Genome-Wide Analysis of Zm4CL Genes Identifies Zm4CL8 Regulating Drought and Salt Tolerance in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of 4CL Genes in Maize
2.2. Phylogenetic Analysis and Prediction of the Physicochemical Properties of Zm4CL Proteins
2.3. Chromosomal Localization and Collinearity Analysis
2.4. Analysis of the Gene Structure and Protein Domain, and Motif Detection of the Zm4CL Gene Family
2.5. Analysis of the cis-Acting Elements in the Promoters of the Zm4CL Gene Family
2.6. Expression Analysis of Zm4CL Genes in Different Tissues
2.7. Prediction of the Upstream Transcription Factors for Zm4CL Genes in Clade 4CL
2.8. Analysis of Zm4CL Genes Expression in Maize Under Abiotic Stresses
2.9. Stress Response Analysis of Selected Zm4CL Genes
2.10. RNA Extraction, Reverse Transcription, and Real-Time Quantitative PCR Analysis
2.11. Construction of VIGS Vectors, Vacuum-Assisted Agroinfiltration, and Co-Cultivation Procedures
2.12. Drought and Salt Stress Tolerance Assays
2.13. Determination of Relative Water Content
3. Results
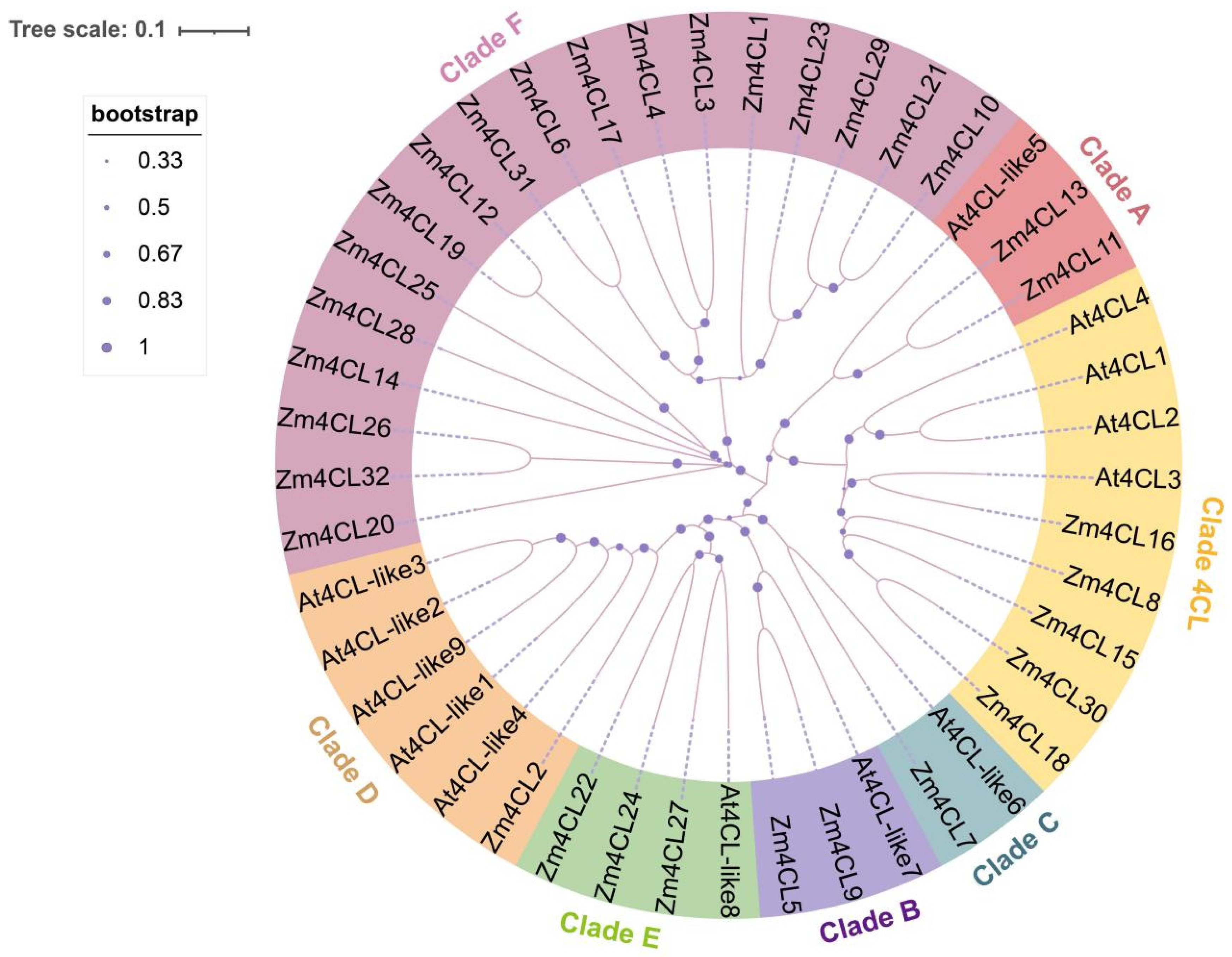
3.1. Identification and Phylogenetic Relationship of 4CL Genes in Maize
3.2. Chromosomal Localization and Syntenic Analysis of Zm4CL Genes
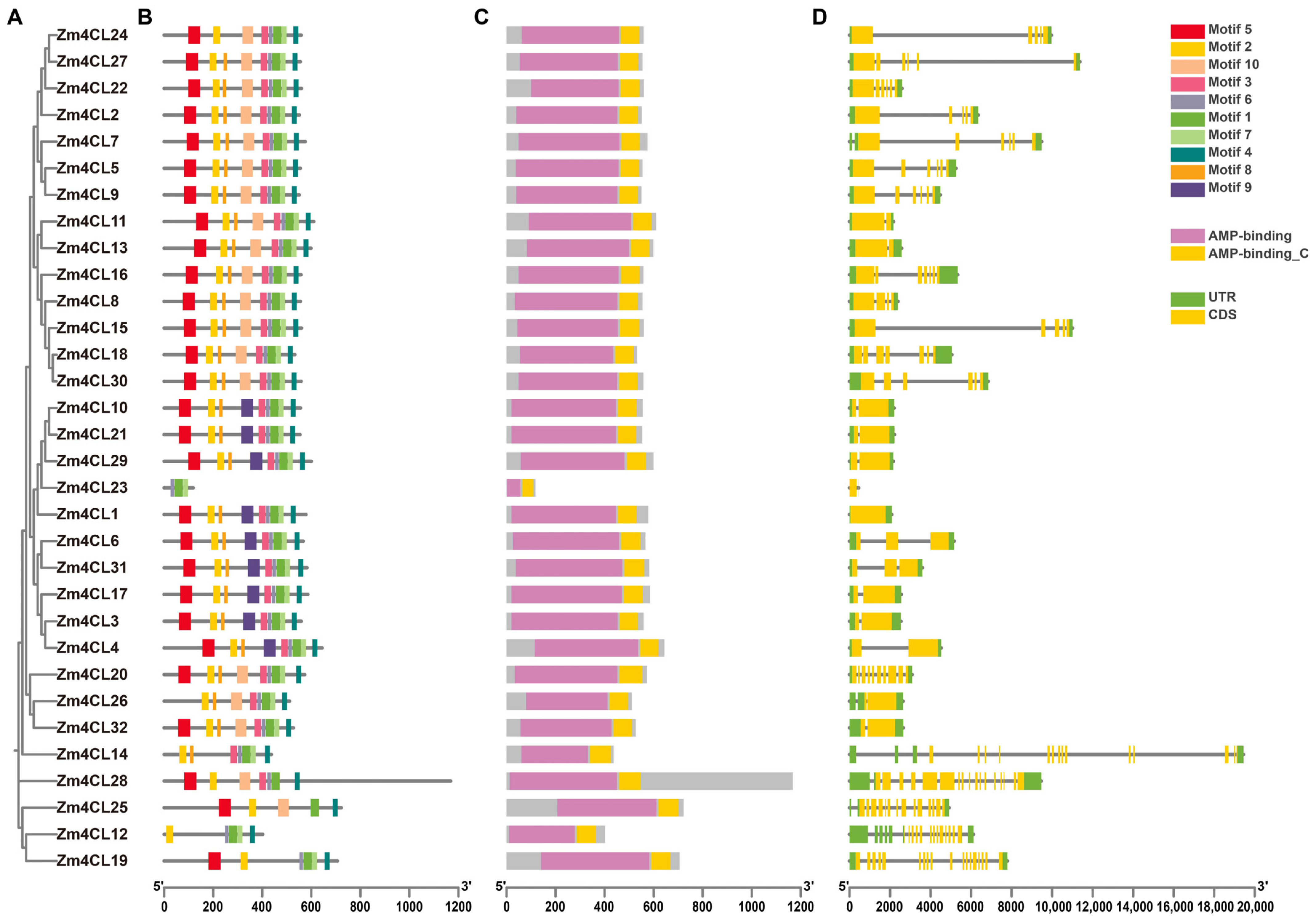
3.3. Conserved Motif, Domain, and Gene Structure Analysis of 4CL Gene Family Members in Maize
3.4. Analysis of Promoter cis-Elements in Zm4CL Genes
