A Comprehensive Analysis of the Multiple AP2/ERF Regulatory Network Unveils Putative Components of the Fatty Acid Pathway for Environmental Adaptation
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomes and Transcriptomes Mining
2.2. Systematic Identification of the AP2/ERF Gene Family
2.3. Orthogroups Identification of Multispecies AP2/ERFs
2.4. Sequence Alignment, TimeTree, and Phylogenetic Tree Construction
2.5. Systematic Analysis of AP2/ERF Gene Structure
2.6. Chromosome Distribution, Syntenic Analysis, and Comparative Genomics
2.7. Identification of AP2/ERFs Gene Types and Development-Related Phenotypic Analysis
2.8. Gene Ontology (GO) Analysis and Correlation Network Construction
2.9. Gene-Module Association Determination (G-MAD)
2.10. Construction of CqAP2/ERFs Expression Profiles
3. Results
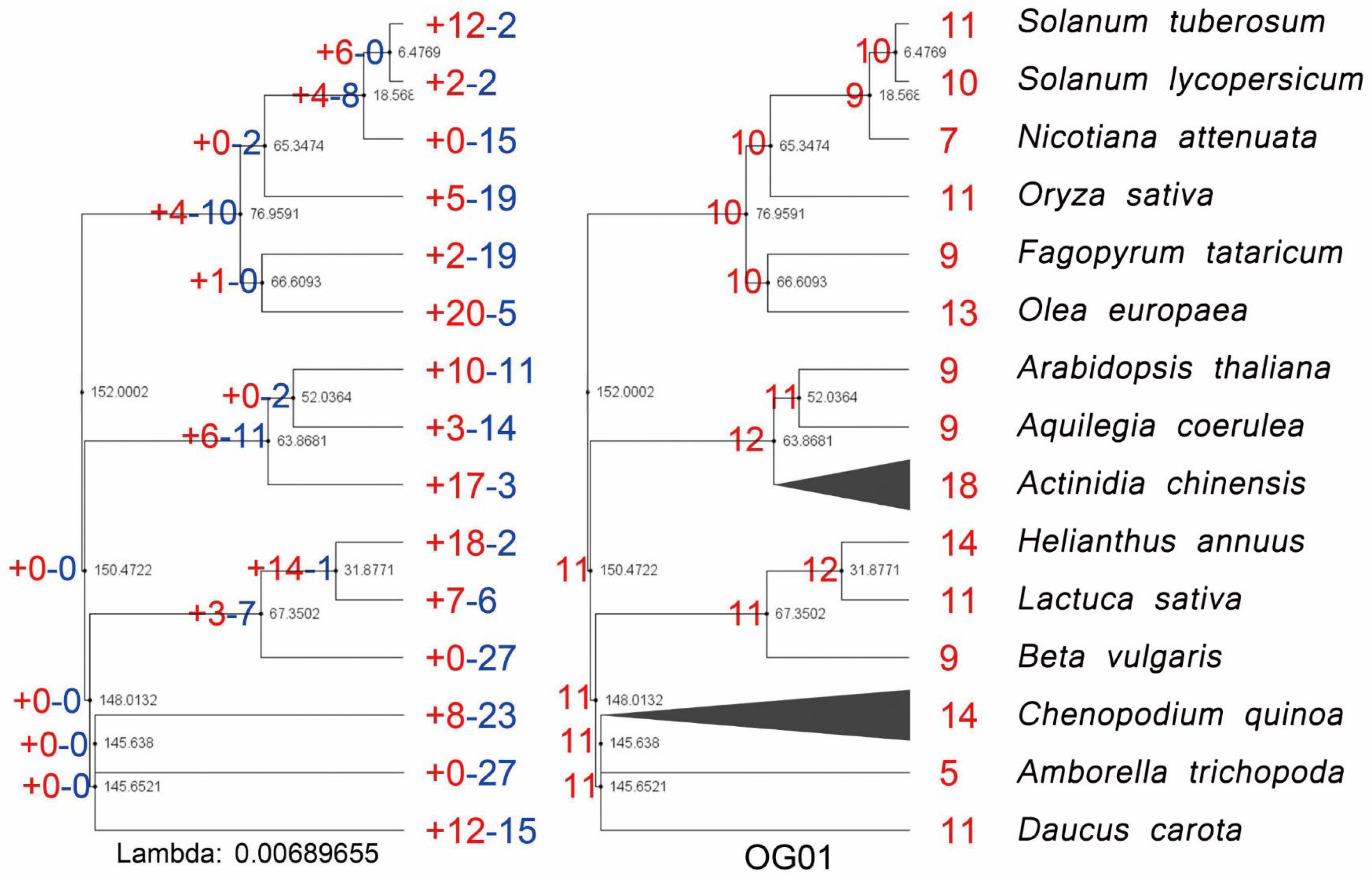
3.1. Phylogenetic Identification, Evolutionary Expansion, and Contraction of AP2/ERFs in Important Plants
3.2. The Expansion of AP2/ERFs in Important Plants Caused by Recursive Amplification
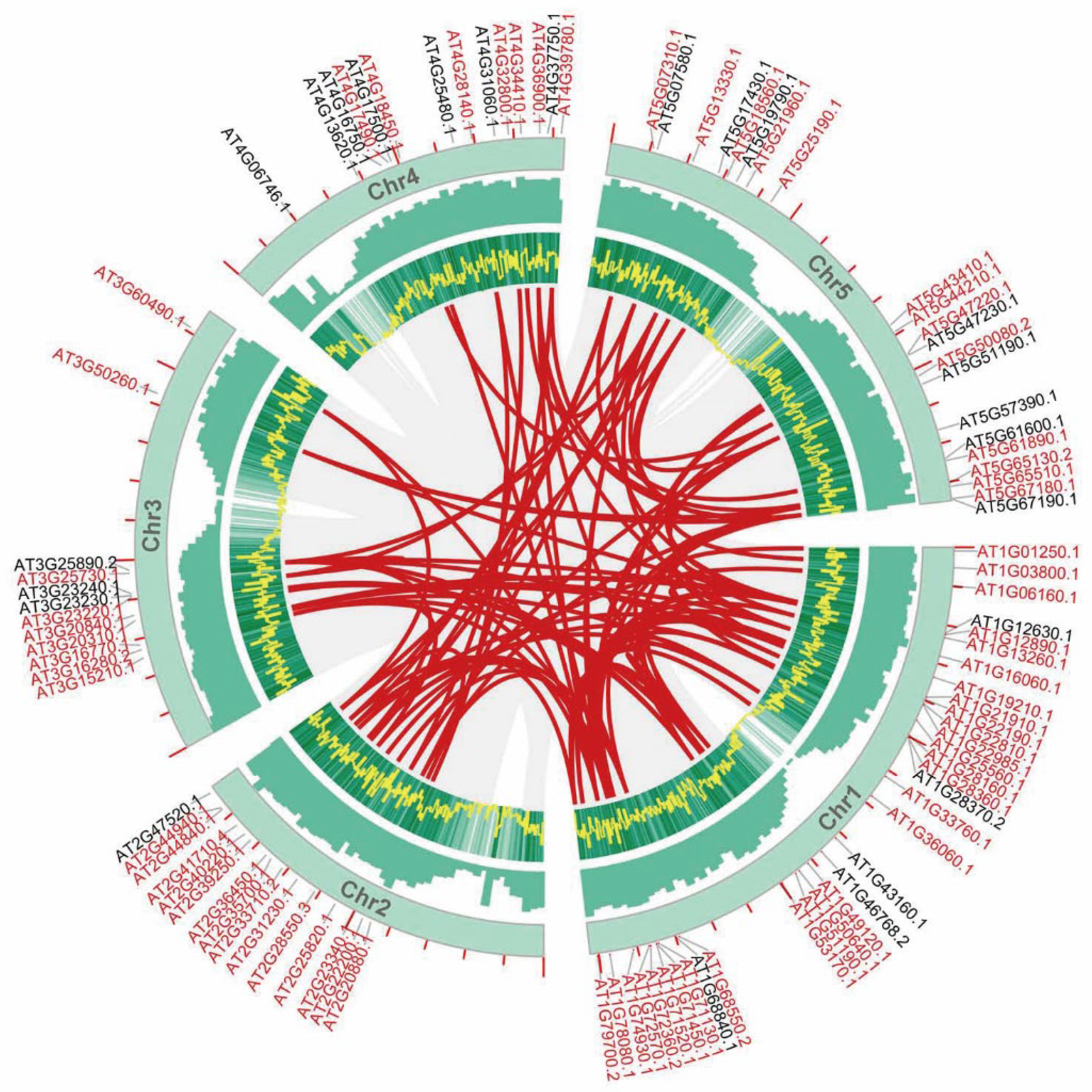
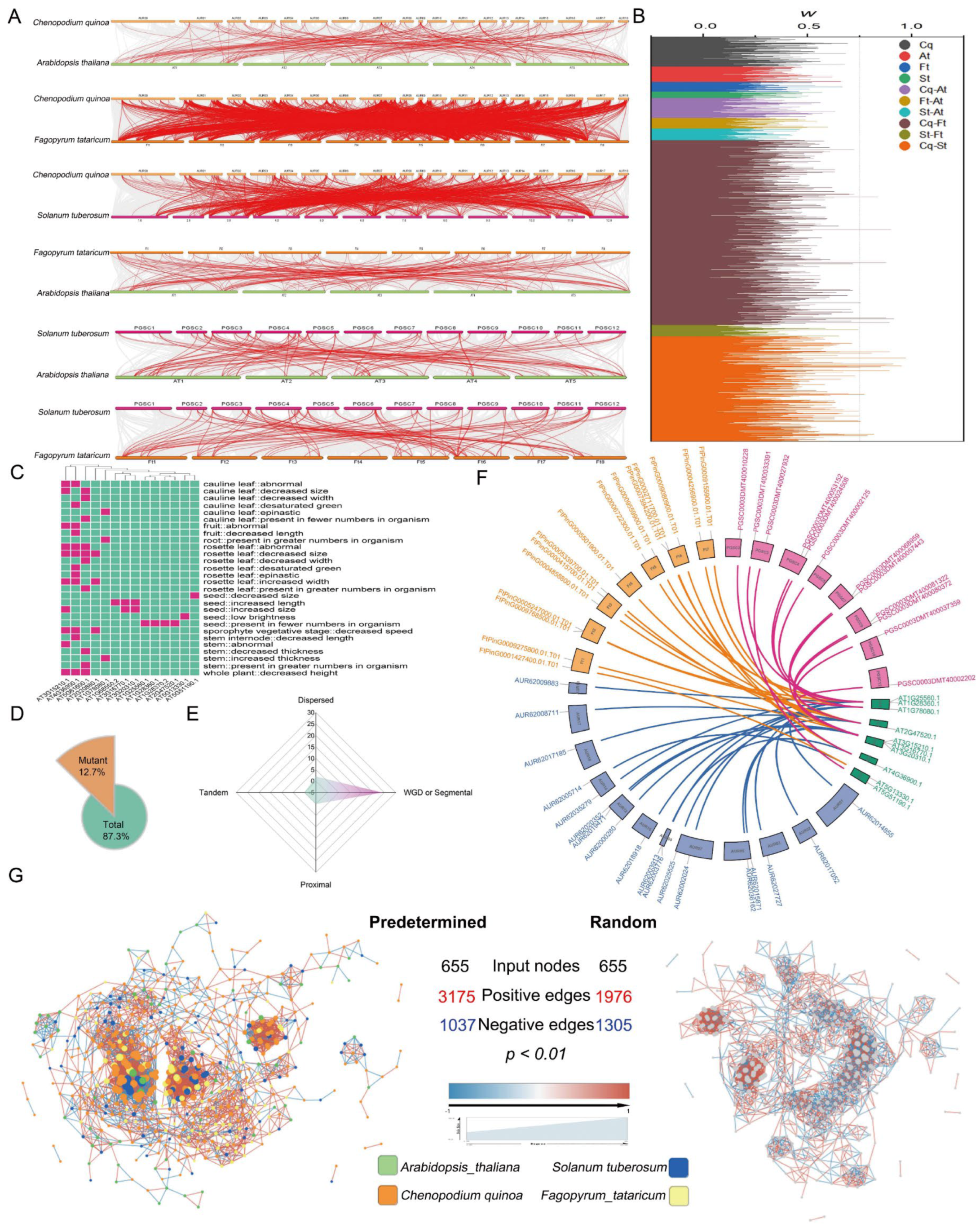
3.3. Genomic Synteny Conservation of AP2/ERFs in Important Plants
3.4. Phenotypic Analysis of AtAP2/ERFs and Mapping to Important AP2/ERFs in Other Species
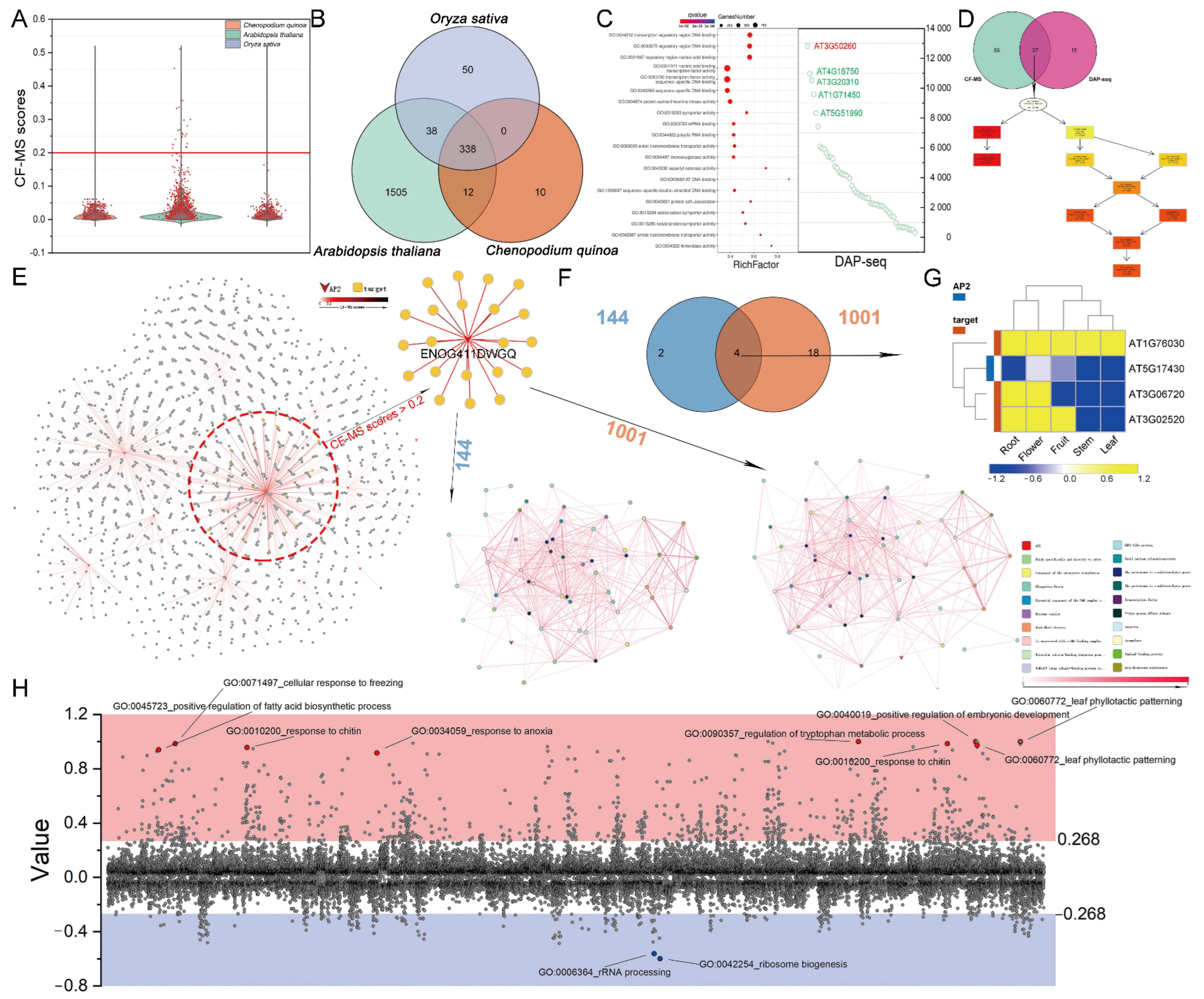
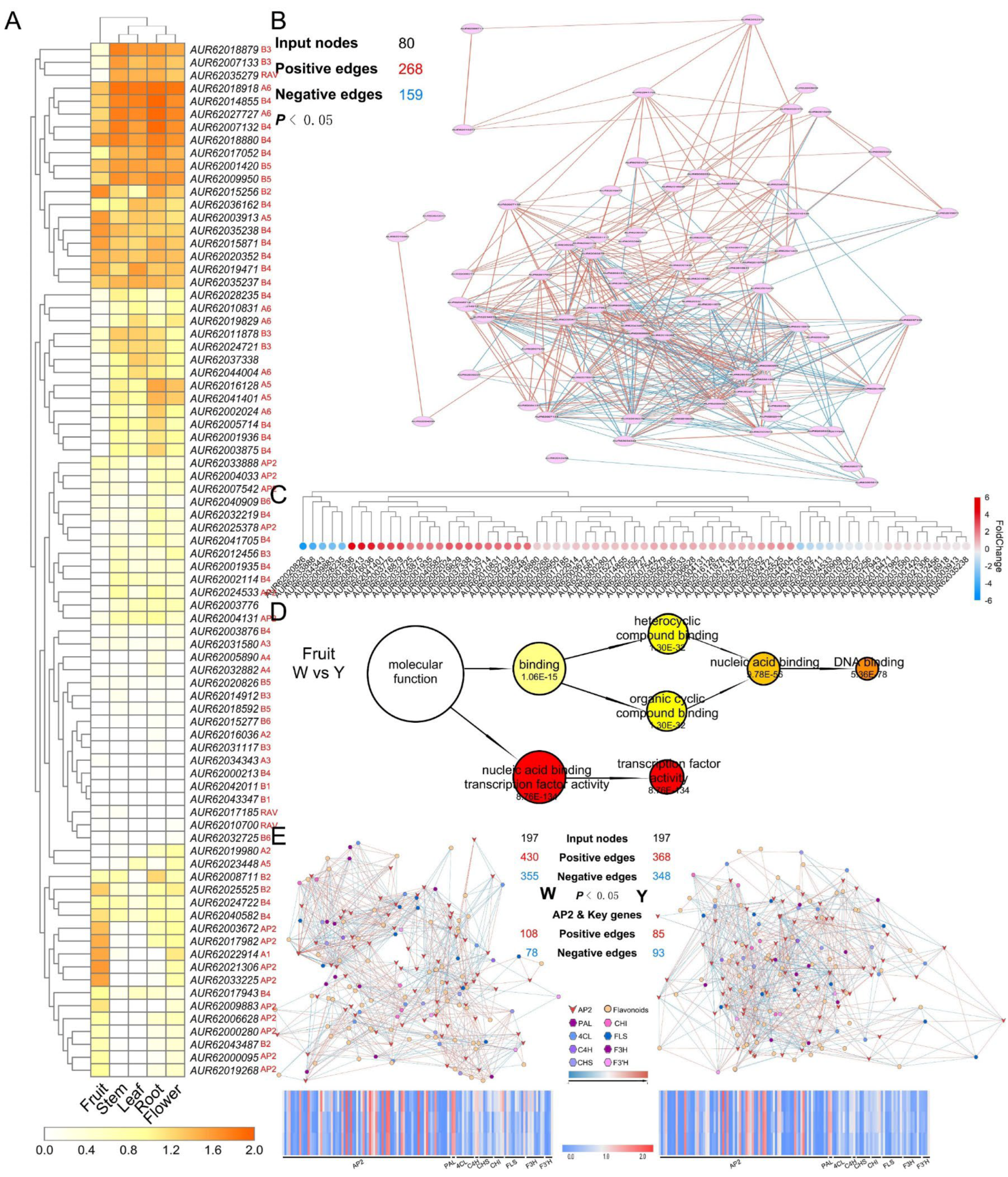
