Green Soybean’s Survival Mechanisms Under Cold Stress: A Transcriptomic Perspective
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Cold Stress Treatment
2.2. cDNA Libsrary Preparation and Sequencing
2.3. De Novo Transcriptome Assembly
2.4. Transcriptome Data Analysis
3. Results
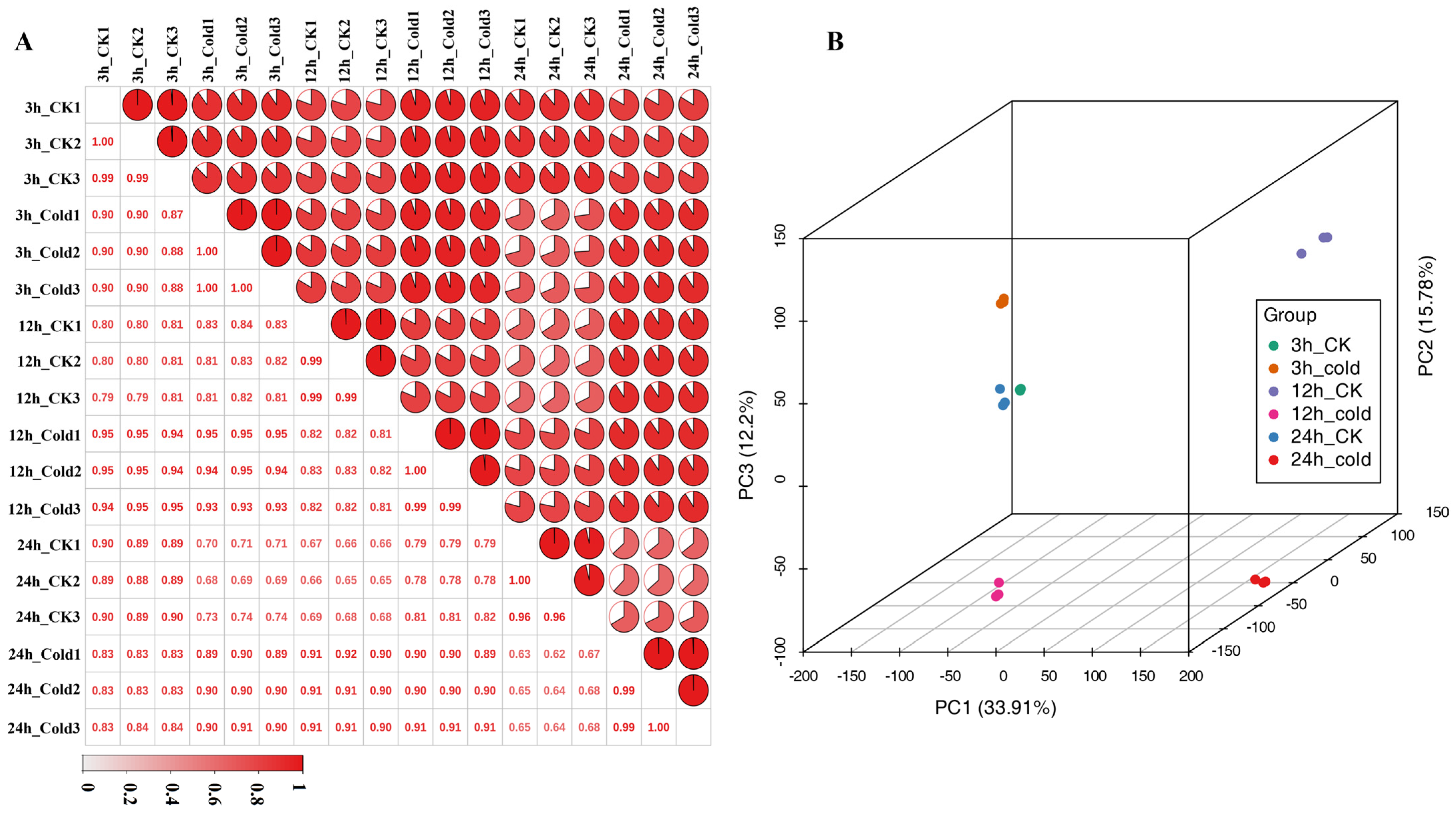
3.1. Transcriptome Sequencing and Annotation of Green Soybean
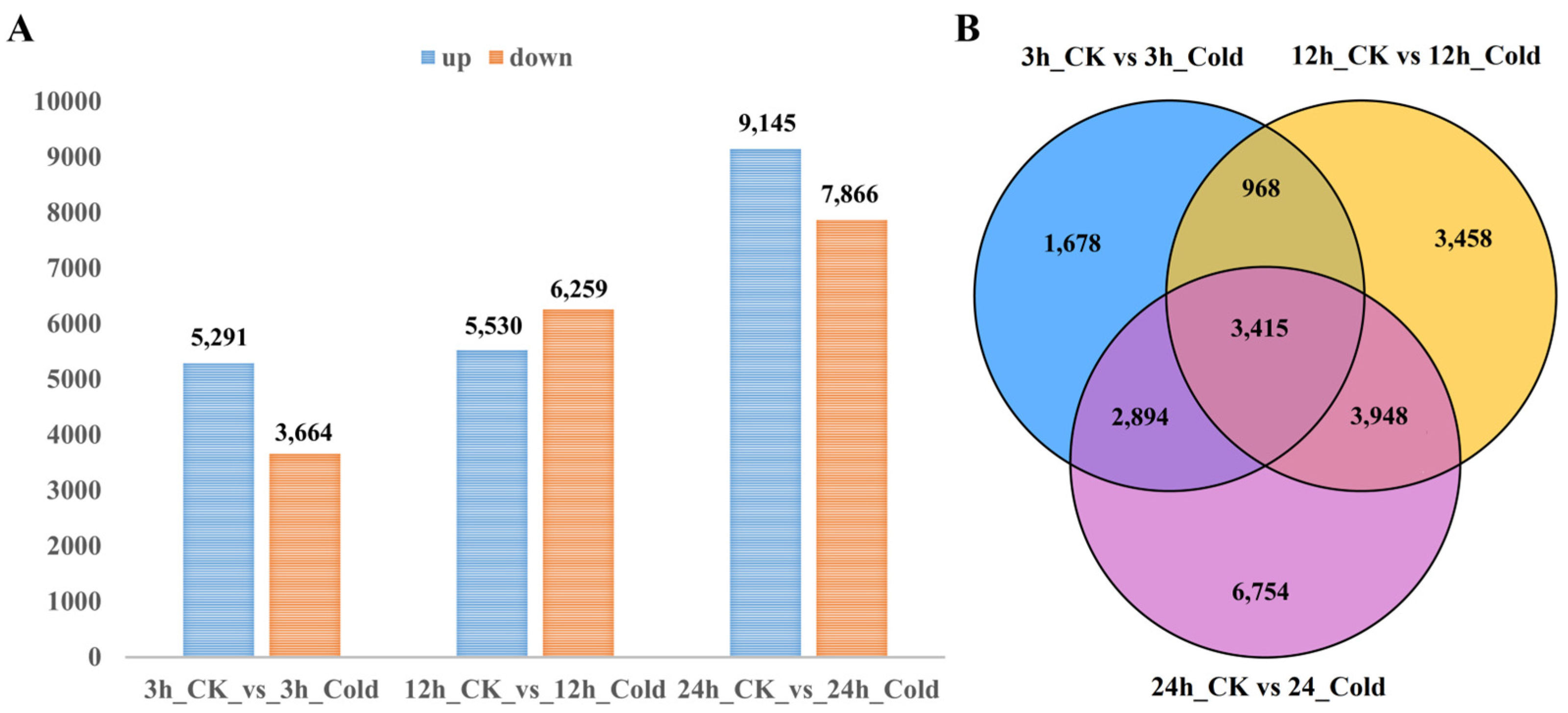
3.2. Differentially Expressed Genes of Soybean Under Cold Stress
3.3. Transcription Factor Analysis
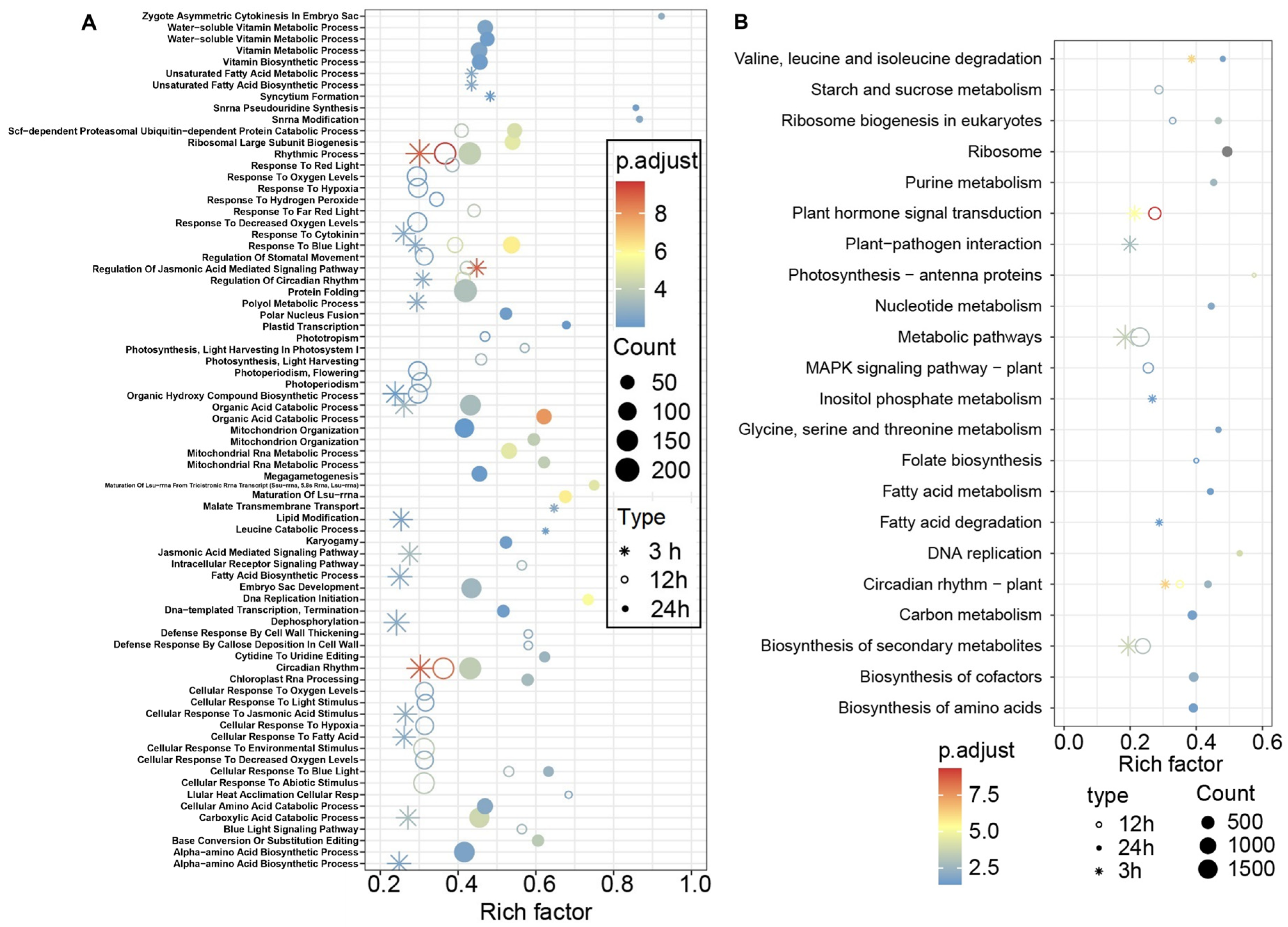
3.4. GO and KEGG Enrichment Analysis of the DEGs
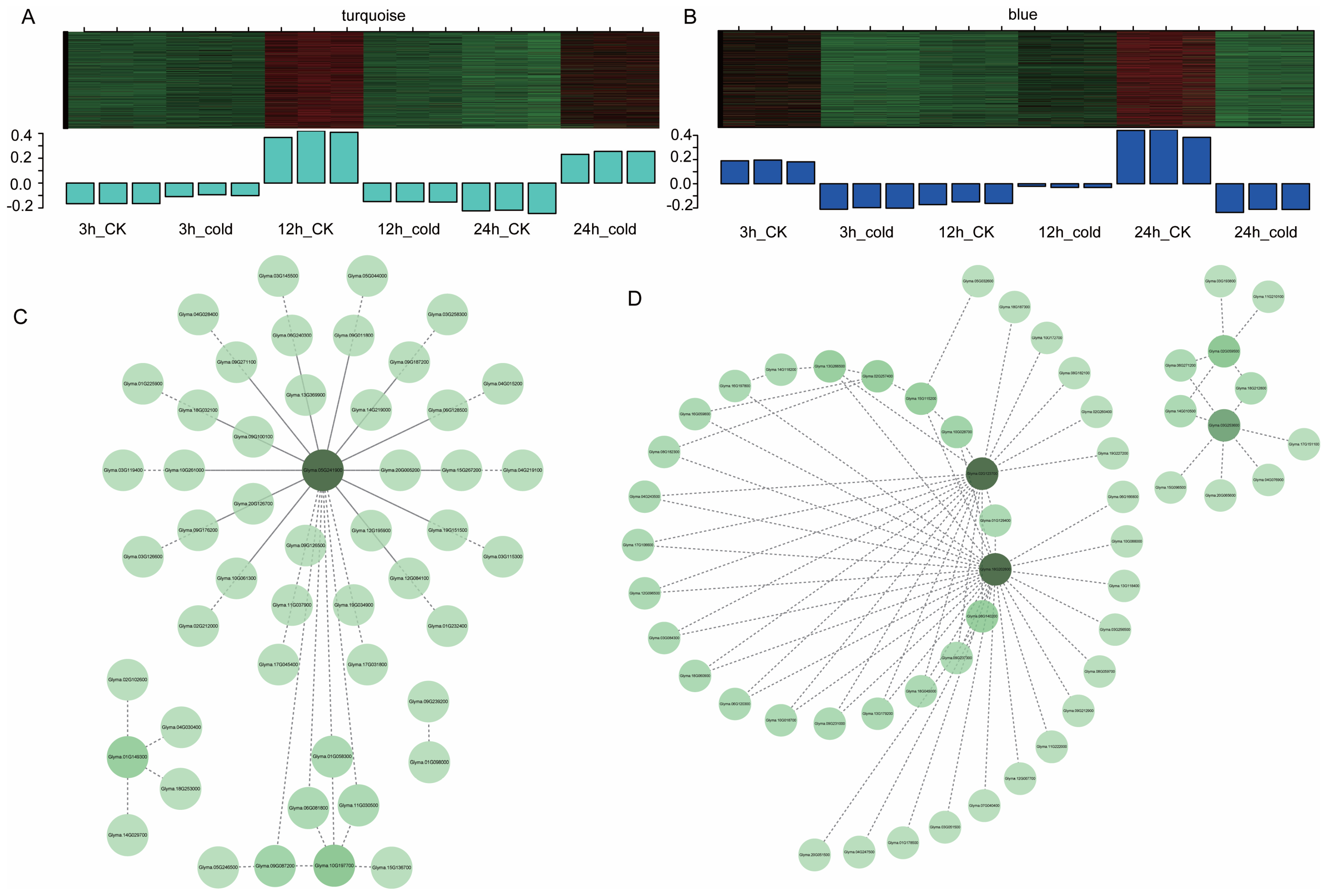
3.5. Gene Co-Expression Modules Responding to Cold Stress
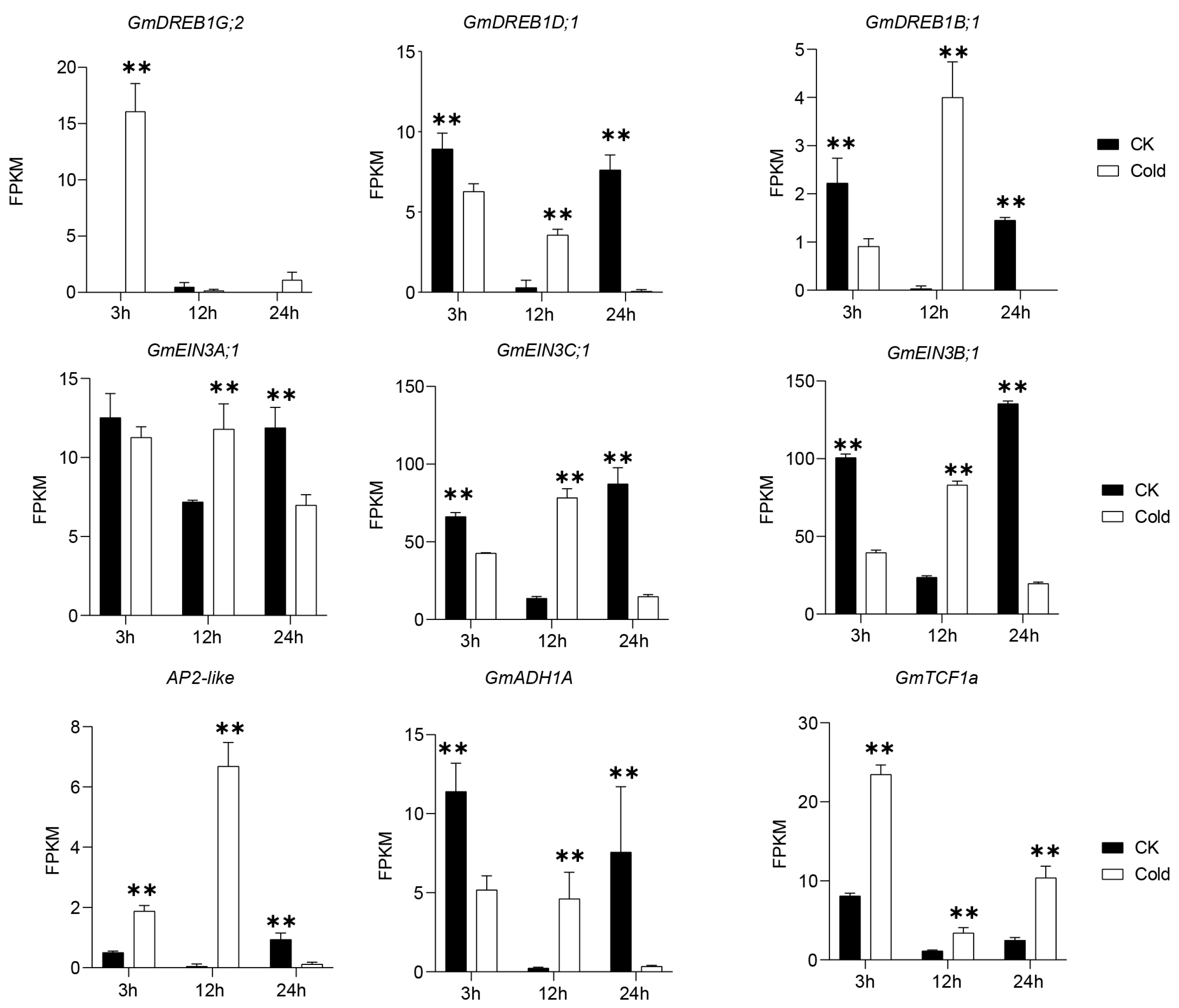
3.6. Identification of Key Genes Involved in Cold Responses in Green Soybean
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Di, D.; He, S.; Zhang, R.; Gao, K.; Qiu, M.; Li, X.; Sun, H.; Xue, S.; Shi, J. Exploring the Dual Role of Anti-Nutritional Factors in Soybeans: A Comprehensive Analysis of Health Risks and Benefits. Crit. Rev. Food Sci. Nutr. 2024, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.M.; Boddepalli, V.N.; Yan, M.-R.; Kumar, V.; Gill, B.; Pan, R.S.; Wang, C.; Hartman, G.L.; Silva E Souza, R.; Somta, P. Global Status of Vegetable Soybean. Plants 2023, 12, 609. [Google Scholar] [CrossRef]
- Bianchini, A.; Dutra de Moraes, P.V.; Domanski Jakubski, J.; Adami, P.F.; Bernardi Rankrape, C.; Rossi, P. Influence of Cover Crops with Allelopathic Potential and Their Reduction of Herbicide Use for Soybean Productivity. J. Environ. Sci. Health Part B 2022, 57, 890–896. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Review: Proteomic Techniques for the Development of Flood-Tolerant Soybean. Int. J. Mol. Sci. 2020, 21, 7497. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, K.; Zhu, Q.; Zhang, B.; Li, M.; Li, S. Analysis of Shoot Architecture Traits in Edamame Reveals Potential Strategies to Improve Harvest Efficiency. Front. Plant Sci. 2021, 12, 614926. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, G.; Wu, F.; Zhou, X.; Zhou, Y.; Xu, K.; Li, Y.; Zhang, W.; Zhao, K.; Jing, Y.; et al. Genome-Wide Association Analysis of Yield-Related Traits and Candidate Genes in Vegetable Soybean. Plants 2024, 13, 1442. [Google Scholar] [CrossRef]
