Abstract
Sesbania cannabina (Retz.) Pers., as a legume, has strong waterlogging tolerance, but the lack of genomic information limits the exploration of key genes and molecular mechanisms. In this study, single-molecule real-time technology was used to sequence stems RNA of two Sesbania varieties at five time points under waterlogging stress through the PacBio Iso-Seq platform. The full-length transcriptome information contained 42 Gb raw data, 32,503 transcripts with an average length of 1912.28 nt, N50 length of 2059 nt and GC content of 42.69%. A total of 32,143 coding sequences (CDSs), 1745 transcription factors (TFs), 282 long non-coding RNAs (LncRNAs), 7497 simple sequence repeats (SSRs) and 202 alternative splicing (AS) events were identified through sequence alignment and software analysis. The analysis showed that 10,075 transcripts were enriched in 137 KEGG pathways, and 519 transcripts were included in plant hormone signal transduction, of which 103 and 123 transcripts were, respectively, involved in the ethylene and auxin pathways. The assembly and annotation of full-length transcriptome data of Sesbania provided reliable and accurate genomic information for the exploration of key genes and the study of molecular mechanisms in stem response to waterlogging stress.
1. Introduction
Sesbania is an annual plant and belongs to the subfamily Faboideae of the family Leguminosae, native to tropical and subtropical areas. It has the characteristics of large biomass, strong nitrogen-fixing ability, and wide adaptability, and is a pioneer plant in improving saline-alkali soil [1]. Sesbania is rich in nutrients and can be used in animal feed [2]. In addition, Sesbania seeds are abundant, and Sesbania gum extracted from them is widely used in industry [3].
The lower oxygen utilization rate is the main factor for plant waterlogging stress. When roots are in a low oxygen environment, aerobic respiration is restricted and anaerobic respiration occurs instead. For most upland plants, long-term anaerobic respiration can disrupt cellular energy supply and affect nutrient absorption, leading to wilting and even necrosis of plant leaves [4,5,6]. Most legumes are sensitive to waterlogging [7]. But Krishnan et al. [8] found that Sesbania grown in waterlogging conditions was better than that in dry land. The waterlogged Sesbania was twice as tall and the biomass had significantly increased. There were a large number of adventitious and spongy roots. However, the molecular mechanism of Sesbania adaptation to waterlogging stress is less studied [9]. To reveal the molecular mechanisms of aerenchyma and adventitious root formation in Sesbania is of great significance for the study of its waterlogging tolerance [10,11]. However, there is currently limited publicly available genome information of Sesbania in databases (NCBI and Phytozome), which limits the development of related research.
So far, RNA sequencing based on high-throughput platforms has obtained a large number of reference transcriptome sequences, which is important for understanding the molecular genetic relationship between the genotypes and phenotypes in plants, when they lack reference genomes [12]. Due to RNA being broken into short fragments (<500 nt), the sequences obtained by second-generation sequencing platforms are difficult to assemble into complete gene transcripts [13,14,15,16]. Single-molecule real-time (SMRT), as a third-generation sequencing technology, can directly obtain high-quality, complete, and accurate cDNA reads, with an average reading length of over 3.5 kb [14], which can effectively overcome the problem of short reading length in second-generation sequencing. The long sequence read length of third-generation sequencing reduces the requirements for sequence assembly, which is beneficial for obtaining higher-quality transcript sequences. This technique helps to identify new transcripts to supplement genome annotation information [17], develop more molecular markers [18], discover more alternative splicing (AS) events [19], and analyze gene families and allelic genes [20,21].
Previous studies found that the epidermis of Sesbania stems under waterlogging stress ruptured and formed sponge stems (aerenchyma) and adventitious roots (Figure S1). An experiment was conducted to treat Sesbania stems with waterlogging and collect mixed samples at different treatment times for transcriptome analysis. The full-length transcriptome data were obtained by SMRT technology of the PacBio sequencing platform. Full-length gene prediction, CDS identification, TF mining, SSR search, LncRNA prediction, AS analysis, and functional annotation were performed. The related pathways such as plant hormone signal transduction, peroxisome, and MAPK signaling pathways were analyzed based on the formation of adventitious roots and aerenchyma during the waterlogging treatment. Therefore, this study laid the foundation for excavating waterlogging tolerance-related genes and revealing the molecular mechanism of Sesbania stem response to waterlogging stress.
2. Materials and Methods
2.1. Plant Materials and Experimental Design
Two Sesbania varieties, YJG01 and YJP06, were planted in 15 cm (diameter) × 21 cm (height) plastic pots containing two-thirds of the volume of field soil. We retained one seedling per pot, with 25 individual plants per variety. Seedlings were cultivated under natural conditions, 32 °C/13 h in the light and 26 °C/11 h in the dark. After 30 days, 15 Sesbania plants with uniform growth were selected for each variety, and the stems of these plants were randomly subjected to waterlogging stress treatments. The water level was kept at least 5 cm above the stem base (Figure S2). The treatment time for waterlogging stress was 0 h, 6 h, 12 h, 24 h, and 48 h. Each treatment was set with three biological replicates. Waterlogged stems (5 cm above the stem base) were rapidly sampled with scissors (Figure S2), wrapped in aluminum foil, and immediately frozen in liquid nitrogen. According to different varieties, the samples were labeled as G-0h, G-6h, G-12h, G-24h, G-48h and P-0h, P-6h, P-12h, P-24h, P-48h. G stood for YJG01, and P stood for YJP06. A total of 30 stem samples were stored at –80 °C for transcriptome sequencing.
