Genome-Wide Identification of BnaPDAT Family in Brassica napus and the Effect of BnaA02.PDAT1 on Seed Oil Content
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Characterization of PDAT Family Members
2.3. Phylogenetic, Conserved Domain, and Conserved Motif Analyses of PDAT Family Members
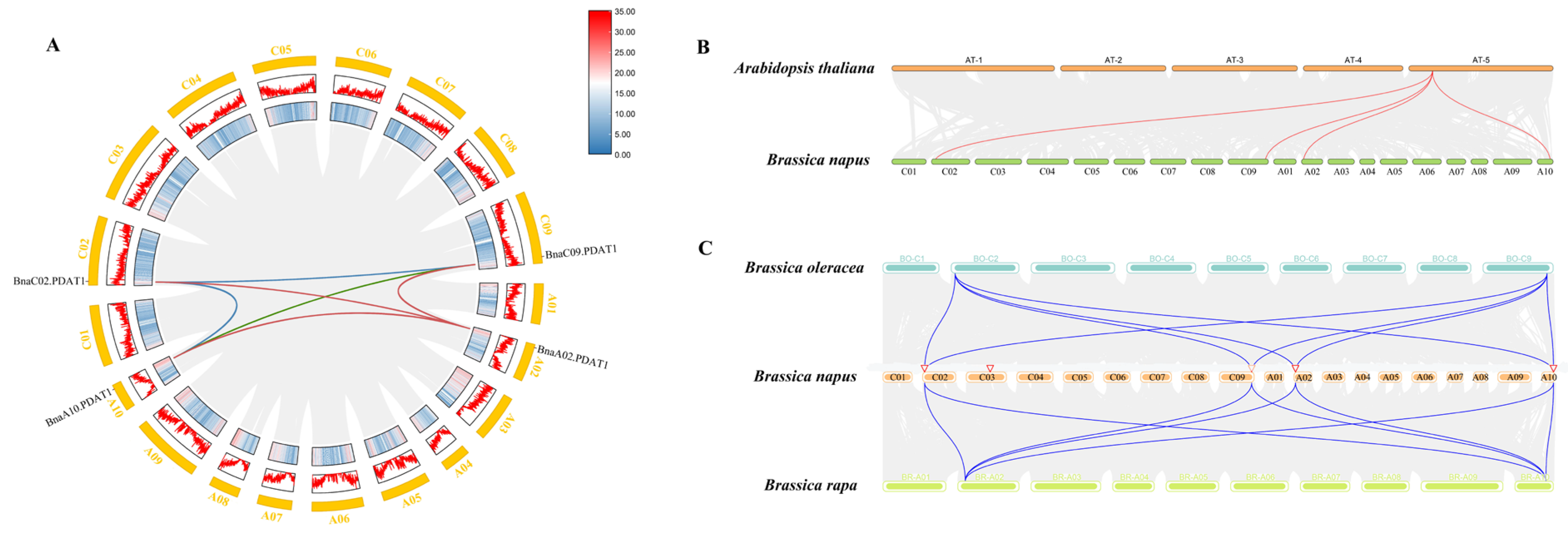
2.4. Chromosome Location and Collinearity Analysis of PDAT Genes
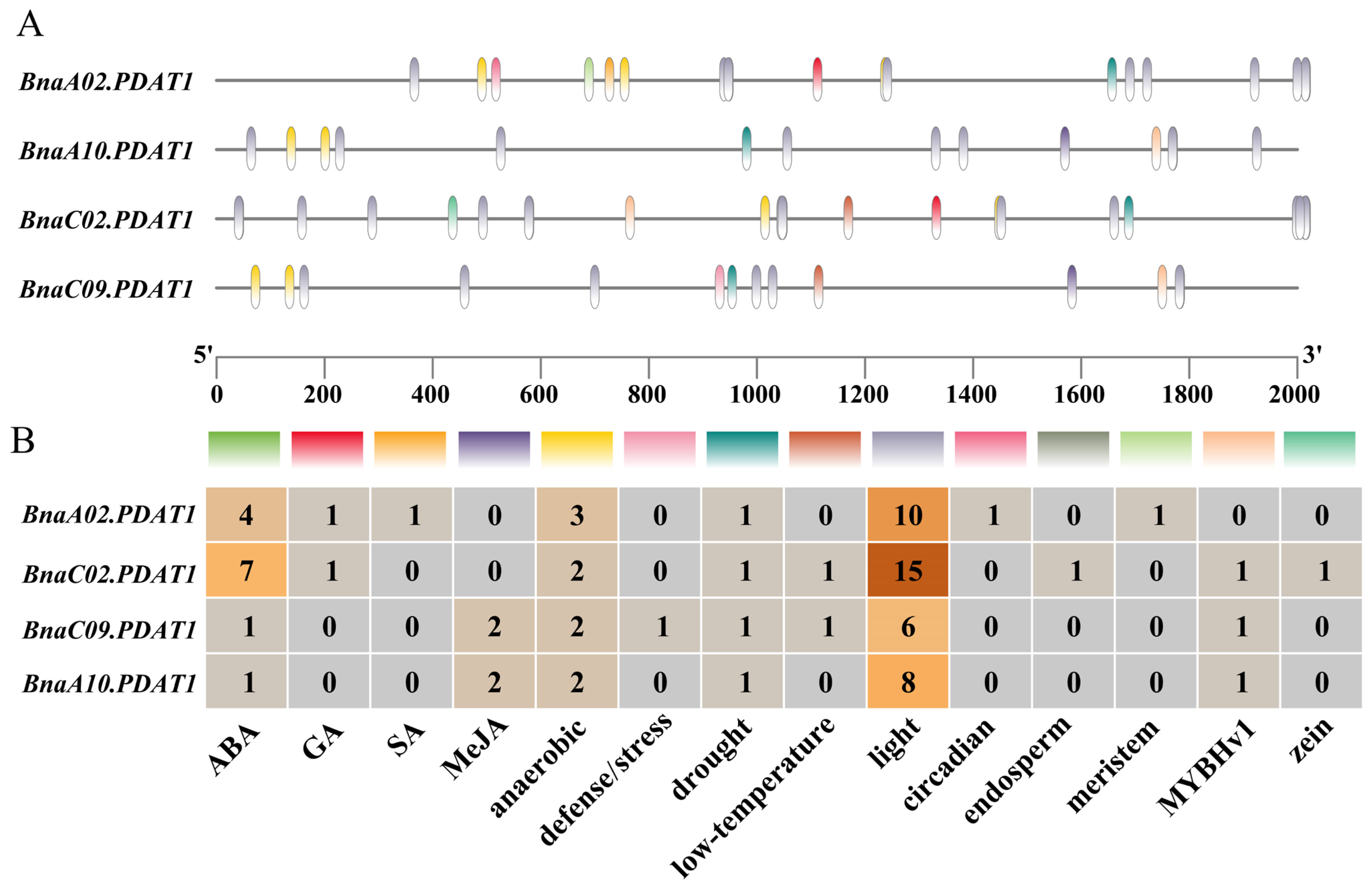
2.5. Promoter cis-Acting Element Analysis of BnaPDAT Genes
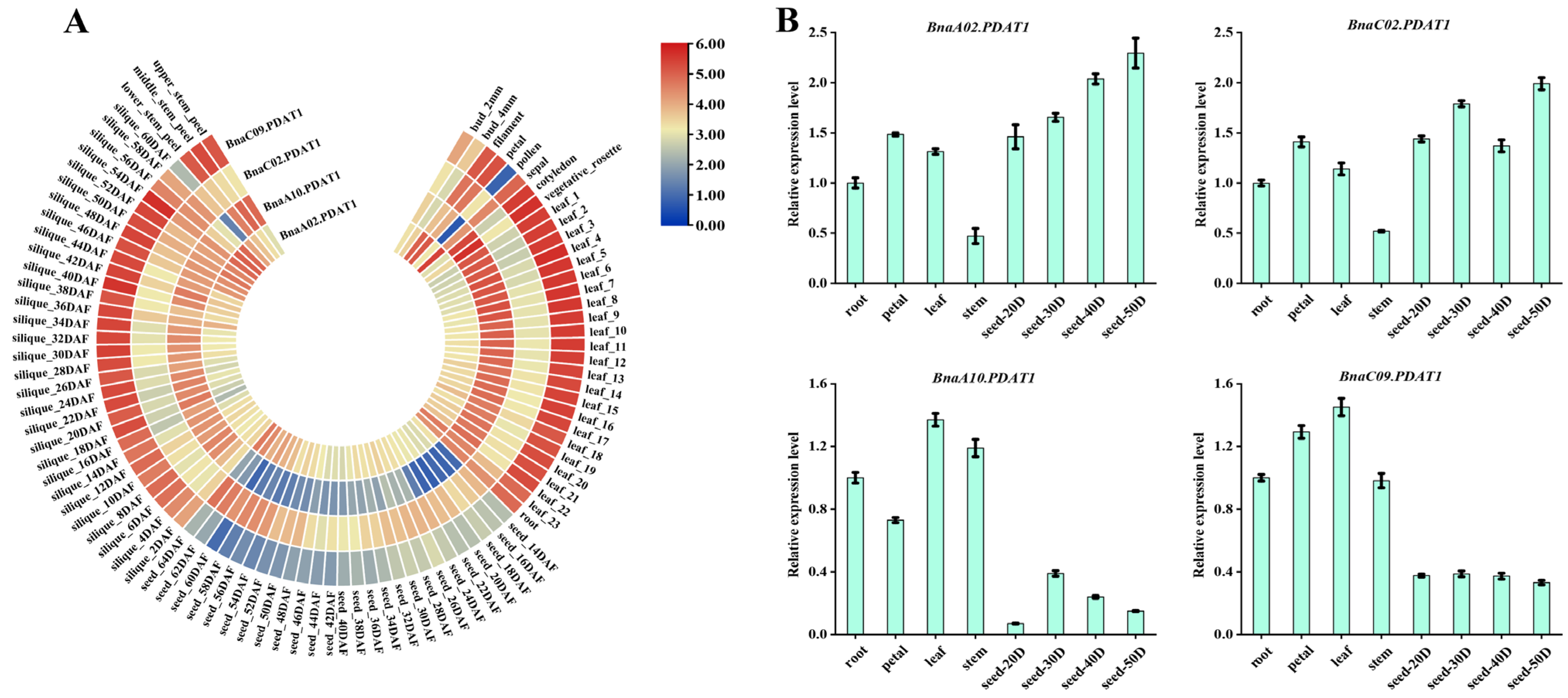
2.6. Transcriptomic Data Acquisition and qRT-PCR Analysis
2.7. Construction of Overexpression Vector and Genetic Transformation in B. napus
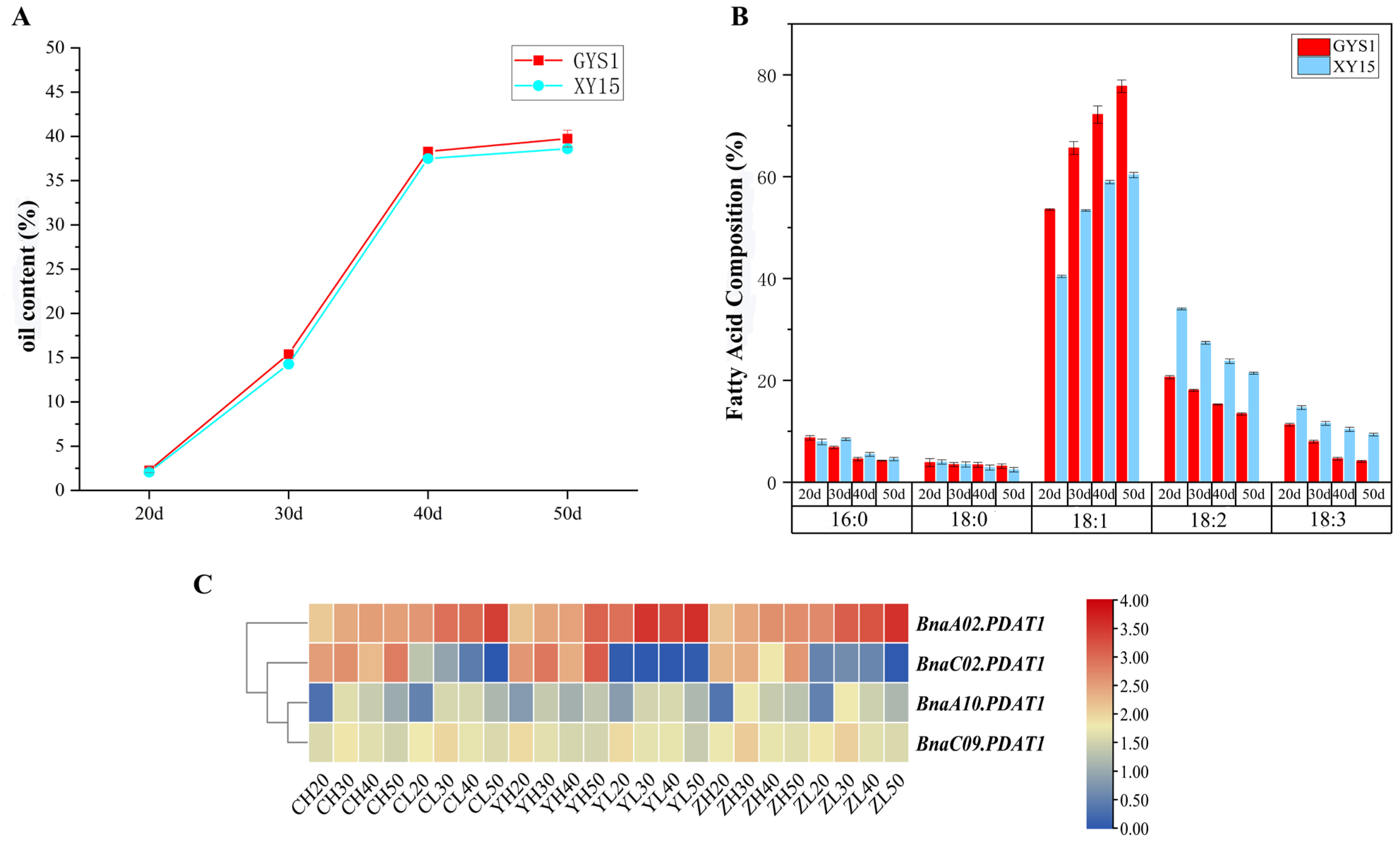
2.8. Determination of Seed Oil Content and Fatty Acid Composition
2.9. Statistical Analysis
3. Results
3.1. Identification and Characterization of PDAT Genes in B. napus, B. rapa, and B. olerace
3.2. Phylogenetic and Gene Structure Characterization of PDAT
3.3. Chromosomal Localization and Collinearity Analysis of PDAT
3.4. cis-Element Analyses of BnaPDAT Genes
3.5. Analysis of Expression Patterns of BnaPDAT Genes in Different Tissues
3.6. Analysis of BnaPDAT Gene Expression Patterns Under Different Abiotic Stress Treatments
3.7. Expression Analysis of BnaPDAT Genes in High- and Low-Oleic Acid Materials
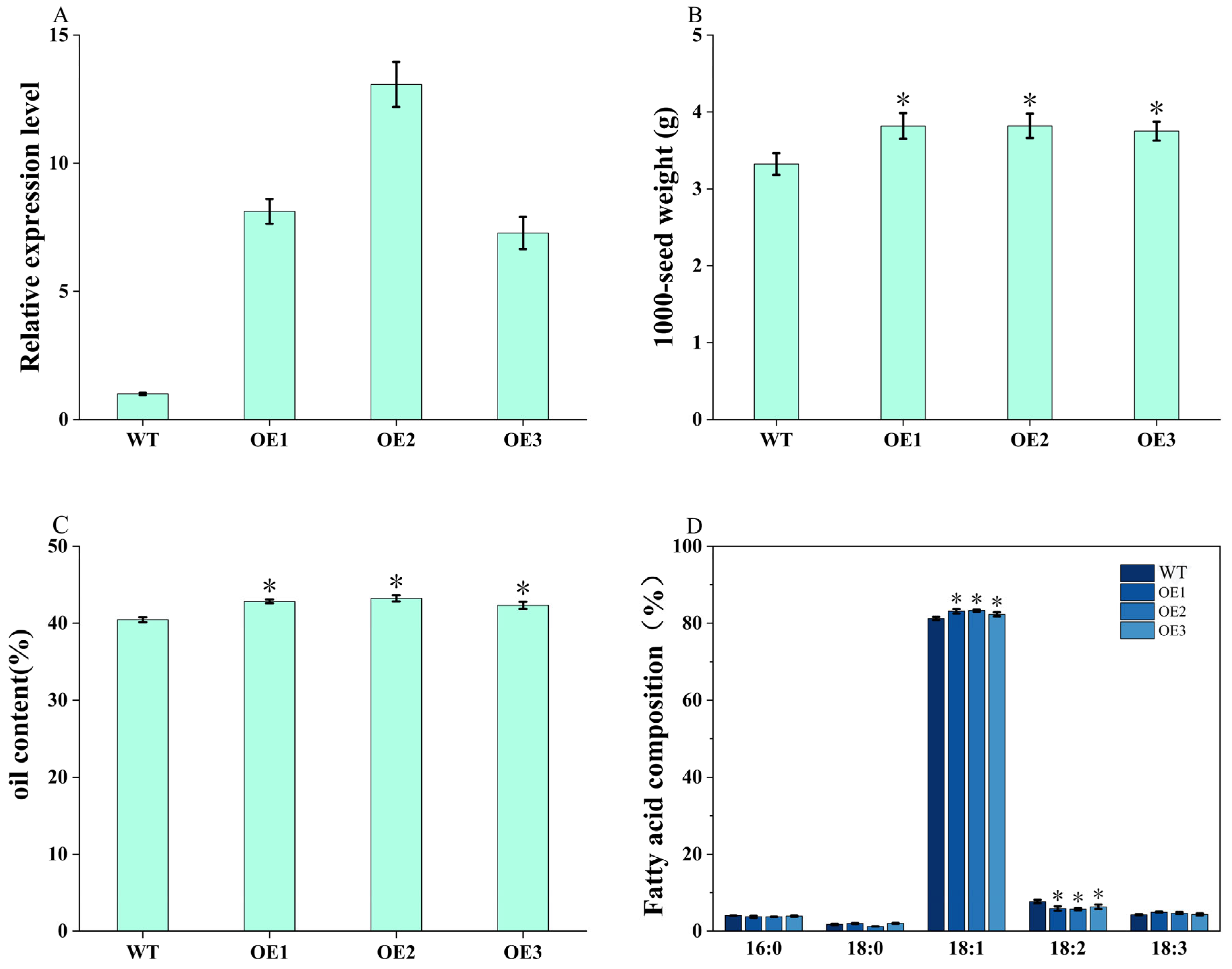