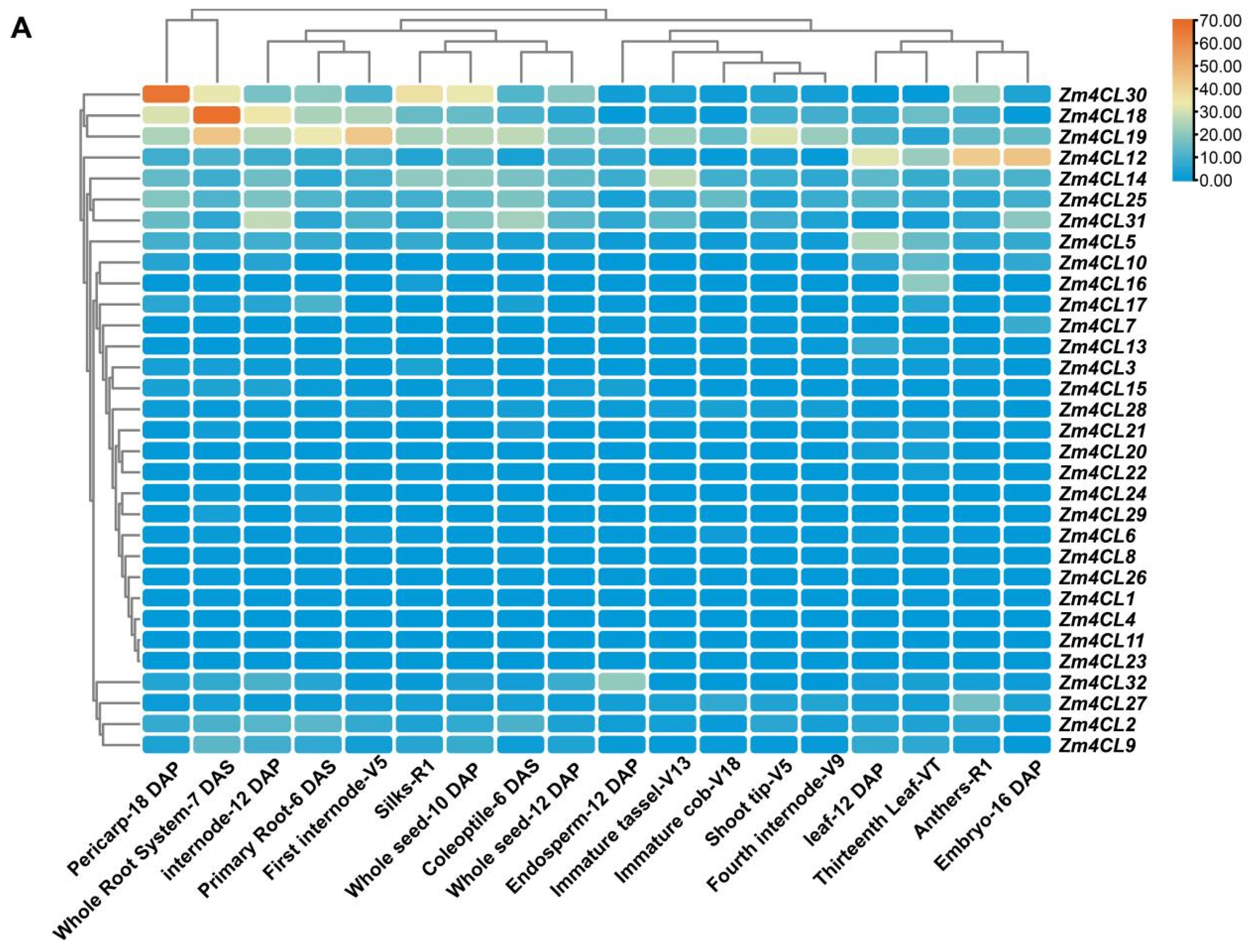
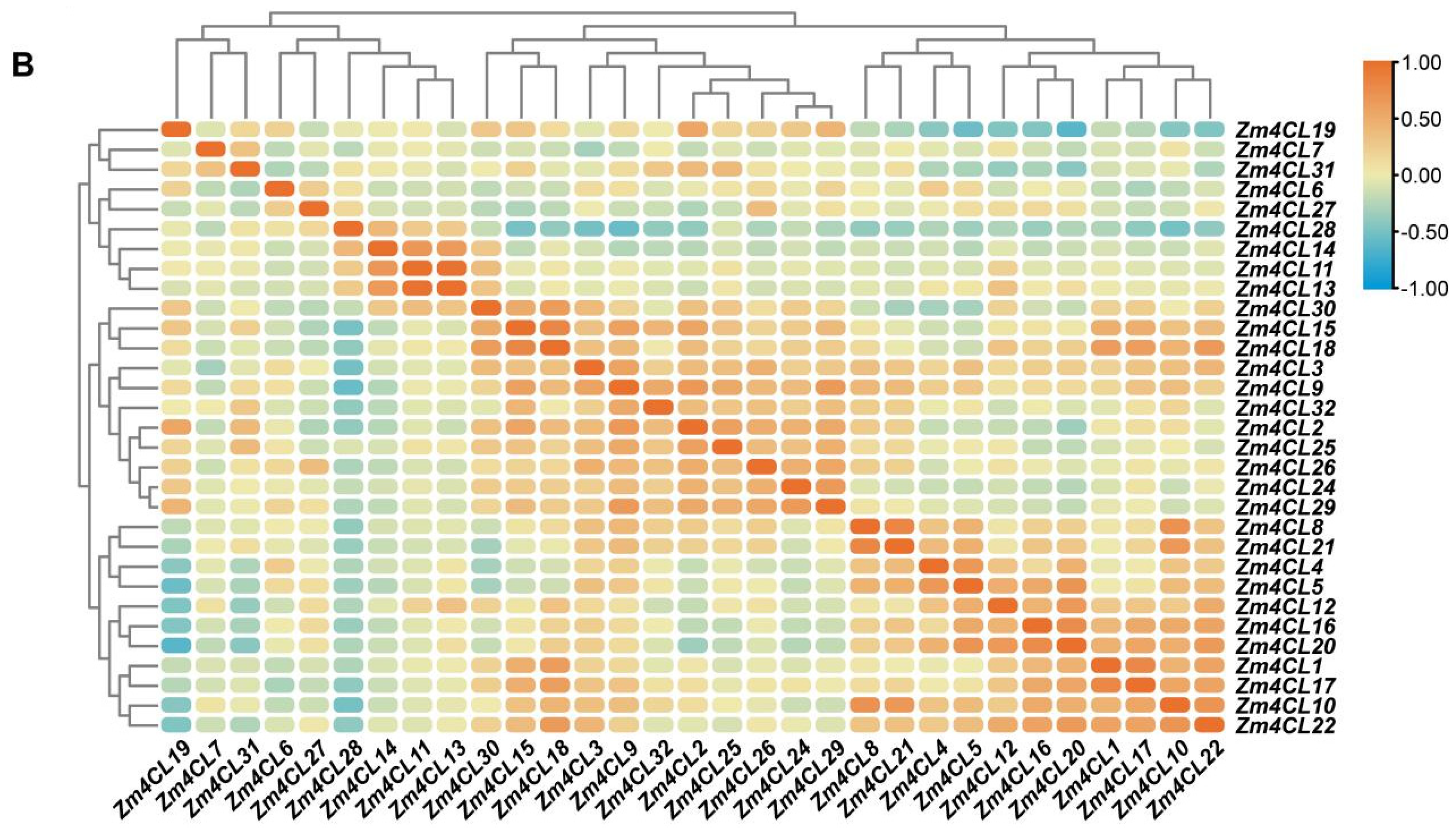
3.5. Expression Patterns and Correlation Analysis of 4CL Genes in Maize
3.6. Prediction of Potential Upstream Regulators for Zm4CL Genes Within the 4CL Clade
3.7. Expression Analysis of Zm4CLs Under Abiotic Stress
3.8. Expression Profiling of Selected Zm4CL Genes Under Stress Conditions Using RT-qPCR
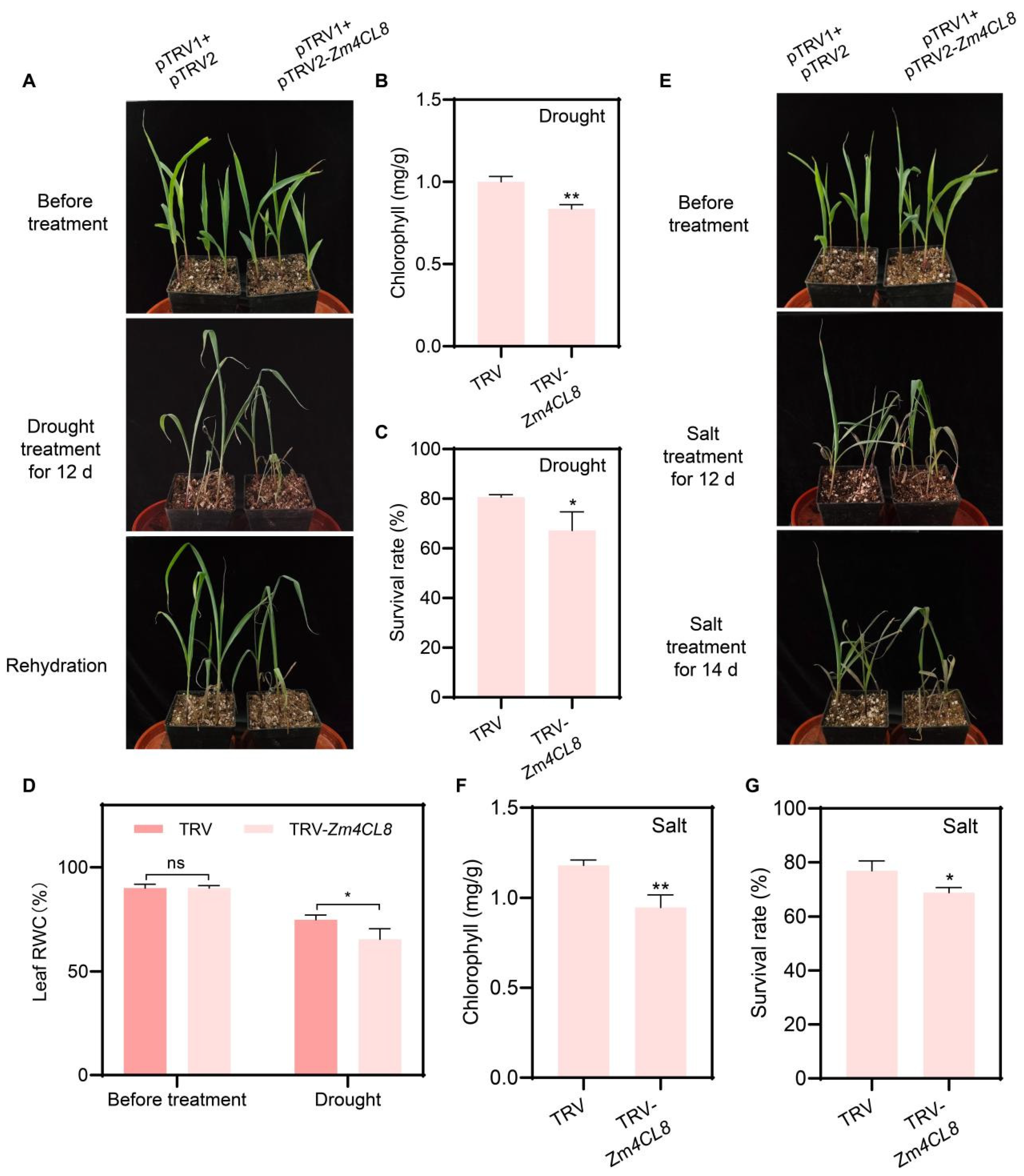
3.9. Silencing of Zm4CL8 Impairs Drought and Salt Stress Tolerance in Maize Seedlings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arabidopsis Book 2010, 8, e0132. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Jayaramaiah, R.H.; Beedkar, S.D.; Singh, P.A.; Joshi, R.S.; Mulani, F.A.; Dholakia, B.B.; Punekar, S.A.; Gade, W.N.; Thulasiram, H.V.; et al. Comparative functional characterization of eugenol synthase from four different Ocimum species: Implications on eugenol accumulation. Biochim. Biophys. Acta 2016, 1864, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar] [CrossRef]
- Allina, S.M.; Pri-Hadash, A.; Theilmann, D.A.; Ellis, B.E.; Douglas, C.J. 4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol. 1998, 116, 743–754. [Google Scholar] [CrossRef]
- Hahlbrock, K.; Scheel, D. Physiology and Molecular Biology of Phenylpropanoid Metabolism. Annu. Rev. Plant Biol. 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.P. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef]