2.2. RNA Extraction and SMRT Sequencing
The RNA of the waterlogging stem samples from YJG01 and YJP06 was extracted using Trizol reagent (Life Technologies, Shanghai, China). RNA integrity was detected by the Agilent 2100 Bioanalyzer and agarose gel electrophoresis. RNA concentration and purity were detected with the Nanodrop micro-spectrophotometer (Thermo Fisher, Shanghai, China). An equal amount of qualified RNA was taken from each sample and mixed. mRNA enriched by Oligo (dT) magnetic beads was reversely transcribed into cDNA using SMRTbell®prep kit 3.0 (PacBio, Shanghai, China, PN:102-396-000). cDNAs obtained by large-scale PCR were DNA damage repaired, end repaired and ligated to sequencing adapters. Then, the full-length transcriptome libraries were constructed and sequenced on the PacBio SequelII platform by Gene Denovo Biotechnology Co. (Guangzhou, China).
2.3. Data Processing and Basic Annotation
Isoform sequencing (IsoSeq) pipeline provided by Pacific Biosciences was used to analysis the raw sequencing of cDNA libraries [22], and high-quality isoforms with prediction accuracy greater than or equal to 0.99 were selected for subsequent analysis. In order to obtain annotation information, isoforms were BLAST analyzed against the NCBI non-redundant protein (Nr) database (http://www.ncbi.nlm.nih.gov, accessed on 25 September 2023), the Swiss-Prot protein database (http://www.expasy.ch/sprot, accessed on 26 September 2023), the Kyoto Encyclopedia of Genes and the Genomes (KEGG) database (http://www.genome.jp/kegg, accessed on 7 October 2023). Isoforms were analyzed against the KOG database (http://www.ncbi.nlm.nih.gov/COG, accessed on 8 October 2023) with BLASTx program (http://www.ncbi.nlm.nih.gov/BLAST/, accessed on 10 October 2023), and an E-value threshold of 1e-5 was used to evaluate sequence similarity with genes of other species. Nr annotation results of isoforms were used to annotate Gene Ontology (GO) using Blast2GO 6.0 software, and the functional classification of isoforms was performed using WEGO 2.0 software.
2.4. Prediction and Analysis of CDS, SSR, TF, LncRNA, AS Event, Functional Unit
CDS, protein sequences, and UTR sequences were obtained by using ANGEL software for the isoform sequences. SSR sequences were detected by MIcroSAtellite (MISA, http://pgrc.ipk-gatersleben.de/misa/, accessed on 8 April 2025). TF families were predicted by aligning protein-coding sequences of isoforms to Plant TFdb (http://planttfdb.cbi.pku.edu.cn/, accessed on 8 April 2025). Potential lncRNAs were mined using Coding-Non-Coding Index (CNCI) (version 2) [23] and Coding Potential Calculator (CPC) [24] to assess the protein-coding potential of transcripts without annotations using default parameters. AS events of transcript isoforms were analyzed by SUPPA. Protein domain analysis was performed using the Pfam (Protein Families Database of Alignments and Hidden Markov Models) database (https://pfam.xfam.org/, accessed on 8 April 2025). The prediction of SMART (Simple Modular Architecture Research Tool) protein domains was performed using the HMMER (profile hidden Markov models for biological sequence analysis) program (http://hmmer.org/, accessed on 8 April 2025). The transmembrane helix structure was predicted using TMHMM (Topology Model of Hidden Markov Model for Membrane Proteins) 2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/, accessed on 8 April 2025). Signal peptide structure prediction was performed using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/, accessed on 8 April 2025).
2.5. Data Statistical Analysis
The data were processed using Microsoft Excel 2021, and the graphs were plotted using Origin 2021.
3. Results
3.1. General Properties of Sesbania Transcriptome
A total of 42,105,099,343 nucleotides and 23,866,027 subreads with average length of 1764 nt and N50 of 1910 nt were obtained from the PacBio SequelII sequencing platform. All subreads were eliminated redundancy and corrected to obtain circular consensus sequence (CCS). The number of CCS and reads were 858,743,388 and 433,270, respectively. The average length of CCS reads was 1982 nt. Read length distribution of CCS was shown in Figure S3a. Full-length non chimeric (FLNC reads) sequence was identified, hierarchical clustered and corrected to obtain 39,214 high quality isoforms (prediction accuracy ≥ 0.99) and 179 low quality isoforms (prediction accuracy ≤ 0.99). After read clustering, merging and alignment, the full-length transcriptome sequence of Sesbania was obtained finally. There were 32,503 transcripts with the total length of 62,154,838 nt, the average length of 1912 nt, the N50 length of 2059 nt, and the GC content of 42.69% (Figure S3b).
3.2. Analysis of CDS, TF, SSR, LncRNA, AS Event, Protein Domain Architecture, and Functional Motif
3.2.1. CDS
A total of 32,143 CDSs were predicted comprising both initial and termination codons. The number of CDSs with length ≤ 2400 nt was 29,809, amounted for 92.74% (Figure 1a). CDS lengths > 3000 nt were 22 and accounted for 0.068%. 1381 CDSs (4.30%) had no 5′UTR, and 394 CDSs (1.23%) had no 3′UTR. The lengths of more than 97% UTRs were less than 1000 nt (Figure 1b,c). The longest 5′UTR and 3′UTR were 3944 nt and 4294 nt, respectively.