3.5. CF-MS and DNA Affinity Purification Sequencing (DAP-Seq) Determine the Functions of AP2/ERFs
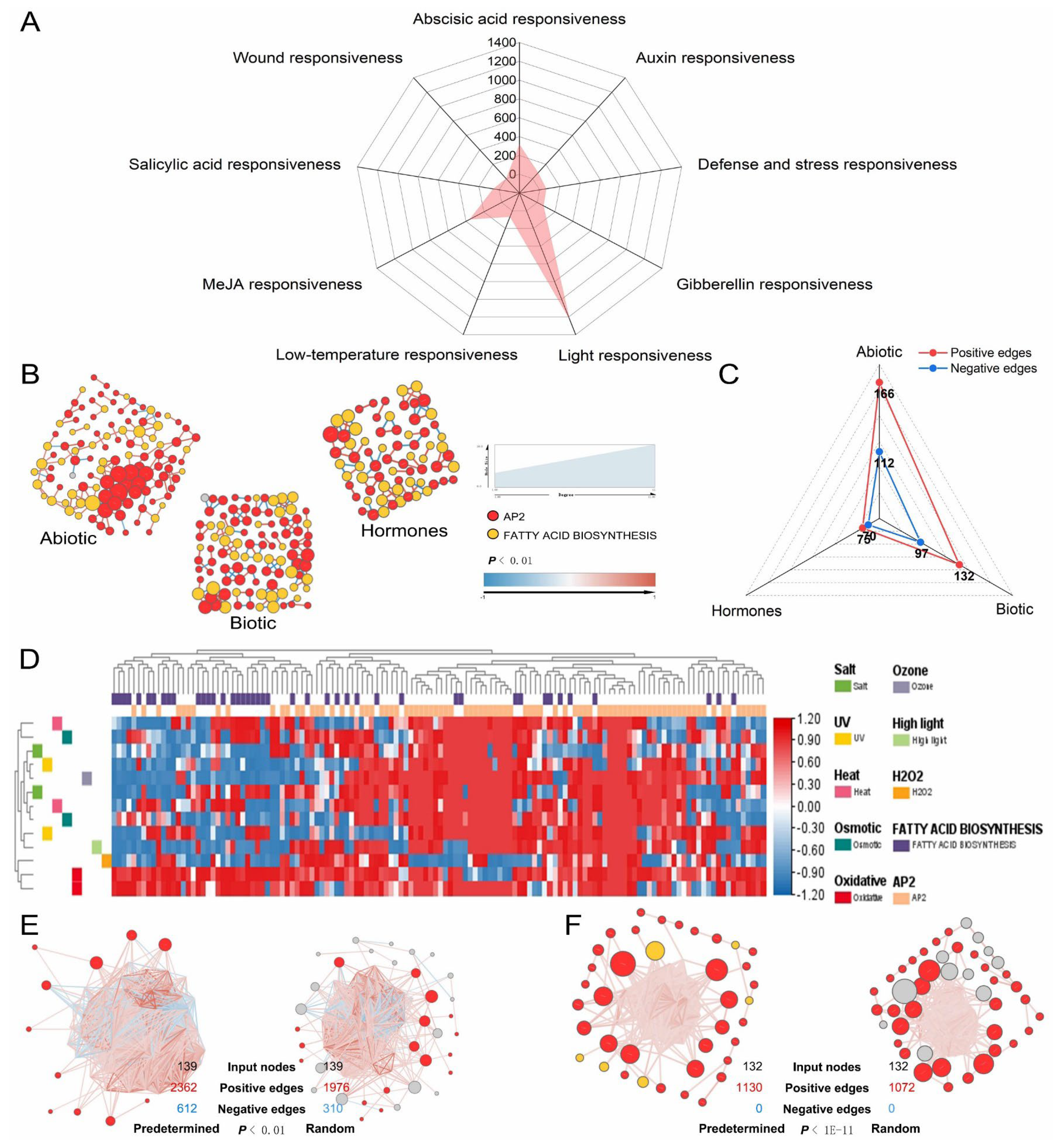
3.6. Polyploidy-Related CqAP2/ERFs May Promote Environmental Adaptability by Regulating the Fatty Acid Synthesis Pathway
3.7. General Applicability Network of AP2/ERFs and Fatty Acid Pathway Genes
4. Discussion
4.1. The Evolution of Auxiliary Motifs in AP2/ERFs and Their Neofunctionalization
4.2. AP2/ERFs Generated by Amplification May Facilitate Environmental Adaptation
4.3. Comparative Perspectives and Breeding Prospects of AP2/ERF Expansion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guyomarc’h, S.; Boutté, Y.; Laplaze, L. AP2/ERF transcription factors orchestrate very long chain fatty acid biosynthesis during Arabidopsis lateral root development. Mol. Plant 2021, 14, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xue, L.; Cheng, J.; Yang, X.; Xie, H.; Song, X.; Qiang, S. Polyploidization-driven differentiation of freezing tolerance in Solidago canadensis. Plant Cell Environ. 2020, 43, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, K.; Dai, X.; Qin, G.; Lu, D.; Gao, Z.; Li, X.; Song, B.; Bian, J.; Ren, D.; et al. A telomere-to-telomere genome assembly coupled with multi-omic data provides insights into the evolution of hexaploid bread wheat. Nat. Genet. 2025, 57, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Liu, M.; Yu, J.; Yang, M.; Cao, L.; Chen, C. Adaptive evolution of chloroplast division mechanisms during plant terrestrialization. Cell Rep. 2024, 43, 113950. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Li, A.; Guo, J.; Wang, H.; Qi, H.; Li, M.; Yang, P.; Song, S. An AP2/ERF transcription factor confers chilling tolerance in rice. Sci. Adv. 2024, 10, eado4788. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, L.; Chen, Y.; Peng, H.; Ni, Z.; Sun, Q.; Guo, W. Deciphering the evolution and complexity of wheat germplasm from a genomic perspective. J. Genet. Genom. 2023, 50, 846–860. [Google Scholar] [CrossRef]
- Yu, H.; Liao, J.; Jiang, Y.; Zhong, M.; Tao, S.; Chai, S.; Wang, L.; Lin, L.; Yang, R.; Deng, X.; et al. Ecotype-specific phenolic acid accumulation and root softness in Salvia miltiorrhiza are driven by environmental and genetic factors. Plant Biotechnol. J. 2025. [Google Scholar] [CrossRef]
- Münzbergová, Z.; Haisel, D. Effects of polyploidization on the contents of photosynthetic pigments are largely population-specific. Photosynth. Res. 2019, 140, 289–299. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, Q.; Xu, Y.; Yang, S.; Gao, M.; Wang, Y. Effects of tung oilseed FAD2 and DGAT2 genes on unsaturated fatty acid accumulation in Rhodotorula glutinis and Arabidopsis thaliana. Mol. Genet. Genom. 2015, 290, 1605–1613. [Google Scholar] [CrossRef]