- Moseley, D.; da Silva, M.P.; Mozzoni, L.; Orazaly, M.; Florez-Palacios, L.; Acuña, A.; Wu, C.; Chen, P. Effect of Planting Date and Cultivar Maturity in Edamame Quality and Harvest Window. Front. Plant Sci. 2021, 11, 585856. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-F.; He, S.-S.; Wang, C.-S.; Lai, Z.-Y.; Lin, D.-G.; Chen, S. A Modified Roger’s Distance Algorithm for Mixed Quantitative-Qualitative Phenotypes to Establish a Core Collection for Taiwanese Vegetable Soybeans. Front. Plant Sci. 2021, 11, 612106. [Google Scholar] [CrossRef]
- Wilkes, J.E.; Kirkpatrick, T.L. The Effects of Meloidogyne Incognita and Heterodera Glycines on the Yield and Quality of Edamame (Glycine max l.) in Arkansas. J. Nematol. 2020, 52, e2020-12. [Google Scholar] [CrossRef]
- Iman, M.N.; Irdiani, R.; Rahmawati, D.; Fukusaki, E.; Putri, S.P. Improvement of the Functional Value of Green Soybean (Edamame) Using Germination and Tempe Fermentation: A Comparative Metabolomics Study. J. Biosci. Bioeng. 2023, 136, 205–212. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Senga, E.F.B.; Wang, D.Y. Vegetable Soy Bean (Glycine max (L.) Merrill) from Production to Processing. Outlook Agric. 2005, 34, 167–172. [Google Scholar] [CrossRef]
- Wszelaki, A.L.; Delwiche, J.F.; Walker, S.D.; Liggett, R.E.; Miller, S.A.; Kleinhenz, M.D. Consumer Liking and Descriptive Analysis of Six Varieties of Organically Grown Edamame-Type Soybean. Food Qual. Prefer. 2005, 16, 651–658. [Google Scholar] [CrossRef]
- Mozzoni, L.A.; Chen, P.; Morawicki, R.O.; Hettiarachchy, N.S.; Brye, K.R.; Mauromoustakos, A. Quality Attributes of Vegetable Soybean as a Function of Boiling Time and Condition. Int. J. Food Sci. Technol. 2009, 44, 2089–2099. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, W.; Shin, D.; Im, H.; Jung, G.; Lee, Y.-B.; Choi, J. Genomic and Metabolomic Analysis of Latilactobacillus sakei DCF0720 for Black Soybean Yogurt Fermentation. Int. J. Food Microbiol. 2024, 425, 110897. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Lan, T.; Liu, Z.; Xu, Z.; Jiang, L.; Zhang, Y.; Sui, X. Shear, Extensional Rheology, and Tribology of Polysaccharide-Thickened Soy Protein-Based Liquid Systems for Dysphagia Management. Food Chem. 2025, 463 Pt 1, 141145. [Google Scholar] [CrossRef]
- Zheng, L.; Regenstein, J.M.; Teng, F.; Li, Y. Tofu Products: A Review of Their Raw Materials, Processing Conditions, and Packaging. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3683–3714. [Google Scholar] [CrossRef]
- Gawda, D.; Nowak, A.; Haliniarz, M.; Woniak, A. Yield and Economic Effectiveness of Soybean Grown Under Different Cropping Systems. Int. J. Plant Prod. 2020, 14, 475–485. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Chin, K.L.; Qi, Y. Vegetable Soybean: Seed Composition and Production Research. Ital. J. Agron. 2017, 12, 872. [Google Scholar] [CrossRef]