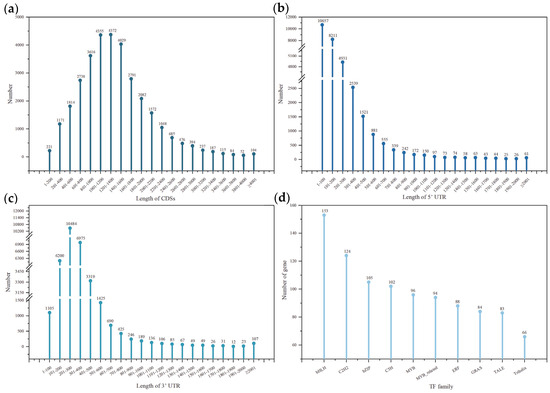
Figure 1.
Number and length of CDSs, 5′UTR, 3′UTR and the top 10 most abundant TF families. (a) CDS; (b) 5′UTR; (c) 3′UTR; (d) TF.3.2.2. TF.
3.2.2. TF
A total of 1745 TFs were found in the stems of Sesbania under waterlogging stress. These TFs were classified into 52 TF families (Table S1). Of which, the top 10 TF families were bHLH, C2H2, bZIP, C3H, MYB, MYB_related, ERF, GRAS, TALE and Trihelix (Figure 1d). They contained 995 TFs, accounted for 57.2%. The top 34 TF families (the number > 10) in the ranking contained 1653 TFs, accounting for a staggering 97%.
3.2.3. SSR
There were 7497 SSRs in 6552 transcripts. The number of SSRs contained in 793 transcripts was greater than or equal to 2. And 550 transcripts contained compound SSRs accounted for 7.34%. Trinucleotide repeats motifs (3106, 41.43%) with 5–19 repeats were the most abundant, and followed by dinucleotide repeats motifs (2410, 32.15%) with 6–22 repeats, tetranucleotide (667, 8.90%) with 4–14 repeats, hexanucleotide repeat motifs (437; 5.83%) with 4–14 repeats, and pentanucleotide repeats motifs (327, 4.36%) with 4–7 repeats, respectively (Figure 2a). As shown in Figure 2b, the proportion of AG/CT (2189, 29.2%), was the highest in the dinucleotide repeats. In the trinucleotide repeats, AAG/CTT (1027, 13.7%) was the most frequent, followed by ACC/GGT (457, 6.1%). Among the tetra-, penta- and hexanucleotide repeats, AAAG/CTTT, AGAGA/TCTCT and AAACCA/TTTGGT were the most abundant, with the repeat number of 150, 10 and 10, respectively.
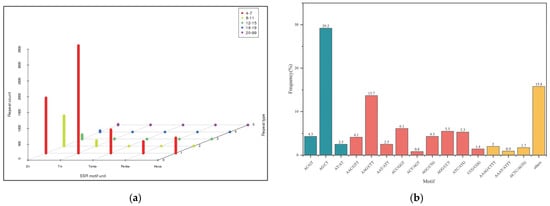
Figure 2.
Nucleotide classification of SSR in Sesbania. (a) Repeat count of five types of SSRs; (b) the proportion of different types of SSRs.
3.2.4. LncRNA and AS Event
The CNCI and CPC identified 320 and 410 LncRNAs, respectively. A total of 282 LncRNAs were simultaneously predicted by both tools and recognized as the final result (Figure 3a). A total of 202 AS events of five types were identified according to the reference sequence assembled by Cogent. RI was the main AS event, with a total of 150, accounting for 50.17%, following by A5 (23.75%) and A3 (22.74%). The proportions of SE and AF were the lowest, both at 1.67% (Figure 3b). The statistical results showed that 2095 genes (31.81%) contained two transcripts. The number of genes containing three transcripts was 1885, accounting for 28.62%, and 147 genes were detected for more than ten transcripts (Figure 3c).
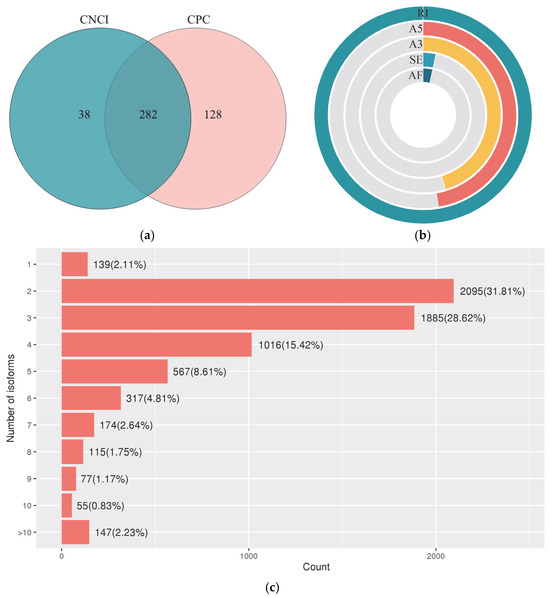
Figure 3.
Identification of LncRNA and analysis of AS events in Sesbania. (a) LncRNA predicted by CNCI and CPC; (b) the proportion of different types of AS event; RI, retained intron; A5, alternative 5′ splice site; A3, alternative 3′ splice site; AF, alternative first exon; SE, skipping exon; (c) quantity statistics of isoforms contained in a gene.
