal variation (SV) plants, with significant differences in methylation patterns between SV plants and cutting seedlings (CK). | [97] |
| Hordeum vulgare (Barley) | Somatic embryos | Tissue culture media with copper and silver ions | Hordeum vulgare | Not specified | In vitro conditions | DNA methylation changes observed in barley regenerants. The study optimized ion concentrations and culture duration to minimize or maximize tissue culture-induced variation. | [98] |
| Dendrocalamus asper (Giant Bamboo) | Shoot tips, plantlets | MS, MSR media with modified phosphorus, sucrose, salts, and mycorrhizal inoculation with Rhizoglomus clarum | Dendrocalamus asper | 3 subcultures | Standard in vitro conditions | BAP promoted shoot multiplication with decreased DNA methylation in the third subculture, while mycorrhization occurred only in MSR and MS/2 media, offering insights into methylation. | [99] |
| Zea mays (Maize) | Embryo tissues | Standard maize tissue culture media, no specific PGRs mentioned | Zea mays A188 | Not specified | Standard in vitro conditions | Altered DNA methylation patterns were found in 39% of progeny lines, primarily due to stable, heritable demethylation. | [100] |
| Lactuca sativa (Lettuce) | Various accessions (major cultivars and wild relatives) | Not specified | Major lettuce cultivars, wild relatives | Not specified | Controlled environment | Lettuce domestication led to a significant increase in DNA methylation, with epigenetic variations linked to leafy and stem types, influencing gene expression and chromatin accessibility and contributing to their divergence. | [101] |
| Coffea canephora (Robusta coffee) | Somatic tissues for Somatic Embryo (SE) | Not specified; pharmacological inhibitor (5-Azacytidine) used | Not specified | Not specified | SE induction and maturation phases; use of epigenetic inhibitor | Dynamic changes in DNA methylation and histone modifications accompany SE progression. | [102] |
| Plant Species | Tissue Culture Method/Explants | Detection Method | Key Findings | Reference |
|---|---|---|---|---|
| Citrus jambhiri (Rough lemon) | Somatic embryogenesis | RAPD markers | Plantlets regenerated from nucellar tissues showed no variation, while those raised from ovaries showed variation in 20–30%. | [31] |
| Populus alba (White Poplar) | Organogenesis | RAPD markers | Highlighting 38.33% genetic instability in tissue culture-derived plants. | [32] |
| Saccharum officinarum (Sugarcane) | Callus and shoot culture | ISSR and RAPD markers | Found genetic variation in sugarcane callus cultures | [33] |
| Dendrobium fimbriatum (Orchid) | Micropropagation | RAPD, ISSR, and SCoT markers | Orchids grown in basal medium without phytohormones showed 100% monomorphism, while those in hormone-enriched media showed low genetic polymorphism. | [81] |
| Vaccinium vitis-idaea (Lingonberry) | Somatic embryogenesis/leaf explants | EST-SSR and GSSR markers | The genetic stability observed | [17] |
| Musa spp. (Banana) | Micropropagation | ISSR and RAPD markers | Detected somaclonal variation among regenerants emphasized the need for genetic fidelity assessment in micropropagation. | [33] |
| Artemisia annua (Sweet wormwood) | Callus culture/apical shoots, nodal segments | qPCR, SCAR, and RAPD markers | Identified genetic differences between in vitro and field-grown plants, affecting high artemisinin production. | [132] |
| Solanum tuberosum (Potato) | Callus culture/Internode explants | qPCR | Enhanced starch accumulation | [133] |
| Glycine max (Soybean) | Root tissue callus culture | RFLP markers | Tissue cultures developed RFLP allelic differences; new alleles matched those in other cultivars, suggesting recombinational events. | [118] |
| Oryza sativa (Rice) | Callus culture/ leaf explants | RFLP markers | Higher genetic instability with more extended callus incubation periods; correlation with methylation changes. | [76] |