- Fulda, M.; Heinz, E.; Wolter, F.P. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol. Gen. Genet. 1994, 242, 241–249. [Google Scholar] [CrossRef]
- Stuible, H.; Büttner, D.; Ehlting, J.; Hahlbrock, K.; Kombrink, E. Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett. 2000, 467, 117–122. [Google Scholar] [CrossRef]
- Stuible, H.P.; Kombrink, E. Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J. Biol. Chem. 2001, 276, 26893–26897. [Google Scholar] [CrossRef]
- Zhang, C.H.; Ma, T.; Luo, W.C.; Xu, J.M.; Liu, J.Q.; Wan, D.S. Identification of 4CL Genes in Desert Poplars and Their Changes in Expression in Response to Salt Stress. Genes 2015, 6, 901–917. [Google Scholar] [CrossRef] [PubMed]
- De Azevedo Souza, C.; Barbazuk, B.; Ralph, S.G.; Bohlmann, J.; Hamberger, B.; Douglas, C.J. Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol. 2008, 179, 987–1003. [Google Scholar] [CrossRef]
- Costa, M.A.; Collins, R.E.; Anterola, A.M.; Cochrane, F.C.; Davin, L.B.; Lewis, N.G. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry 2003, 64, 1097–1112. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.M.; Fulda, M.S.; Browse, J. Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme a synthetases. Plant Physiol. 2003, 132, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef]