3.2.5. Functional Unit Prediction and Analysis
Using the Pfam database, we predicted a total of 47,889 functional units across 28,363 isoforms. These functional units were classified into four types: Domain (46.65%), Family (41.86%), Motif (0.80%), and Repeat (10.70%). 7 isoforms contained the highest number of functional units (12 units each). But the majority of isoforms (58.50%) contained only one functional unit (Figure 4a). SMART protein domain analysis identified 22,151 domains in 11,391 isoforms. The analysis revealed an inverse relationship between domain number and isoform frequency. Isoforms containing a single domain were most prevalent (50.54%) (Figure 4b). Transmembrane helix prediction detected helical structures in 32,143 isoforms, with each isoform containing exactly one predicted helix. Similarly, signal peptide prediction identified single peptide structures in 4311 isoforms. Integrated analysis showed that 1251 isoforms were identified across all four analyses (Pfam/SMART domains, transmembrane helices, and signal peptides).
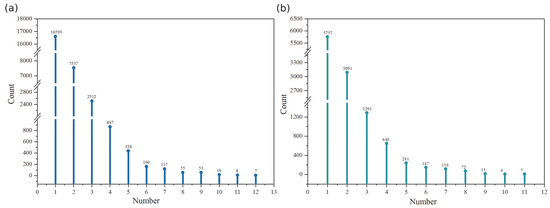
Figure 4.
Relationship between the number of functional units per isoform and isoform counts in Pfam and SMART protein structure predictions. (a) Pfam protein structure prediction; X-axis: Number of functional units per isoform; Y-axis: Number of isoforms containing a specific quantity (X-axis value) of functional units; (b) SMART protein structure prediction; X-axis: Number of domains per isoform; Y-axis: Number of isoforms containing a specific quantity (X-axis value) of domains.
3.3. Functional Annotation
A total of 32,503 transcripts were identified by blast isoform sequences from databases Nr, SwissProt, KEGG and KOG, of which 481 were not annotated. A total of 32,022 transcripts were annotated by at least one database, accounting for 98.52% (Table 1), and 20,666 transcripts were annotated by all four databases, accounting for 63.58% (Figure 5a). Statistical analysis revealed that 32,002 sequences from 226 species showed homology with isoforms in Sesbania (Table S2). The result showed that Abrus precatorius (9732, 29.94%) had the largest number of homologous sequences in Sesbania, followed by Spatholobus suberectus (5580, 17.17%), Glycine max (2435, 7.49%), Cicer arietinum (2376, 7.31%) and Cajanus cajan (2041, 6.28%) (Figure 5b). There were a total of 22,164 homologous sequences among these five species, accounting for 69.26%.

Table 1.
Statistical results of functional annotation of transcripts.
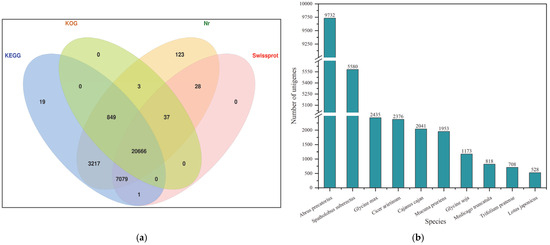
Figure 5.
Database for functional annotation of isoforms and the number of homologous sequences in species. (a) Venn diagram of annotated transcripts in the Nr, Swiss-Prot, KEGG and KOG databases; (b) the top ten species with the highest number of homologous sequences.
3.4. KOG Annotation
Results showed that these transcripts were divided into 25 clusters in total (Figure 6). General function prediction only (cluster R) had the highest number of transcripts (4095). The next was signal transduction mechanisms (cluster T), which contained 3235 transcripts. Then, cluster O (posttranslational modification, protein turnover, chaperones) contained 2985 transcripts ranking third, followed by cluster G (carbohydrate transport and metabolism, 1903 transcripts) and cluster J (translation, ribosomal structure and biogenesis, 1679 transcripts). Cell motility (Group N, 16 transcripts) had the fewest transcripts, and the functions of 1176 transcripts were unknown.
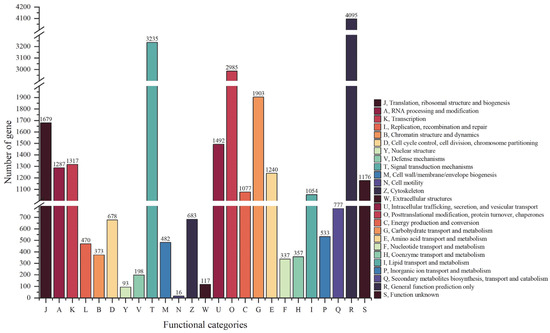
Figure 6.
KOG function annotation of transcript isoforms.
3.5. GO Annotation
There were 26,039 transcripts enriched by GO annotation analysis. Biological process (BP), cellular component (CC) and molecular function (MF) contained 29, 3 and 20 subgroups, respectively (Figure 7 and Table S3). Transcripts in the BP category were mainly enriched in the cellular process (19,325), metabolic process (16,524) and response to stimulus (5807), etc. The CC category included three subgroups: cellular anatomical entity (13,585), protein-containing complex (5163) and virion component (96). Transcripts involved in the MF category were mainly associated with binding (16,130), catalytic activity (15,156) and transporter activity (2406), etc.