| Malus domestica (Apple) | Organogenesis | ISSR and SCoT markers | High genetic fidelity with 98.43% monomorphic bands; minimal somaclonal variation detected. | [83] |
| Citrus madurensis (Calamondin) | Callus culture | SSR, RAPD markers | Diphenylurea derivatives enhance somatic embryogenesis in Citrus madurensis but compromise genetic stability, unlike BAP or Hormone Free media, which maintain clonal fidelity. | [70] |
| Prunus persica (Peach) | Callus culture/ cotyledon | RAPD markers | High somaclonal variation was observed in regenerants. | [121] |
| Fragaria × ananassa (Strawberry) | Micropropagation/shoot tips | qPCR | Somaclonal variation was observed | [134] |
| Ficus carica (Fig) | Callus culture/leaf and stem explants | ISSR markers | High genetic variation observed | [78] |
| Elaeis guineensis (African oil palm) | Callus culture | SSR and RAPD markers | Genetic variation found in oil palm regenerants. SSR and RAPD markers detected polymorphisms linked to culture conditions and subculturing practices. | [135] |
| Bixa Orellana (lipstick tree) | Organogenesis/ nodal explants | ISSR and RAPD markers | The results of the RAPD marker system revealed the genetic stability among the micropropagated plants. | [136] |
| Rubus ideals (Raspberry) | Direct organogenesis through bioreactor leaf segments | SSR markers | Genetically stable. | [82] |
| Aloe vera (Aloe) | Organogenesis/axillary shoot buds callus culture/inflorescence axis | ISSR markers, and RAPD | Plantlets produced through indirect organogenesis exhibited considerable variation, while those generated via direct organogenesis showed complete uniformity. | [84] |
| Coffea arabica (Coffee) | Somatic embryogenesis | SRAP and SCoT markers | 98% and 99% genetic similarities, respectively, between the regenerated and mother plants. | [94] |
| Phoenix dactylifera (Date palm) | Callus culture/ shoot tips and axillary shoot meristems | RAPD markers | No variation observed. | [44] |
| Corylus avellana (Hazel) | Embryogenesis | ISSR and RAPD markers | Identified genetic differences between in vitro and field-grown hazelnuts, affecting nut quality and tree height. | [137] |
| Caladium × hortulanum (Caladium) | Callus culture/leaf culture | SSR markers, and flow cytometry analysis | Somaclonal variation was high in in vitro-cultured caladium aneuploids, with tetraploid aneuploid caladium exhibiting the greatest variability | [138] |
| Carica papaya (Papaya) | Somatic embryogenesis | RAPD and ISSR markers | RAPD and ISSR markers for detecting genetic fidelity | [85] |
| Tylophora indica (Indian Ipecac) | indirect, direct and somatic embryo | SCoT marker, and flow cytometry | Determination of 2C DNA content and verification of genetic uniformity using SCoT molecular markers. | [131] |
| Cannabis sativa (Hemp) | Callus culture | SSR markers | No variation observed. | [128] |
| Musa rubra (Bronze Banana) | Micropropagation/shoot tips | ISSR markers | Genetic stability observes. | [139] |
| Vaccinium vitis-idaea (Lingonberry) | Micropropagation/shoot tips | EST and SSR primers | Genetic equality observed. | [129] |
| Hevea brasiliensis (Rubber) | Somatic embryogenesis/ flower explants | RAPD and SSR markers | No genetic variation was detected between the mother and in vitro plantlets, as indicated by RAPD and SSR marker analysis. | [88] |
| Psidium guajava (guava) | Somatic embryogenesis/zygotic embryo | RAPD, ISSR or SSR markers | Approximately 99% of bands were monomorphic. | [122] |
| Vanilla planifolia (Vanilla) | Callus culture/immature capsules | ISSR marker and Phenotypic observation | Molecular analysis of regenerated plantlets revealed 71.66% genetic polymorphism. | [91] |