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic Stress in Crop Production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, J.; Zhao, L.; Wu, H.; Zhu, Y.; Ahmad, I.; Zhou, G. Abiotic Stress Responses in Crop Plants: A Multi-Scale Approach. J. Integr. Agric. 2024, in press. [CrossRef]
- Esmaeili, N.; Shen, G.; Zhang, H. Genetic Manipulation for Abiotic Stress Resistance Traits in Crops. Front. Plant Sci. 2022, 13, 1011985. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant Abiotic Stress Response and Nutrient Use Efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Villalobos-López, M.A.; Arroyo-Becerra, A.; Quintero-Jiménez, A.; Iturriaga, G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. Int. J. Mol. Sci. 2022, 23, 12053. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and Challenges in Uncovering Cold Tolerance Regulatory Mechanisms in Plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Ermilova, E. Cold Stress Response: An Overview in Chlamydomonas. Front. Plant Sci. 2020, 11, 569437. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, D.; Yan, R.; Xu, Y.; Xing, M.; Liao, S.; Wan, C.; Chen, C.; Zhu, L.; Kai, W.; et al. Amelioration of Chilling Injury by Fucoidan in Cold-Stored Cucumber via Membrane Lipid Metabolism Regulation. Foods 2023, 12, 301. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and Biotic Stress Combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory Networks in Plant Responses to Drought and Cold Stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef]
- Kao, P.-H.; Baiya, S.; Lai, Z.-Y.; Huang, C.-M.; Jhan, L.-H.; Lin, C.-J.; Lai, Y.-S.; Kao, C.-F. An Advanced Systems Biology Framework of Feature Engineering for Cold Tolerance Genes Discovery from Integrated Omics and Non-Omics Data in Soybean. Front. Plant Sci. 2022, 13, 1019709. [Google Scholar] [CrossRef]
- Wang, X.; Ding, Y.; Li, Z.; Shi, Y.; Wang, J.; Hua, J.; Gong, Z.; Zhou, J.-M.; Yang, S. PUB25 and PUB26 Promote Plant Freezing Tolerance by Degrading the Cold Signaling Negative Regulator MYB15. Dev. Cell 2019, 51, 222–235. [Google Scholar] [CrossRef]
- Mehrotra, S.; Verma, S.; Kumar, S.; Kumari, S.; Mishra, B.N. Transcriptional Regulation and Signalling of Cold Stress Response in Plants: An Overview of Current Understanding. Environ. Exp. Bot. 2020, 180, 104243. [Google Scholar] [CrossRef]