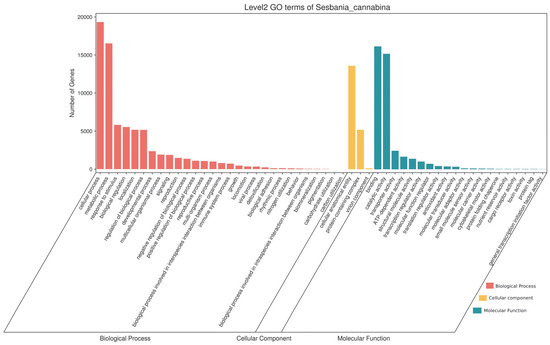
Figure 7.
Classification of transcripts annotated by Gene Ontology.
3.6. KEGG Annotation
The analysis results of the KEGG annotation indicated that 10,075 transcripts were enriched in 137 pathways, including five major categories and 20 subcategories (Table S4). Among the five major categories, metabolism was the largest category, containing 11 subclasses and 106 pathways. In the KEGG pathway, the metabolic pathway contained the most isoforms, followed by the biosynthesis of secondary metabolites, carbon metabolism and biosynthesis of amino acids (Figure 8).
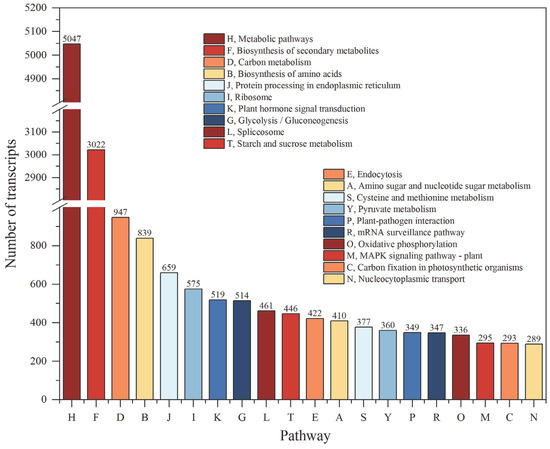
Figure 8.
The top 20 KEGG pathways with the highest number of transcripts.
Multiple hormone signaling pathways were enriched in the KEGG pathway under the waterlogging stress of Sesbania stems. Auxin, ethylene, abscisic acid, brassinosteroid, jasmonic acid, gibberellin, salicylic acid and cytokinin pathways contained 123, 103, 81, 61, 44, 42, 40 and 25 transcripts, respectively, and involved 40 related genes. In the MAPK signaling pathway, abscisic acid promoted the production of H2O2 by inducing the expression of MPK6 (2 transcripts). H2O2 promoted cell death and the further production of H2O2 through the signaling pathway ANP1-MKK4/5-MPK3/6-MRKY22/29, involving 44 transcripts. The antioxidant system in peroxisome utilized CAT (10 transcripts) and SOD (nine transcripts) to participate in the metabolism of H2O2 in tissues.
4. Discussion
The formation of aerenchyma and adventitious roots plays a critical role in plant adaptation to waterlogging stress. It was reported that wheat developed well-formed aerenchyma at 10 mm from the root tip after 72 h of waterlogging treatment [25]. In barley, aerenchyma formation could be observed at 6 cm from the root tip following 7 days of waterlogging [26]. Rice possessed aerenchyma in its roots, stems, and leaves. Under submergence conditions, rice responded to rising water levels by elongating its stem internodes to keep leaves above the water surface, thereby maintaining respiration and photosynthesis. The adventitious roots formed on submerged stem nodes were believed to function primarily in nutrient absorption rather than respiration [27]. In cucumbers, adventitious root primordia began to form from the stem cambium within 48 h of waterlogging and penetrated the hypocotyl epidermis by 96 h [28]. Overall, studies on plant responses to waterlogging stress primarily focused on the underground parts (roots) [25]. However, the stems of Sesbania were treated with waterlogging stress in this study. Both varieties YJG01 and YJP06 formed aerenchyma in stems and exhibited epidermal penetration by adventitious roots after 24 h of waterlogging stress. Under the same treatment duration, YJP06 demonstrated faster aerenchyma formation as well as a greater number and longer length of adventitious roots compared to YJG01. The response of Sesbania to waterlogging treatment was remarkably rapid compared with plants such as wheat and cucumber.
We pooled the stem RNAs from different varieties subjected to varying durations of waterlogging treatment and performed third-generation full-length transcriptome sequencing. The raw data were obtained through the PacBio Iso-Seq platform to analyze genetic information. After redundancy removal and error correction, 32,503 high-quality transcripts were obtained with an N50 length of 2059 nt. However, the transcriptome data obtained by second-generation sequencing were analyzed for Sesbania root samples under waterlogging stress, resulting in 213,990 unigenes with an N50 length of 1055 nt [9]. The comparative results indicated the advantages of long read length and good continuity of SMRT sequencing. Therefore, full-length transcriptome data may reflect more complete information about mRNA [29,30].
After comparing the isoform sequences with the Nr database in this study, the homologous sequences mapped were almost all existed in leguminous plants. This was consistent with the results of Ren et al. [9]. The species with the highest number of homologous sequences to the isoforms was Abrus precatorius, while that in Sesbania roots under waterlogging stress was Glycine max [9]. This difference could be due to the different organs under waterlogging stress treatment.