3.8. Overexpression of BnaA02.PDAT1 Enhances Seed Oil Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and Temporal Mapping of Key Lipid Species in Brassica napus Seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.S.; Yilmaz, J.L.; Green, A.G.; Stymne, S.; Hofvander, P. Replacing fossil oil with fresh oil—With what and for what? Eur. J. Lipid Sci. Technol. 2011, 113, 812–831. [Google Scholar] [CrossRef]
- Biermann, U.; Bornscheuer, U.; Meier, M.A.R.; Metzger, J.O.; Schäfer, H.J. Oils and Fats as Renewable Raw Materials in Chemistry. Angew. Chem.-Int. Ed. 2011, 50, 3854–3871. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.H.C. Plant Lipid Droplets and Their Associated Proteins: Potential for Rapid Advances. Plant Physiol. 2017, 176, 1894–1918. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.-L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef]
- Shockey, J.; Regmi, A.; Cotton, K.; Adhikari, N.; Browse, J.; Bates, P.D. Identification of Arabidopsis GPAT9 (At5g60620) as an Essential Gene Involved in Triacylglycerol Biosynthesis. Plant Physiol. 2015, 170, 163–179. [Google Scholar] [CrossRef]
- Lewin, T.M.; Wang, P.; Coleman, R.A. Analysis of Amino Acid Motifs Diagnostic for the sn-Glycerol-3-phosphate Acyltransferase Reaction. Biochemistry 1999, 38, 5764–5771. [Google Scholar] [CrossRef]
- Weiss, S.B.; Kennedy, E.P. The Enzymatic Synthesis of Triglycerides. J. Am. Chem. Soc. 1956, 78, 3550. [Google Scholar] [CrossRef]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.-C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung Tree DGAT1 and DGAT2 Have Nonredundant Functions in Triacylglycerol Biosynthesis and Are Localized to Different Subdomains of the Endoplasmic Reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef]