- Zuo, S.; Guo, X.; Mandáková, T.; Edginton, M.; Al-Shehbaz, I.A.; Lysak, M.A. Genome diploidization associates with cladogenesis, trait disparity, and plastid gene evolution. Plant Physiol. 2022, 190, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Magar, M.M.; Liu, H.; Yan, G. Genome-Wide Analysis of AP2/ERF Superfamily Genes in Contrasting Wheat Genotypes Reveals Heat Stress-Related Candidate Genes. Front. Plant Sci. 2022, 13, 853086. [Google Scholar] [CrossRef]
- Xiao, L.; Ren, J.-Z.; Li, Q.; Yang, B.; Liu, Z.-J.; Chen, R.-B.; Zhang, L. Genome-wide analysis of AP2/ERF superfamily in Isatis indigotica. J. Integr. Med. 2023, 21, 77–88. [Google Scholar] [CrossRef]
- Ghorbani, R.; Zakipour, Z.; Alemzadeh, A.; Razi, H. Genome-wide analysis of AP2/ERF transcription factors family in Brassica napus. Physiol. Mol. Biol. Plants 2020, 26, 1463–1476. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Zhao, M.; Haxim, Y.; Liang, Y.; Qiao, S.; Gao, B.; Zhang, D.; Li, X. Genome-wide investigation of AP2/ERF gene family in the desert legume Eremosparton songoricum: Identification, classification, evolution, and expression profiling under drought stress. Front. Plant Sci. 2022, 13, 885694. [Google Scholar] [CrossRef]
- Liu, M.; Xu, Y.; Song, Y.; Fan, D.; Li, J.; Zhang, Z.; Wang, L.; He, J.; Chen, C.; Ma, C. Hierarchical Regulatory Networks Reveal Conserved Drivers of Plant Drought Response at the Cell-Type Level. Adv. Sci. 2025, e2415106. [Google Scholar] [CrossRef]