TFs can bind to specific regions of DNA, affecting gene expression and regulating cell growth, differentiation, and function. Under waterlogging stress, ERF13 exhibited high expression levels in both citrus roots and leaves. Heterologous expression of PjERF13 in transgenic tobacco reduced the accumulation of H₂O₂ and malondialdehyde, thereby significantly enhancing waterlogging tolerance [31]. Integrated RNA-seq and RT-qPCR analyses revealed that bHLH083 and bHLH108 in ginger played pivotal roles in mediating salt stress and waterlogging stress, respectively [32]. In this study, a total of 1745 TFs were found and classified into 52 families. Among them, a substantial number of genes were classified into the bHLH and ERF families. These TFs might regulate gene expression in Sesbania stems, contributing to the formation of adventitious roots and aerenchyma.
SSRs are widely distributed in genomes and have the characteristics of high repeatability, rich polymorphism, and codominant inheritance. SSR markers are widely used in gene linkage map construction, molecular marker-assisted breeding, variety identification, and genetic resource conservation [33]. In Sesbania stems under waterlogging stress, a total of 7497 SSRs were identified in the full-length transcriptome data. Dinucleotide and trinucleotide repeats accounted for 36.0% and 43.7%, respectively, which constituted the majority of SSR loci. In Nitraria sibirica Pall., the proportions of the two were 24.21% and 14.9%, respectively [34]. In Platycladus orientalis, dinucleotide and trinucleotide repeats of SSR types accounted for 29.6% and 52.7%, respectively, in third-generation sequencing data, but they accounted for 70.31% and 28.7%, respectively, in second-generation sequencing data [33]. It followed that the proportion of SSR types could be different among species, and the number of SSRs obtained through different sequencing techniques would also vary greatly.
LncRNAs play an important role in many life activities, such as dosage compensation effect, epigenetic regulation, and cell cycle regulation. Previous studies showed that lncRNA MtCIR1 negatively regulated the response of Medicago truncatula to salt stress [35]. LncRNA TCONS00065739 was an endogenous competitive target of miR530 and improved the adaptability of Ammopiptanthus nanus to cold stress by regulating TZP [36]. Combining the results of CNCI and CPC, a total of 282 lncRNAs were identified in this study, which should be some lncRNAs related to the response of Sesbania stems to waterlogging stress.
Under waterlogging stress, aerobic respiration of Sesbania stems was weakened, and the epidermis cracked to form white sponge-like tissue and adventitious roots. Numerous KEGG pathways were involved in these processes. The formation of aerenchyma and adventitious roots was closely related to the regulation of reactive oxygen species and hormones. The reactive oxygen species and ethylene induced the formation of aerenchyma by initiating programmed cell death in specific cells [37]. The direction of adventitious root growth was determined by the hormone-auxin gradient at the rice root tip [38]. In this study, H₂O₂ participated in the MAPK signaling pathway and activated the transcription factors WRKY22/29 through phosphorylation, thus promoting cell death. The generation of reactive oxygen species also promoted the synthesis of ethylene by inducing the expression of MPK3/6. There were 123 and 103 transcripts related to ethylene and auxin in the plant hormone signal transduction pathway, which were much higher than other hormones. Plant hormones played an important role in resisting waterlogging stress and regulating Sesbania growth.
In this study, we enriched the third-generation full-length transcriptome dataset through comprehensive analyses including: (1) prediction of CDSs and TF genes; (2) functional annotation of genes; (3) detection of SSRs; (4) characterization of lncRNAs and AS events; (5) enrichment analysis of metabolic pathways. Leveraging the advantages of third-generation sequencing technology, including its long read length and good continuity, we successfully obtained accurate and complete mRNA information related to Sesbania’s response to waterlogging stress. This laid a foundation for future research on waterlogging-tolerant gene mining, gene family analysis, molecular marker development, and metabolic pathway investigation in Sesbania.
5. Conclusions
Since no public reference genome sequence was available for Sesbania, full-length transcriptome data of Sesbania stems in response to waterlogging stress at the different times was obtained by using SMRT technology of PacBio Iso-Seq platform in this study. Ultimately, 32,503 transcripts, 32,143 CDSs, 1745 TFs, 282 LncRNAs, 7497 SSRs and 202 AS events were identified through sequence alignment and software analysis. A total of 10,075 transcripts were enriched in 137 KEGG pathways, and 519 and 295 transcripts were enriched in plant hormone signal transduction and MAPK signaling pathway, respectively. These metabolic pathways might be beneficial for the formation of aerenchyma and adventitious roots in plants under waterlogging stress. This study enriched the genome information of Sesbania and provided a reference transcriptome for gene expression. The results laid a foundation for the mining of key genes, the molecular mechanism research of Sesbania response to waterlogging stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15051197/s1. Figure S1: The status of Sesbania stems under waterlogging stress at 24 h and 48 h. Aer, aerenchyma; AR, adventitious root. After 24 h of waterlogging treatment, white sponge-like aerenchyma formed in the stems of Sesbania, and adventitious roots emerged by breaking through the epidermis. After 48 h, the area of aerenchyma in Sesbania stems expanded, and the number of adventitious roots increased with enhanced elongation. Figure S2: Schematic diagram of the waterlogging stress treatment and sampling site in Sesbania. Figure S3: Length distribution of circular consensus sequences and isoform sequences. (a) X-axis: length of circular consensus sequences; Left Y-axis (bar plot scale): number of CCS within specific length ranges (X-axis values); Right Y-axis (line plot scale): cumulative number of CCS exceeding each length threshold (X-axis values). (b) X-axis: isoform length; Left Y-axis (bar plot): number of isoforms within specific length ranges (X-axis values); Right Y-axis (line plot): number of isoforms longer than a given value (X-axis values). Table S1: Classification of transcription factor families and number of isoforms. Table S2: Species and number of Sesbania homologous sequences. Table S3: GO annotation analysis of transcript isoforms of Sesbania. Table S4: KEGG enrichment of the Sesbania transcriptome.