- Stahl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.; Banaś, W.; Banaś, A.; Stymne, S. Cloning and Functional Characterization of a Phospholipid:Diacylglycerol Acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 Acyltransferases Have Overlapping Functions in Arabidopsis Triacylglycerol Biosynthesis and Are Essential for Normal Pollen and Seed Development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.P.; Ohlrogge, J.B. A Novel Pathway for Triacylglycerol Biosynthesis Is Responsible for the Accumulation of Massive Quantities of Glycerolipids in the Surface Wax of Bayberry (Myrica pensylvanica) Fruit. Plant Cell 2016, 28, 248–264. [Google Scholar] [CrossRef]
- Fan, J.; Yan, C.; Zhang, X.; Xu, C. Dual Role for Phospholipid:Diacylglycerol Acyltransferase: Enhancing Fatty Acid Synthesis and Diverting Fatty Acids from Membrane Lipids to Triacylglycerol in Arabidopsis Leaves. Plant Cell 2013, 25, 3506–3518. [Google Scholar] [CrossRef]
- Pan, X.; Siloto, R.M.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a Pair of Phospholipid:Diacylglycerol Acyltransferases from Developing Flax (Linum usitatissimum L.) Seed Catalyzing the Selective Production of Trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Kim, H.U.; Lee, K.