- Chowdhury, M.E.K.; Choi, B.; Cho, B.-K.; Kim, J.B.; Park, S.U.; Natarajan, S.; Lim, H.-S.; Bae, H. Regulation of 4CL, encoding 4-coumarate: Coenzyme A ligase, expression in kenaf under diverse stress conditions. Plant Omics 2013, 6, 254–262. [Google Scholar]
- Sun, H.; Li, Y.; Feng, S.; Zou, W.; Guo, K.; Fan, C.; Si, S.; Peng, L. Analysis of five rice 4-coumarate:coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 2013, 430, 1151–1156. [Google Scholar] [CrossRef]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C.; Xiong, X.P.; Zhang, X.L.; Feng, H.J.; Zhu, Q.H.; Sun, J.; Li, Y.J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Qing, J.; Wang, Q.; Liu, C.; Du, H.; Liu, P.; Du, Q.; Du, L.; Wang, L. Genome-Wide Identification and Expression Analyses of the 4-Coumarate: CoA Ligase (4CL) Gene Family in Eucommia ulmoides. Forests 2022, 13, 1253. [Google Scholar] [CrossRef]
- Nie, T.; Sun, X.; Wang, S.; Wang, D.; Ren, Y.; Chen, Q. Genome-Wide Identification and Expression Analysis of the 4-Coumarate: CoA Ligase Gene Family in Solanum tuberosum. Int. J. Mol. Sci. 2023, 24, 1642. [Google Scholar] [CrossRef]
- Fan, J.; Luo, Z.; Wang, Y.; Jiao, P.; Wang, Q.; Dai, Y.; Guan, S.; Ma, Y.; Yu, H.; Liu, S. Maize 4-coumarate coenzyme A ligase Zm4CL-like9 gene positively regulates drought stress response in Arabidopsis thaliana. GM Crops Food 2025, 16, 199–215. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, D.; Zhang, Y.; Liu, K.; Xu, K.; Zhang, F.; Wang, J.; Tan, G.; Nie, X.; Ji, Q.; et al. Vacuum and Co-cultivation Agroinfiltration of (Germinated) Seeds Results in Tobacco Rattle Virus (TRV) Mediated Whole-Plant Virus-Induced Gene Silencing (VIGS) in Wheat and Maize. Front. Plant Sci. 2017, 8, 393. [Google Scholar] [CrossRef] [PubMed]
- Sade, N.; Vinocur, B.J.; Diber, A.; Shatil, A.; Ronen, G.; Nissan, H.; Wallach, R.; Karchi, H.; Moshelion, M. Improving plant stress tolerance and yield production: Is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 2009, 181, 651–661. [Google Scholar] [CrossRef]
- Lee, D.; Ellard, M.; Wanner, L.A.; Davis, K.R.; Douglas, C.J. The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: Stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol. Biol. 1995, 28, 871–884. [Google Scholar] [CrossRef]
- Douglas, C.J. Phenylpropanoid metabolism and lignin biosynthesis: From weeds to trees. Trends Plant Sci. 1996, 1, 171–178. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic Diversity, Conservation, and Utilization of Plant Genetic Resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Schrell, S.; Peterson, P.A.; Saedler, H.; Wienand, U. Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm 1 and Zm 38. Plant J. 1994, 6, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Wang, D. AP2/ERF Transcription Factors for Tolerance to Both Biotic and Abiotic Stress Factors in Plants. Trop. Plant Biol. 2023, 16, 105–112. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Salika, R.; Riffat, J. Abiotic stress responses in maize: A review. Acta Physiol. Plant. 2021, 43, 130. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Li, X.; Ma, K.; Zhan, Y.; Zeng, F. Molecular cloning and functional analysis of 4-Coumarate:CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis. BMC Plant Biol. 2019, 19, 231. [Google Scholar] [CrossRef]





| Gene | Gene ID | Chromosome | No. of Amino Acids | Mol. Wt (Da) | Isoelectric Point (pI) | Instability Index (II) | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|
| Zm4CL1 | Zm00001eb002380 | Chr1 | 578 | 62,669.17 | 8.84 | 33.93 | 93.17 | −0.012 |
| Zm4CL2 | Zm00001eb002570 | Chr1 | 551 | 59,442.89 | 9.07 | 39.26 | 98.09 | 0.011 |
| Zm4CL3 | Zm00001eb002700 | Chr1 | 559 | 60,395.25 | 8.6 | 36.06 | 83.61 | −0.084 |
| Zm4CL4 | Zm00001eb002720 | Chr1 | 644 | 70,392.47 | 7.96 | 42.02 | 79.41 | −0.13 |
| Zm4CL5 | Zm00001eb004210 | Chr1 | 555 | 59,456.5 | 7 | 39.3 | 94.38 | 0.16 |
| Zm4CL6 | Zm00001eb014500 | Chr1 | 567 | 61,693.77 | 6.58 | 25.3 | 87.62 | −0.051 |
| Zm4CL7 | Zm00001eb025180 | Chr1 | 575 | 60,860.43 | 8.35 | 48.9 | 99.11 | 0.253 |
| Zm4CL8 | Zm00001eb040790 | Chr1 | 555 | 58,828.15 | 5.89 | 34.95 | 100.56 | 0.199 |
| Zm4CL9 | Zm00001eb048810 | Chr1 | 550 | 58,196.46 | 8.57 | 33.78 | 103.6 | 0.257 |