- Aanniz, T.; El Baaboua, A.; Aboulaghras, S.; Bouyahya, A.; Benali, T.; Balahbib, A.; El Omari, N.; Butnariu, M.; Muzammil, K.; Yadav, K.K.; et al. Impact of water stress to plant epigenetic mechanisms in stress and adaptation. Physiol. Plant. 2025, 177, e70058. [Google Scholar] [CrossRef]
- Landis, J.B.; Soltis, D.E.; Li, Z.; Marx, H.E.; Barker, M.S.; Tank, D.C.; Soltis, P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018, 105, 348–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gao, M.; Zhao, Y.; Chen, Y.; Wu, L.; Yin, H.; Xiong, S.; Wang, S.; Wang, J.; Yang, Y.; et al. LcERF19, an AP2/ERF transcription factor from Litsea cubeba, positively regulates geranial and neral biosynthesis. Hortic. Res. 2022, 9, uhac093. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular Events of Rice AP2/ERF Transcription Factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Singh, S.K.; Patra, B.; Liu, X.; Pattanaik, S.; Yuan, L. Mutually Regulated AP2/ERF Gene Clusters Modulate Biosynthesis of Specialized Metabolites in Plants. Plant Physiol. 2020, 182, 840–856. [Google Scholar] [CrossRef]
- Kandhol, N.; Jain, M.; Tripathi, D.K. Nanoparticles as potential hallmarks of drought stress tolerance in plants. Physiol. Plant. 2022, 174, e13665. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.F.R.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.-H.; Reuzeau, C.; Kim, J.-K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef]