-R.; Go, Y.S.; Jung, J.H.; Suh, M.-C.; Kim, J.B. Endoplasmic Reticulum-Located PDAT1-2 from Castor Bean Enhances Hydroxy Fatty Acid Accumulation in Transgenic Plants. Plant Cell Physiol. 2011, 52, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Mao, X.; Zhao, K.; Ji, X.; Ji, C.; Xue, J.; Li, R. Characterisation of phospholipid: Diacylglycerol acyltransferases (PDATs) from Camelina sativa and their roles in stress responses. Biol. Open 2017, 6, 1024–1034. [Google Scholar] [CrossRef]
- Demski, K.; Łosiewska, A.; Jasieniecka-Gazarkiewicz, K.; Klińska, S.; Banaś, A. Phospholipid:Diacylglycerol Acyltransferase1 Overexpression Delays Senescence and Enhances Post-heat and Cold Exposure Fitness. Front. Plant Sci. 2020, 11, 611897. [Google Scholar] [CrossRef]
- Parkin, I.A.; Sharpe, A.G.; Keith, D.J.; Lydiate, D.J. Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 1995, 38, 1122–1131. [Google Scholar] [CrossRef]
- Palmer, J.D.; Shields, C.R.; Cohen, D.B.; Orton, T.J. Chloroplast DNA evolution and the origin of amphidiploid Brassica species. Theor. Appl. Genet. 1983, 65, 181–189. [Google Scholar] [CrossRef]
- Yates, A.D.; Allen, J.; Amode, R.M.; Azov, A.G.; Barba, M.; Becerra, A.; Bhai, J.; Campbell, L.I.; Martinez, M.C.; Chakiachvili, M.; et al. Ensembl Genomes 2022: An expanding genome resource for non-vertebrates. Nucleic Acids Res. 2022, 50, D996–D1003. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Abbas, A.; Ye, F. Computational methods and key considerations for in silico design of proteolysis targeting chimera (PROTACs). Int. J. Biol. Macromol. 2024, 277, 134293. [Google Scholar] [CrossRef] [PubMed]