- Williams, M.M.; Zhang, B.; Fu, X.; Ross, J. Editorial: Everything Edamame: Biology, Production, Nutrition, Sensory and Economics. Front. Plant Sci. 2022, 13, 976008. [Google Scholar] [CrossRef] [PubMed]
- Bandara, A.Y.; Weerasooriya, D.K.; Bell, T.H.; Esker, P.D. Prospects of alleviating early planting-associated cold susceptibility of soybean using microbes: New insights from microbiome analysis. J. Agron. Crop Sci. 2021, 207, 171–185. [Google Scholar] [CrossRef]
- Tsegaw, M.; Zegeye, W.A.; Jiang, B.; Sun, S.; Yuan, S.; Han, T.; Wu, T. Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants 2023, 12, 459. [Google Scholar] [CrossRef] [PubMed]
- Muncan, J.; Jinendra, B.M.S.; Kuroki, S.; Tsenkova, R. Aquaphotomics Research of Cold Stress in Soybean Cultivars with Different Stress Tolerance Ability: Early Detection of Cold Stress Response. Molecules 2022, 27, 744. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Shi, Y.; Liu, Z.; Zhang, X.; Gong, Z.; Yang, S. Strigolactones Promote Plant Freezing Tolerance by Releasing the WRKY41-Mediated Inhibition of CBF/DREB1 Expression. EMBO J. 2023, 42, e112999. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Han, D.; Yang, Q.; Li, C.; Shi, X.; Zhang, M.; Yang, C.; Qiu, L.; Jia, H.; et al. Cold Tolerance SNPs and Candidate Gene Mining in the Soybean Germination Stage Based on Genome-Wide Association Analysis. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2024, 137, 178. [Google Scholar] [CrossRef]
- Hussain, M.A.; Li, S.; Gao, H.; Feng, C.; Sun, P.; Sui, X.; Jing, Y.; Xu, K.; Zhou, Y.; Zhang, W.; et al. Comparative Analysis of Physiological Variations and Genetic Architecture for Cold Stress Response in Soybean Germplasm. Front. Plant Sci. 2023, 13, 1095335. [Google Scholar] [CrossRef]
- Robison, J.D.; Yamasaki, Y.; Randall, S.K. The Ethylene Signaling Pathway Negatively Impacts CBF/DREB-Regulated Cold Response in Soybean (Glycine max). Front. Plant Sci. 2019, 10, 121. [Google Scholar] [CrossRef]
- Yang, M.; Teng, Y.; Yue, T.; Wang, Z.; Feng, G.; Ruan, J.; Yan, S.; Zheng, Y.; Zhang, L.; Chen, Q.; et al. The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance. Plants 2023, 12, 2928. [Google Scholar] [CrossRef]
- Shen, J.; Xu, Y.; Yuan, S.; Jin, F.; Huang, Y.; Chen, H.; Shan, Z.; Yang, Z.; Chen, S.; Zhou, X.; et al. Genome-Wide Identification of GmSPS Gene Family in Soybean and Expression Analysis in Response to Cold Stress. Int. J. Mol. Sci. 2023, 24, 12878. [Google Scholar] [CrossRef] [PubMed]