| Musa acuminata (Banana) | Organogenesis/shoot apices | RAPD markers | Molecular analysis showed 23.46% polymorphism, whereas transverse sections confirmed genetic uniformity with the parent plants. | [90] |
| Hypericum gaitii (St. John’s wort) | Organogenesis/apical and axillary meristems | ISSR markers | No polymorphism among the micropropagated plants and mother plants. | [140] |
| Saccharum officinarum (Sugarcane) | Organogenesis/shoot tips | SCoT markers | Genetic stability observed. | [141] |
| Rhaponticum carthamoides (Maral root) | Direct organogenesis/leaf explants | flow cytometry, RAPD, and ISSR markers | Genetic stability observed. | [89] |
| Curcuma angustifolia (Indian Arrowroot) | Organogenesis/shoot tips | EST-ISSR markers | No variation observed. | [124] |
| Artemisia vulgaris (Mugwort) | Organogenesis/ nodal explants | SCoT, ISSR markers and DNA barcoding | Genetic stability observed. | [142] |
| Cicer arietinum (Chickpea) | Organogenesis/ embryo axis, half-seed, axillary meristem, and cotyledonary node explant | SCoT and ISSR markers | No polymorphism was detected. | [125] |
| Magnolia dealbata | Somatic embryogenesis from zygotic embryo | SSR | Genetic integrity 90%. | [87] |
| Cymbidium aloifolium (Cymbidium) | Organogenesis/seeds | SCoT, DAMD, and ISSR markers | The analysis of the in vitro-derived plantlets showed 86.87% genetic monomorphism and 13.13% polymorphism. | [143] |
| Rhynchostylis retusa (Punjab fig) | Organogenesis/ Capsule | RAPD | High uniformity was observed among regenerated plants and mother plants. | [144] |
| Plant Species | Tissue Culture Method | DNA Methylation Detection | Variation Observed | Reference |
|---|---|---|---|---|
| Zea mays (Maize) | Embryogenic callus culture | Bisulfite sequencing | Variation observed | [92] |
| Allium sativum (Garlic) | Direct Organogenesis, meristem tissue | AFLP, and MASP | Genetic and epigenetic polymorphism under field growing conditions | [34] |
| Vaccinium vitis-idaea (Lingonberry) | Organogenesis/ Stem and leaf explants | MSAP | More methylation events are observed in vitro-derived plants than in those derived from cuttings of plants | [93] |
| Coffea arabica (Coffee) | Somatic embryogenesis regeneration/ Nodal segments | MASP | Genetically stable | [94] |
| X Triticosecale spp. (Winter triticale) | Another Culture, Anther | metAFLP | 51% of tissue culture-induced variation | [156] |
| Fragaria x ananassa (Strawberry) | Indirect organogenesis, runner tips | WGS | Variation observed | [96] |
| Oryza sativa (Rice) | Callus culture | Bisulfite sequencing | Variation observed | [80] |
| Ananas comosus (Pineapple) | Callus culture | WGBS | Variation observed | [97] |
| Malus domestica (Apple) | Shoot culture | Bisulfite sequencing | Variation observed | [153] |
| Humulus lupulus (Hop) | Direct organogenesis | MASP | No variation observed | [157] |
| Hordeum vulgare (Barley) | Somatic embryogenesis | Met-AFLP | Polymorphism observed | [98] |
| Manihot esculenta (Cassava) | Meristem micropropagation | MASP | Variation observed | [158] |
| Hordeum vulgare (Barley) | Micropropagation | HPLC | No variation observed | [159] |
| Cannabis sativa | Organogenesis | Global epigenetic analysis | DNA hypomethylation progressively occurred in micropropagated shoots compared to plants grown in the greenhouse, with the level of hypomethylation rising over extended periods of in vitro culture, potentially impacting gene expression and plant development | [160] |
| Vaccinium angustifolium (Lowbush Blueberry) | Micropropagation via softwood cuttings and tissue culture | MSAP | Variation observed | [95] |
| Saccharum spp. (Sugarcane) | Micropropagation | MSAP | Variation observed | [146] |
| Salix purpurea (Basket willow) | Regenerated shoots (in vitro) | WGBS | Extensive methylation reprogramming observed during regeneration | [161] |