- Bernhofer, M.; Dallago, C.; Karl, T.; Satagopam, V.; Heinzinger, M.; Littmann, M.; Olenyi, T.; Qiu, J.; Schütze, K.; Yachdav, G.; et al. PredictProtein—Predicting Protein Structure and Function for 29 Years. Nucleic Acids Res. 2021, 49, W535–W540. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Nystrom, S.L.; McKay, D.J. Memes: A motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, D.; Li, X.; Zhou, B.; Chang, T.; Hong, B.; Guan, C.; Guan, M. Integrated Analysis of lncRNA–mRNA Regulatory Networks Related to Lipid Metabolism in High-Oleic-Acid Rapeseed. Int. J. Mol. Sci. 2023, 24, 6277. [Google Scholar] [CrossRef]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Li, Y.; Li, L.; Du, Z.; Lin, S.; Tian, X.; Li, S.; Yang, B.; Yao, W.; Wang, J.; et al. An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol. Breed. 2020, 40, 96. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the Wax Ester Synthase/Acyl-Coenzyme A:Diacylglycerol Acyltransferase WSD1 Required for Stem Wax Ester Biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [PubMed]
- López-Bascón, M.A.; Luque de Castro, M.D. Chapter 11—Soxhlet Extraction. In Liquid-Phase Extraction; Poole, C.F., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 327–354. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, K.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef]