- Haidar, S.; Lackey, S.; Charette, M.; Yoosefzadeh-Najafabadi, M.; Gahagan, A.C.; Hotte, T.; Belzile, F.; Rajcan, I.; Golshani, A.; Morrison, M.J.; et al. Genome-Wide Analysis of Cold Imbibition Stress in Soybean, Glycine max. Front. Plant Sci. 2023, 14, 1221644. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Wang, H.; Zhou, Z.; Jia, J.; Zhang, Q.; Liang, C.; Li, W.; Zhang, Y.; Yu, G. Genome-Wide Identification of the B3 Gene Family in Soybean and the Response to Melatonin under Cold Stress. Front. Plant Sci. 2023, 13, 1091907. [Google Scholar] [CrossRef]
- Bai, W.; Salih, H.; Yang, R.; Yang, Q.; Jin, P.; Liang, Y.; Zhang, D.; Li, X. ScDREBA5 Enhances Cold Tolerance by Regulating Photosynthetic and Antioxidant Genes in the Desert Moss Syntrichia caninervis. Plant Cell Environ. 2025, 48, 3293–3313. [Google Scholar] [CrossRef]
- Han, L.; Wang, Z.; Wang, Q.; Sun, D.W.; He, X.; Wang, H.; Ma, L.; Bai, C.; Watkins, C.B.; Zuo, J. Multiomics Comprehensive Analysis of Pre-Storage Low-Temperature on Cowpea Metabolism. Postharvest Biol. Technol. 2024, 216, 113056. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, F.; Lyu, X.; Chen, J.; Zeng, H.; Xu, N.; Wu, Y.; Zhu, Q. Transcriptome Profiling of Bergenia purpurascens under Cold Stress. BMC Genom. 2023, 24, 754. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Randall, S.K. Functionality of Soybean CBF/DREB1 Transcription Factors. Plant Sci. Int. J. Exp. Plant Biol. 2016, 246, 80–90. [Google Scholar] [CrossRef]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-Induced Modulation and Functional Analyses of the DRE-Binding Transcription Factor Gene, GmDREB3, in Soybean (Glycine max L.). J. Exp. Bot. 2009, 60, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-T.; Li, J.-W.; Liu, D.-Q.; Zhai, Y.; Zhang, H.-J.; Li, X.-W.; Zhang, Q.-L.; Wang, Y.; Wang, Q.-Y. A Novel MYB Transcription Factor, GmMYBJ1, from Soybean Confers Drought and Cold Tolerance in Arabidopsis thaliana. Gene 2014, 538, 46–55. [Google Scholar] [CrossRef]
- Pan, W.-J.; Tao, J.-J.; Cheng, T.; Shen, M.; Ma, J.-B.; Zhang, W.-K.; Lin, Q.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Soybean NIMA-Related Kinase1 Promotes Plant Growth and Improves Salt and Cold Tolerance. Plant Cell Physiol. 2017, 58, 1268–1278. [Google Scholar] [CrossRef]
- Li, J.; Sun, M.; Liu, Y.; Sun, X.; Yin, K. Genome-Wide Identification of Wild Soybean Mitochondrial Calcium Uniporter Family Genes and Their Responses to Cold and Carbonate Alkaline Stresses. Front. Plant Sci. 2022, 13, 867503. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Wang, H.; Li, X.; Ji, H. Enhancement of Plant Cold Tolerance by Soybean RCC1 Family Gene GmTCF1a. BMC Plant Biol. 2021, 21, 369. [Google Scholar] [CrossRef] [PubMed]