| Zm4CL10 | Zm00001eb066390 | Chr2 | 556 | 60,128.97 | 6.95 | 39.36 | 84.87 | −0.072 |
| Zm4CL11 | Zm00001eb083110 | Chr2 | 610 | 65,701.33 | 5.64 | 39.92 | 94.92 | 0.062 |
| Zm4CL12 | Zm00001eb084450 | Chr2 | 402 | 44,535.59 | 6.71 | 23.87 | 75.65 | −0.247 |
| Zm4CL13 | Zm00001eb119050 | Chr3 | 599 | 64,131.9 | 6.21 | 48.1 | 96.21 | 0.108 |
| Zm4CL14 | Zm00001eb152410 | Chr3 | 437 | 48,320.47 | 6.14 | 35.74 | 86.38 | −0.221 |
| Zm4CL15 | Zm00001eb178360 | Chr4 | 560 | 60,274.48 | 5.6 | 38.64 | 97.7 | 0.031 |
| Zm4CL16 | Zm00001eb187910 | Chr4 | 558 | 58,623.7 | 5.61 | 30.45 | 98.51 | 0.242 |
| Zm4CL17 | Zm00001eb230220 | Chr5 | 586 | 62,142.25 | 6.52 | 35.06 | 89.66 | 0.091 |
| Zm4CL18 | Zm00001eb233720 | Chr5 | 533 | 57,520.36 | 5.13 | 35.3 | 97.71 | 0.104 |
| Zm4CL19 | Zm00001eb242380 | Chr5 | 706 | 78,020.65 | 5.47 | 29.46 | 79.8 | −0.219 |
| Zm4CL20 | Zm00001eb261830 | Chr6 | 573 | 60,668.2 | 6.68 | 43.9 | 85.86 | −0.002 |
| Zm4CL21 | Zm00001eb280520 | Chr6 | 554 | 60,226.29 | 6.97 | 40.14 | 87.44 | −0.064 |
| Zm4CL22 | Zm00001eb298620 | Chr7 | 560 | 59,152.98 | 7.72 | 56.41 | 96.07 | 0.129 |
| Zm4CL23 | Zm00001eb305920 | Chr7 | 118 | 13,004.53 | 4.58 | 53.21 | 75.93 | −0.236 |
| Zm4CL24 | Zm00001eb308860 | Chr7 | 559 | 59,085.01 | 8.59 | 48.24 | 92.68 | 0.118 |
| Zm4CL25 | Zm00001eb311650 | Chr7 | 722 | 78,336.76 | 5.64 | 39.5 | 89.47 | −0.023 |
| Zm4CL26 | Zm00001eb365520 | Chr8 | 511 | 54,147.49 | 6.08 | 36.75 | 93.01 | 0.006 |