- Parks, M.B.; Nakov, T.; Ruck, E.C.; Wickett, N.J.; Alverson, A.J. Phylogenomics reveals an extensive history of genome duplication in diatoms (Bacillariophyta). Am. J. Bot. 2018, 105, 330–347. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Ma, N.; Sun, P.; Li, Z.-Y.; Zhang, F.-J.; Wang, X.-F.; You, C.-X.; Zhang, C.-L.; Zhang, Z. Plant disease resistance outputs regulated by AP2/ERF transcription factor family. Stress Biol. 2024, 4, 2. [Google Scholar] [CrossRef]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- He, M.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Multiple Roles in Stress Response. Front. Plant Sci. 2020, 11, 562785. [Google Scholar] [CrossRef]
- Feng, D.; Cheng, J.; Yang, X.; Tian, Z.; Liu, Y.; Zhang, Y.; Qiang, S. Polyploidization-enhanced effective clonal reproduction endows the successful invasion of Solidago canadensis. Ecol. Appl. 2024, 34, e2738. [Google Scholar] [CrossRef] [PubMed]
- Clo, J. Polyploidization: Consequences of genome doubling on the evolutionary potential of populations. Am. J. Bot. 2022, 109, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Polyploidy and interspecific hybridization: Partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Liu, M.; Yang, M.; Liang, H.; Luo, B.; Deng, J.; Cao, L.; Zheng, D.; Chen, C. Polyploidy drives autophagy to participate in plant-specific functions. iMeta 2024, 3, e252. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S.P.; Schwarzacher, T.; Liu, Q. Polyploidy: Its consequences and enabling role in plant diversification and evolution. Ann. Bot. 2023, 131, 1–10. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Su, Y.; Shen, H. Rice Responses to Abiotic Stress: Key Proteins and Molecular Mechanisms. Int. J. Mol. Sci. 2025, 26, 896. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, X.; Wei, L.; Wansee, S.; Rabbani Nasab, H.; Chen, L.; Kang, Z.; Wang, J. TaAP2-10, an AP2/ERF transcription factor, contributes to wheat resistance against stripe rust. J. Plant Physiol. 2023, 288, 154078. [Google Scholar] [CrossRef]
- Zhang, Y.; Ji, A.; Xu, Z.; Luo, H.; Song, J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 2019, 100, 83–93. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, R.; Xiang, C.; Zhang, R.; Wang, Q.; Wang, T.; Li, X.; Lu, X.; Gao, S.; Liu, Z.; et al. Transcriptomic and Physiological Analysis Reveal That α-Linolenic Acid Biosynthesis Responds to Early Chilling Tolerance in Pumpkin Rootstock Varieties. Front. Plant Sci. 2021, 12, 669565. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, S.; Chang, X.; Wang, X.; Zhao, Y.; Xia, Y.; Trigiano, R.N.; Jiao, Y.; Chen, F. The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant Cell Environ. 2020, 43, 2847–2856. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tan, N.W.K.; Chung, F.Y.; Yamaguchi, N.; Gan, E.