- Román, Á.; Andreu, V.; Hernández, M.L.; Lagunas, B.; Picorel, R.; Martínez-Rivas, J.M.; Alfonso, M. Contribution of the Different Omega-3 Fatty Acid Desaturase Genes to the Cold Response in Soybean. J. Exp. Bot. 2012, 63, 4973–4982. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, Q.; Wang, Z.; Li, J.; Liu, S.; Chang, R.; Chen, H.; Liu, G. Integrated Analysis of Transcriptomics and Metabolomics of Peach under Cold Stress. Front. Plant Sci. 2023, 14, 1153902. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, J.; Zheng, S.; Wang, Z.; Xu, C.; Zhang, Q.; Wu, J.; Lou, H. Metabolite Profiling and Transcriptome Analyses Reveal Novel Regulatory Mechanisms of Melatonin Biosynthesis in Hickory. Hortic. Res. 2021, 8, 196. [Google Scholar] [CrossRef]
- Sun, J.; Qiu, C.; Ding, Y.; Wang, Y.; Sun, L.; Fan, K.; Gai, Z.; Dong, G.; Wang, J.; Li, X.; et al. Fulvic Acid Ameliorates Drought Stress-Induced Damage in Tea Plants by Regulating the Ascorbate Metabolism and Flavonoids Biosynthesis. BMC Genom. 2020, 21, 411. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Shomo, Z.D.; Li, F.; Smith, C.N.; Edmonds, S.R.; Roston, R.L. From Sensing to Acclimation: The Role of Membrane Lipid Remodeling in Plant Responses to Low Temperatures. Plant Physiol. 2024, 196, 1737–1757. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, L.; Sanchez-Ballesta, M.T.; Romojaro, F.; Flores, F.B. Physiological, Hormonal and Molecular Mechanisms Regulating Chilling Injury in Horticultural Species. Postharvest Technologies Applied to Reduce Its Impact. J. Sci. Food Agric. 2009, 89, 555–573. [Google Scholar] [CrossRef]
- Winfield, M.O.; Lu, C.; Wilson, I.D.; Coghill, J.A.; Edwards, K.J. Plant Responses to Cold: Transcriptome Analysis of Wheat. Plant Biotechnol. J. 2010, 8, 749–771. [Google Scholar] [CrossRef]
- Radhakrishnan, V.S.; Reddy Mudiam, M.K.; Kumar, M.; Dwivedi, S.P.; Singh, S.P.; Prasad, T. Silver Nanoparticles Induced Alterations in Multiple Cellular Targets, Which Are Critical for Drug Susceptibilities and Pathogenicity in Fungal Pathogen (Candida albicans). Int. J. Nanomed. 2018, 13, 2647–2663. [Google Scholar] [CrossRef]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid Signalling in Plant Responses to Abiotic Stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]
- Knapp, B.D.; Huang, K.C. The Effects of Temperature on Cellular Physiology. Annu. Rev. Biophys. 2022, 51, 499–526. [Google Scholar] [CrossRef]