| Zm4CL27 | Zm00001eb365590 | Chr8 | 555 | 58,787.98 | 6.49 | 46.5 | 98.63 | 0.185 |
| Zm4CL28 | Zm00001eb378990 | Chr9 | 1168 | 128,452.95 | 6.11 | 36.21 | 89.38 | 0.002 |
| Zm4CL29 | Zm00001eb380130 | Chr9 | 600 | 65,108.57 | 7.26 | 40.21 | 84.63 | −0.109 |
| Zm4CL30 | Zm00001eb389420 | Chr9 | 558 | 59,417.37 | 5.29 | 34.11 | 98.08 | 0.131 |
| Zm4CL31 | Zm00001eb396330 | Chr9 | 582 | 63,238.8 | 8.28 | 28.05 | 86.32 | −0.053 |
| Zm4CL32 | Zm00001eb434030 | Chr10 | 527 | 55,132.85 | 6.25 | 32.86 | 96.24 | 0.118 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Wang, Y.; Li, R. Genome-Wide Analysis of Zm4CL Genes Identifies Zm4CL8 Regulating Drought and Salt Tolerance in Maize. Agronomy 2025, 15, 1100. https://doi.org/10.3390/agronomy15051100
Zhang Z, Wang Y, Li R. Genome-Wide Analysis of Zm4CL Genes Identifies Zm4CL8 Regulating Drought and Salt Tolerance in Maize. Agronomy. 2025; 15(5):1100. https://doi.org/10.3390/agronomy15051100
Chicago/Turabian StyleZhang, Ze, Yanbin Wang, and Rong Li. 2025. "Genome-Wide Analysis of Zm4CL Genes Identifies Zm4CL8 Regulating Drought and Salt Tolerance in Maize" Agronomy 15, no. 5: 1100. https://doi.org/10.3390/agronomy15051100
APA StyleZhang, Z., Wang, Y., & Li, R. (2025). Genome-Wide Analysis of Zm4CL Genes Identifies Zm4CL8 Regulating Drought and Salt Tolerance in Maize. Agronomy, 15(5), 1100. https://doi.org/10.3390/agronomy15051100