- Zang, X.; Geng, X.; Ma, L.; Wang, N.; Pei, W.; Wu, M.; Zhang, J.; Yu, J. A genome-wide analysis of the phospholipid: Diacylglycerol acyltransferase gene family in Gossypium. BMC Genom. 2019, 20, 402. [Google Scholar] [CrossRef]
- Cittadino, G.M.; Andrews, J.; Purewal, H.; Estanislao Acuña Avila, P.; Arnone, J.T. Functional Clustering of Metabolically Related Genes Is Conserved across Dikarya. J. Fungi 2023, 9, 523. [Google Scholar] [CrossRef]
- Fan, J.; Yan, C.; Xu, C. Phospholipid:diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of Arabidopsis. Plant J. 2013, 76, 930–942. [Google Scholar] [CrossRef]
- Hernández, M.L.; Moretti, S.; Sicardo, M.D.; García, Ú.; Pérez, A.; Sebastiani, L.; Martínez-Rivas, J.M. Distinct Physiological Roles of Three Phospholipid: Diacylglycerol Acyltransferase Genes in Olive Fruit with Respect to Oil Accumulation and the Response to Abiotic Stress. Front. Plant Sci. 2021, 12, 751959. [Google Scholar] [CrossRef]
- Mueller, S.P.; Unger, M.; Guender, L.; Fekete, A.; Mueller, M.J. Phospholipid:Diacylglycerol Acyltransferase-Mediated Triacylglyerol Synthesis Augments Basal Thermotolerance. Plant Physiol. 2017, 175, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Gasulla, F.; vom Dorp, K.; Dombrink, I.; Zähringer, U.; Gisch, N.; Dörmann, P.; Bartels, D. The role of lipid metabolism in the acquisition of desiccation tolerance in raterostigma