- Fagiani, F.; Di Marino, D.; Romagnoli, A.; Travelli, C.; Voltan, D.; Di Cesare Mannelli, L.; Racchi, M.; Govoni, S.; Lanni, C. Molecular Regulations of Circadian Rhythm and Implications for Physiology and Diseases. Signal Transduct. Target. Ther. 2022, 7, 41. [Google Scholar] [CrossRef]
- Liang, T.; Yu, S.; Pan, Y.; Wang, J.; Kay, S.A. The Interplay between the Circadian Clock and Abiotic Stress Responses Mediated by ABF3 and CCA1/LHY. Proc. Natl. Acad. Sci. USA 2024, 121, e2316825121. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yuan, L.; Yang, X.; Zhang, X.; Wang, L.; Xie, Q. Circadian Clock in Plants: Linking Timing to Fitness. J. Integr. Plant Biol. 2022, 64, 792–811. [Google Scholar] [CrossRef]
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Maurya, R.; Sharma, D.; Pandey, P.; Singh, K.N. Role of Circadian Rhythm in Plant System: An Update from Development to Stress Response. Environ. Exp. Bot. 2019, 162, 256–271. [Google Scholar] [CrossRef]
- Cano-Ramirez, D.L.; Panter, P.E.; Takemura, T.; de Fraine, T.S.; de Barros Dantas, L.L.; Dekeya, R.; Barros-Galvão, T.; Paajanen, P.; Bellandi, A.; Batstone, T.; et al. Low-Temperature and Circadian Signals Are Integrated by the Sigma Factor SIG5. Nat. Plants 2023, 9, 661–672. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Cold-Stress Responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Sharma, M.; Irfan, M.; Kumar, A.; Kumar, P.; Datta, A. Recent Insights into Plant Circadian Clock Response Against Abiotic Stress. J. Plant Growth Regul. 2022, 41, 3530–3543. [Google Scholar] [CrossRef]
- Hüner, N.P.A.; Bode, R.; Dahal, K.; Hollis, L.; Rosso, D.; Krol, M.; Ivanov, A.G. Chloroplast Redox Imbalance Governs Phenotypic Plasticity: The “Grand Design of Photosynthesis” Revisited. Front. Plant Sci. 2012, 3, 255. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, M.; Kim, R.J.-A.; Moore, C.M.; Chen, M. Daytime Temperature Is Sensed by Phytochrome B in Arabidopsis through a Transcriptional Activator HEMERA. Nat. Commun. 2019, 10, 140. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Klose, C.; Burgie, E.S.; Costigliolo Rojas, C.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schaefer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B Integrates Light and Temperature Signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Li, M.; Fu, D.; Wu, S.; Li, J.; Gong, Z.; Liu, H.; Yang, S. The CRY2-COP1-HY5-BBX7/8 Module Regulates Blue Light-Dependent Cold Acclimation in Arabidopsis. Plant Cell 2021, 33, 3555–3573. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2024. Nucleic Acids Res. 2024, 52, D18–D32. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Clean Base (G) | Q30 (%) | GC Content (%) | Reads Mapped |