-S.; Ito, T. Transcriptional Regulators of Plant Adaptation to Heat Stress. Int. J. Mol. Sci. 2023, 24, 13297. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Yang, M.; Liang, H.; Zheng, D.; Zhu, G.; Ye, Z.; Lai, X.; Liu, M. A Comprehensive Analysis of the Multiple AP2/ERF Regulatory Network Unveils Putative Components of the Fatty Acid Pathway for Environmental Adaptation. Agronomy 2025, 15, 1112. https://doi.org/10.3390/agronomy15051112
Deng J, Yang M, Liang H, Zheng D, Zhu G, Ye Z, Lai X, Liu M. A Comprehensive Analysis of the Multiple AP2/ERF Regulatory Network Unveils Putative Components of the Fatty Acid Pathway for Environmental Adaptation. Agronomy. 2025; 15(5):1112. https://doi.org/10.3390/agronomy15051112
Chicago/Turabian StyleDeng, Junjie, Ming Yang, Heng Liang, Daojun Zheng, Guangshun Zhu, Zhenpei Ye, Xinjie Lai, and Moyang Liu. 2025. "A Comprehensive Analysis of the Multiple AP2/ERF Regulatory Network Unveils Putative Components of the Fatty Acid Pathway for Environmental Adaptation" Agronomy 15, no. 5: 1112. https://doi.org/10.3390/agronomy15051112
APA StyleDeng, J., Yang, M., Liang, H., Zheng, D., Zhu, G., Ye, Z., Lai, X., & Liu, M. (2025). A Comprehensive Analysis of the Multiple AP2/ERF Regulatory Network Unveils Putative Components of the Fatty Acid Pathway for Environmental Adaptation. Agronomy, 15(5), 1112. https://doi.org/10.3390/agronomy15051112