plantagineum: A comparative approach. Plant J. 2013, 75, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Han, D.; Li, Y.; Sommerfeld, M.; Hu, Q. Phospholipid:Diacylglycerol Acyltransferase Is a Multifunctional Enzyme Involved in Membrane Lipid Turnover and Degradation While Synthesizing Triacylglycerol in the Unicellular Green Microalga Chlamydomonas reinhardtii. Plant Cell 2012, 24, 3708–3724. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Park, M.-E.; Park, B.Y.; Kim, H.U.; Seo, P.J. The Arabidopsis MYB96 Transcription Factor Mediates ABA-Dependent Triacylglycerol Accumulation in Vegetative Tissues under Drought Stress Conditions. Plants 2019, 8, 296. [Google Scholar] [CrossRef]
- Zhou, B.; Fei, W.; Yang, S.; Yang, F.; Qu, G.; Tang, W.; Ou, J.; Peng, D. Alteration of the fatty acid composition of Brassica napus L. via overexpression of phospholipid: Diacylglycerol acyltransferase 1 from Sapium sebiferum (L.) Roxb. Plant Sci. 2020, 298, 110562. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Length | PI | Molecular Weight (kDa) | Instability Index | Predicted Subcellular Localization | Hydrophobicity Index |
|---|---|---|---|---|---|---|---|
| BnaA10.PDAT1 | BnaA10T0220200ZS | 666 | 6.08 | 73.75 | 36.18 | periplasm | −0.32 |
| BnaC09.PDAT1 | BnaC09T0524800ZS | 667 | 5.98 | 73.94 | 35.89 | periplasm | −0.323 |
| BnaA02.PDAT1 | BnaA02T0049700ZS | 665 | 6.61 | 73.53 | 36.55 | periplasm | −0.302 |