Author Contributions
Conceptualization, T.H. and J.X.; methodology, T.H.; formal analysis, T.H. and G.S.; investigation, T.H., S.H. and J.D. (Jing Dong); resources, J.X.; data curation, X.Z., J.D. (Jinying Dai) and K.W.; writing—original draft preparation, T.H.; writing—review and editing, T.H. and Z.Z.; project administration, Z.Z. and J.X.; funding acquisition, Z.Z. and J.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Jiangsu Provincial Crop Germplasm Resource Bank (Salt-tolerant plants) of Yancheng Municipal Bureau of Agriculture and Rural Affairs (JS-ZW-K12) and the Scientific and Technological Innovation Fund of Carbon Emissions Peak and Neutrality of Jiangsu Provincial Department of Science and Technology (BE2022304).
Data Availability Statement
Source data used for analysis in this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank all the participants involved in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Singh, K.; Gera, R.; Sharma, R.; Maithani, D.; Chandra, D.; Bhat, M.A.; Kumar, R.; Bhatt, P. Mechanism and application of Sesbania root-nodulating bacteria: An alternative for chemical fertilizers and sustainable development. Arch. Microbiol. 2021, 203, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Bunma, S.; Balslev, H. A review of the economic botany of Sesbania (Leguminosae). Bot. Rev. 2019, 85, 185–251. [Google Scholar] [CrossRef]
- Jiang, P.; Tang, H.; Li, Y.; Liu, X. Effect of particle size of Sesbania gum on its modification, structure and performances. Int. J. Biol. Macromol. 2024, 262 Pt 2, 129719. [Google Scholar] [CrossRef] [PubMed]
- Daniel, K.; Hartman, S. How plant roots respond to waterlogging. J. Exp. Bot. 2024, 75, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Olorunwa, O.J.; Adhikari, B.; Shi, A.; Barickman, T.C. Screening of cowpea (Vigna unguiculata (L.) Walp.) genotypes for waterlogging tolerance using morpho-physiological traits at early growth stage. Plant Sci. 2022, 315, 111136. [Google Scholar] [CrossRef]
- Molla, M.; Rohman, M.; Islam, M.; Hasanuzzaman, M.; Hassan, L. Screening and evaluation of chilli (Capsicum annuum L.) genotypes for waterlogging tolerance at seedling stage. Biocell 2022, 46, 1613–1627. [Google Scholar] [CrossRef]
- Rajendran, A.; Ramlal, A.; Harika, A.; Subramaniam, S.; Raju, D.; Lal, S.K. Waterlogging stress mechanism and membrane transporters in soybean (Glycine max (L.) Merr.). Plant Physiol. Biochem. 2025, 220, 109579. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Oehrle, N.W.; Alaswad, A.A.; Stevens, W.G.; Mari John, K.M.; Luthria, D.L.; Natarajan, S.S. Biochemical and anatomical investigation of Sesbania herbacea (Mill.) McVaugh nodules grown under flooded and non–flooded conditions. Int. J. Mol. Sci. 2019, 20, 1824. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Yan, K.; Zhang, H.; Luo, Y.M.; Xie, Z.H. Elucidation of the molecular responses to waterlogging in Sesbania cannabina roots by transcriptome profiling. Sci. Rep. 2017, 7, 9256. [Google Scholar] [CrossRef]
- Geng, S.; Lin, Z.; Xie, S.; Xiao, J.; Wang, H.; Zhao, X.; Zhou, Y.; Duan, L. Ethylene enhanced waterlogging tolerance by changing root architecture and inducing aerenchyma formation in maize seedlings. J. Plant Physiol. 2023, 287, 154042. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, C.; Pang, J.; Niu, Y.; Liu, H.; Zhang, W.; Zhou, M. Genome-wide association study reveals quantitative trait loci for waterlogging-triggered adventitious roots and aerenchyma formation in common wheat. Front. Plant Sci. 2022, 13, 1066752. [Google Scholar] [CrossRef] [PubMed]
- Elena, L.; Hans, V.; Pierdomenico, P. Plant responses to fooding stress. Curr. Opin. Plant Biol. 2016, 33, 64–71. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Xie, Z.H. Role of abscisic acid in strigolactone-induced salt stress tolerance in arbuscular mycorrhizal Sesbania cannabina seedlings. BMC Plant Biol. 2018, 18, 74. [Google Scholar] [CrossRef]
- Nakano, K.; Shiroma, A.; Shimoji, M.; Tamotsu, H.; Ashimine, N.; Ohki, S.; Shinzato, M.; Minami, M.; Nakanishi, T.; Teruya, K.; et al. Advantages of genome sequencing by long-read sequencer using SMRT technology in medical area. Hum. Cell 2017, 30, 149–161. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Vollger, M.R.; Eichler, E.E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 2020, 21, 597–614. [Google Scholar] [CrossRef]
- Hu, X.F.; Jin, M.J.; Gong, Z.X.; Lin, Z.L.; Zhang, L.Z.; Zeng, Z.J.; Wang, Z.L. Full-length transcriptome profile of Apis cerana revealed by nanopore sequencing. Int. J. Mol. Sci. 2024, 25, 10833. [Google Scholar] [CrossRef]