| Dendrocalamus asper (Giant Bamboo) | In vitro shoots (multiple passages) | ELISA (5-mC quantification) | Methylation decreased with subculturing, linked to somaclonal variation | [99] |
| Zea mays (Maize) | Callus culture | MSAP and HPLC | No variation observed | [100] |
| Camellia sinensis (Tea) | Various tissues (leaves, roots, etc.) | HPLC with UV detection | Observed tissue-specific variations in DNA methylation levels across different tea plant tissues | [148] |
| Lactuca sativa (lettuce) | Callus culture | Global epigenetic analysis | Polymorphism observed | [101] |
| × Triticosecale (Triticale) | Anther culture | metAFLP | Variation observed | [162] |
| Common Name | Scientific Name | Mode of Regeneration | Somaclonal Variants | References |
|---|---|---|---|---|
| Rice | Oryza sativa | Indirect organogenesis | Drought resistant (cv PR113) | [35] |
| Tomato | Solanum lycopersicum | Indirect organogenesis | High-yielding variety (SE10, SE1, SS5) | [36] |
| Eggplant | Solanum melongena | Indirect organogenesis | Salt stress-tolerant variant | [37] |
| Cucumber | Cucumis sativus | Indirect organogenesis | More number of lateral shoots and the highest main shoot length (MSC 28) | [38] |
| Carrot | Daucus carota | Indirect organogenesis | Drought resistant | [39] |
| Pomegranate | Punica granatum | Indirect organogenesis (Callus formation) | Better fruit quality | [170] |
| Garden Tulip | Tulipa suaveolens | Direct organogenesis | Disease resistant | [169] |
| Banana | Musa sp. | Indirect organogenesis Callus formation (shoot tips) | Fusarium wilt-resistant variety | [40] |
| Japanese Mint | Mentha arvensis | Indirect organogenesis | Increased quality | [167] |
| Banana | Musa sp. | Direct organogenesis | Improved resistance to Fusarium wilt and better bunch quality | [171] |
| Cotton | Gossypium hirsutum | Direct organogenesis | Drought-tolerant variety | [172] |
| Potato | Solanum tuberosum | Direct organogenesis | Increased resistance to viral diseases and improved tuber quality | [168] |
| Tomato | Solanum lycopersicum | Indirect organogenesis | Improved fruit size, shape, and resistance to Fusarium wilt | [173] |
| Rice | Oryza sativa | Direct organogenesis | Enhanced resistance to rice blast and better grain quality | [35] |
| Sugarcane | Saccharum officinarum | Indirect organogenesis | Increased sugar yield, fiber quality, stalk length, internode length, and disease resistance | [165] |
| Grape | Vitis vinifera | Direct Organogenesis | Seedless fruit variety | [41] |
| Pineapple | Ananas comosus | Somatic embryogenesis | Improved fruit size and resistance to pests | [174] |
| Wheat | Triticum aestivum | Somatic embryogenesis | Increased resistance to wheat rust and spot blotch disease, enhanced grain size | [175] |
| Carnation | Dianthus caryophyllus | Indirect organogenesis | Resistant to Fusarium oxysporum | [176] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumder, S.; Igamberdiev, A.U.; Debnath, S.C. Somaclonal Variation and Clonal Fidelity in Commercial Micropropagation: Challenges and Perspectives. Agronomy 2025, 15, 1489. https://doi.org/10.3390/agronomy15061489
Majumder S, Igamberdiev AU, Debnath SC. Somaclonal Variation and Clonal Fidelity in Commercial Micropropagation: Challenges and Perspectives. Agronomy. 2025; 15(6):1489. https://doi.org/10.3390/agronomy15061489
Chicago/Turabian StyleMajumder, Sweety, Abir U. Igamberdiev, and Samir C. Debnath. 2025. "Somaclonal Variation and Clonal Fidelity in Commercial Micropropagation: Challenges and Perspectives" Agronomy 15, no. 6: 1489. https://doi.org/10.3390/agronomy15061489
APA StyleMajumder, S., Igamberdiev, A. U., & Debnath, S. C. (2025). Somaclonal Variation and Clonal Fidelity in Commercial Micropropagation: Challenges and Perspectives. Agronomy, 15(6), 1489. https://doi.org/10.3390/agronomy15061489