| BnaC02.PDAT1 | BnaC02T0057200ZS | 660 | 7.04 | 73.1 | 36.86 | periplasm | −0.288 |
| BraA02.PDAT1 | Bra023426.1 | 597 | 6.38 | 65.69 | 35.83 | periplasm | −0.251 |
| BraA10.PDAT1 | Bra008812.1 | 666 | 6.11 | 73.74 | 37.59 | periplasm | −0.315 |
| BoC09.PDAT1 | Bo9g166600.1 | 667 | 6.08 | 73.82 | 35.51 | periplasm | −0.304 |
| BoC02.PDAT1 | Bo2g011450.1 | 660 | 6.71 | 73.11 | 38.25 | periplasm | −0.287 |
| AtPDAT2 | AT3G44830.1 | 665 | 8.69 | 73.65 | 49.02 | outer membrane | −0.226 |
| AtPDAT1 | AT5G13640.1 | 671 | 6.5 | 74.15 | 35.95 | periplasm | −0.301 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Guan, C.; Guan, M. Genome-Wide Identification of BnaPDAT Family in Brassica napus and the Effect of BnaA02.PDAT1 on Seed Oil Content. Agronomy 2025, 15, 1204. https://doi.org/10.3390/agronomy15051204
Chen H, Guan C, Guan M. Genome-Wide Identification of BnaPDAT Family in Brassica napus and the Effect of BnaA02.PDAT1 on Seed Oil Content. Agronomy. 2025; 15(5):1204. https://doi.org/10.3390/agronomy15051204
Chicago/Turabian StyleChen, Hu, Chunyun Guan, and Mei Guan. 2025. "Genome-Wide Identification of BnaPDAT Family in Brassica napus and the Effect of BnaA02.PDAT1 on Seed Oil Content" Agronomy 15, no. 5: 1204. https://doi.org/10.3390/agronomy15051204
APA StyleChen, H., Guan, C., & Guan, M. (2025). Genome-Wide Identification of BnaPDAT Family in Brassica napus and the Effect of BnaA02.PDAT1 on Seed Oil Content. Agronomy, 15(5), 1204. https://doi.org/10.3390/agronomy15051204