|---|---|---|---|---|---|---|
| 3h_CK_1 | 43,871,184 | 43,211,288 | 6.48 | 92.67 | 45.07 | 41,622,905 (96.32%) |
| 3h_CK_2 | 43,948,954 | 43,235,366 | 6.49 | 92.5 | 45.37 | 41,588,504 (96.19%) |
| 3h_CK_3 | 61,577,250 | 60,025,770 | 9.00 | 90.2 | 44.64 | 57,794,718 (96.28%) |
| 3h_cold_1 | 51,789,686 | 50,931,456 | 7.64 | 92.51 | 45.43 | 49,039,983 (96.29%) |
| 3h_cold_2 | 43,213,704 | 42,391,064 | 6.36 | 92.69 | 45.01 | 40,910,763 (96.51%) |
| 3h_cold_3 | 49,329,766 | 48,449,944 | 7.27 | 92.38 | 45.32 | 46,613,834 (96.21%) |
| 12h_CK_1 | 43,301,638 | 42,618,514 | 6.39 | 92.7 | 44.83 | 40,996,780 (96.19%) |
| 12h_CK_2 | 50,526,542 | 49,545,272 | 7.43 | 92.55 | 44.52 | 47,752,758 (96.38%) |
| 12h_CK_3 | 48,957,654 | 48,167,518 | 7.23 | 92.99 | 44.49 | 46,529,026 (96.60%) |
| 12h_cold_1 | 48,718,240 | 48,086,168 | 7.21 | 92.5 | 44.96 | 46,329,279 (96.35%) |
| 12h_cold_2 | 45,488,068 | 44,703,392 | 6.71 | 92.26 | 45.05 | 43,147,507 (6.52%) |
| 12h_cold_3 | 55,215,940 | 53,451,410 | 8.02 | 90.5 | 44.26 | 51,525,397 (96.40%) |
| 24h_CK_1 | 44,086,618 | 43,341,330 | 6.5 | 92.41 | 44.81 | 41,568,022 (95.91%) |
| 24h_CK_2 | 55,132,572 | 53,774,908 | 8.07 | 92.58 | 44.55 | 51,604,386 (95.96%) |
| 24h_CK_3 | 44,103,032 | 43,475,426 | 6.52 | 92.67 | 44.7 | 41,275,933 (94.94%) |
| 24h_cold_1 | 44,451,846 | 43,693,100 | 6.55 | 92.75 | 44.74 | 42,095,358 (96.34%) |
| 24h_cold_2 | 47,111,752 | 46,262,290 | 6.94 | 92.38 | 44.57 | 44,302,151 (95.76%) |
| 24h_cold_3 | 43,195,264 | 42,491,908 | 6.37 | 91.91 | 44.53 | 40,733,667 (95.86%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, G.; Lin, Y.; Xu, J.; Zhu, H.; Liu, L. Green Soybean’s Survival Mechanisms Under Cold Stress: A Transcriptomic Perspective. Agronomy 2025, 15, 1456. https://doi.org/10.3390/agronomy15061456
Cao G, Lin Y, Xu J, Zhu H, Liu L. Green Soybean’s Survival Mechanisms Under Cold Stress: A Transcriptomic Perspective. Agronomy. 2025; 15(6):1456. https://doi.org/10.3390/agronomy15061456
Chicago/Turabian StyleCao, Guangping, Yanhui Lin, Jing Xu, Honglin Zhu, and Ling Liu. 2025. "Green Soybean’s Survival Mechanisms Under Cold Stress: A Transcriptomic Perspective" Agronomy 15, no. 6: 1456. https://doi.org/10.3390/agronomy15061456
APA StyleCao, G., Lin, Y., Xu, J., Zhu, H., & Liu, L. (2025). Green Soybean’s Survival Mechanisms Under Cold Stress: A Transcriptomic Perspective. Agronomy, 15(6), 1456. https://doi.org/10.3390/agronomy15061456