- Xiong, Y.; Yang, J.; Xiong, Y.; Zhao, J.; Liu, L.; Liu, W.; Sha, L.; Zhou, J.; You, M.; Li, D.; et al. Full-length transcriptome sequencing analysis and characterization, development and validation of microsatellite markers in Kengyilia melanthera. Front. Plant Sci. 2022, 13, 959042. [Google Scholar] [CrossRef]
- Liao, T.; Zhang, L.; Wang, Y. Full-length transcriptome characterization of Platycladus orientalis based on the PacBio platform. Front. Genet. 2024, 18, 1345039. [Google Scholar] [CrossRef]
- Wang, L.; Sa, W.; Yin, W.; Wei, W.; Wang, Y.; Sa, W.; Liang, J. Single-molecule real-time sequencing of the full-length transcriptome of purple garlic (Allium sativum L. cv. Leduzipi) and identification of serine O-acetyltransferase family proteins involved in cysteine biosynthesis. J. Sci. Food Agric. 2022, 102, 2864–2873. [Google Scholar] [CrossRef]
- Zhang, X.; Li, G.; Zhou, J.; Lv, M.; Li, L.; Chen, J. Full-length gonad transcriptome analysis of Amur sturgeon Dmrt family genes: Identification, characterization, and expression patterns during gonadal differentiation. Fish Physiol. Biochem. 2022, 48, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.; Meng, X.; Zhao, Z.; Kang, D.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Xu, Q.T.; Yang, L.; Zhou, Z.Q.; Mei, F.Z.; Qu, L.H.; Zhou, G.S. Process of aerenchyma formation and reactive oxygen species induced by waterlogging in wheat seminal roots. Planta 2013, 238, 969–982. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Tuan, P.A.; Mukherjee, S.; Son, S.; Ayele, B.T. Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat. J. Exp. Bot. 2018, 69, 4065–4082. [Google Scholar] [CrossRef]
- Nagai, K.; Ashikari, M. Molecular mechanism of internode elongation in rice. Breed Sci. 2023, 73, 108–116. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Hu, Q.; Yan, W.; Pan, J.; Yan, Y.; Chen, X. A CsEIL3-CsARN6.1 module promotes waterlogging-triggered adventitious root formation in cucumber by activating the expression of CsPrx5. Plant J. 2023, 114, 824–835. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, J.; Wang, Z.; Li, X.; Kan, J.; Li, J. SMRT sequencing of full-length transcriptome of birch-leaf pear (Pyrus betulifolia Bunge) under drought stress. J. Genet. 2021, 100, 29. [Google Scholar] [CrossRef]
- Zhu, L.; Lu, L.; Yang, L.; Hao, Z.; Chen, J.; Cheng, T. The full-length transcriptome sequencing and identification of Na+/H+ antiporter genes in halophyte Nitraria tangutorum Bobrov. Genes 2021, 12, 836. [Google Scholar] [CrossRef]
- He, W.; Luo, L.; Xie, R.; Chai, J.; Wang, H.; Wang, Y.; Chen, Q.; Wu, Z.; Yang, S.; Li, M.; et al. Genome-Wide identification and functional analysis of the AP2/ERF transcription factor family in citrus rootstock under waterlogging stress. Int. J. Mol. Sci. 2023, 24, 8989. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, P.; Zhou, T.; Wu, Y.; Yuan, M.; Zhang, X.; Liu, Y. Genome-wide characterization and expression analysis of the bHLH gene family in response to abiotic stresses in Zingiber officinale Roscoe. BMC Genom. 2025, 26, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Yang, L.; Niu, J.; Huang, R.; Yuan, F.; Liang, Q. Development of SSR and SNP markers for identifying opium poppy. Int. J. Legal Med. 2022, 136, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Z.; Hu, A.; Wu, H.; Zhu, J.; Wang, F.; Cao, P.; Yang, X.; Zhang, H. Full-length transcriptome analysis of the halophyte Nitraria sibirica Pall. Genes 2022, 13, 661. [Google Scholar] [CrossRef]
- Tian, R.; Sun, X.; Liu, C.; Chu, J.; Zhao, M.; Zhang, W.H. A Medicago truncatula lncRNA MtCIR1 negatively regulates response to salt stress. Planta 2023, 257, 32. [Google Scholar] [CrossRef]
- Zhu, Z.; Dong, Q.; Bing, J.; Songbuerbatu; Zheng, L.; Dorjee, T.; Liu, Q.; Zhou, Y.; Gao, F. Combined lncRNA and mRNA expression profiles identified the lncRNA-miRNA-mRNA modules regulating the cold stress response in Ammopiptanthus nanus. Int. J. Mol. Sci. 2023, 24, 6502. [Google Scholar] [CrossRef]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.Q.; Jia, Y.; Li, C. Opportunities for improving waterlogging tolerance in cereal crops-physiological traits and genetic mechanisms. Plants 2021, 10, 1560. [Google Scholar] [CrossRef]
- Lin, C.; Sauter, M. Polar auxin transport determines adventitious root emergence and growth in rice. Front. Plant Sci. 2019, 10, 444. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).