Phenotypic Plasticity of Maize Flowering Time and Plant Height Using the Interactions Between QTNs and Meteorological Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Germplasm and Phenotype Evaluation
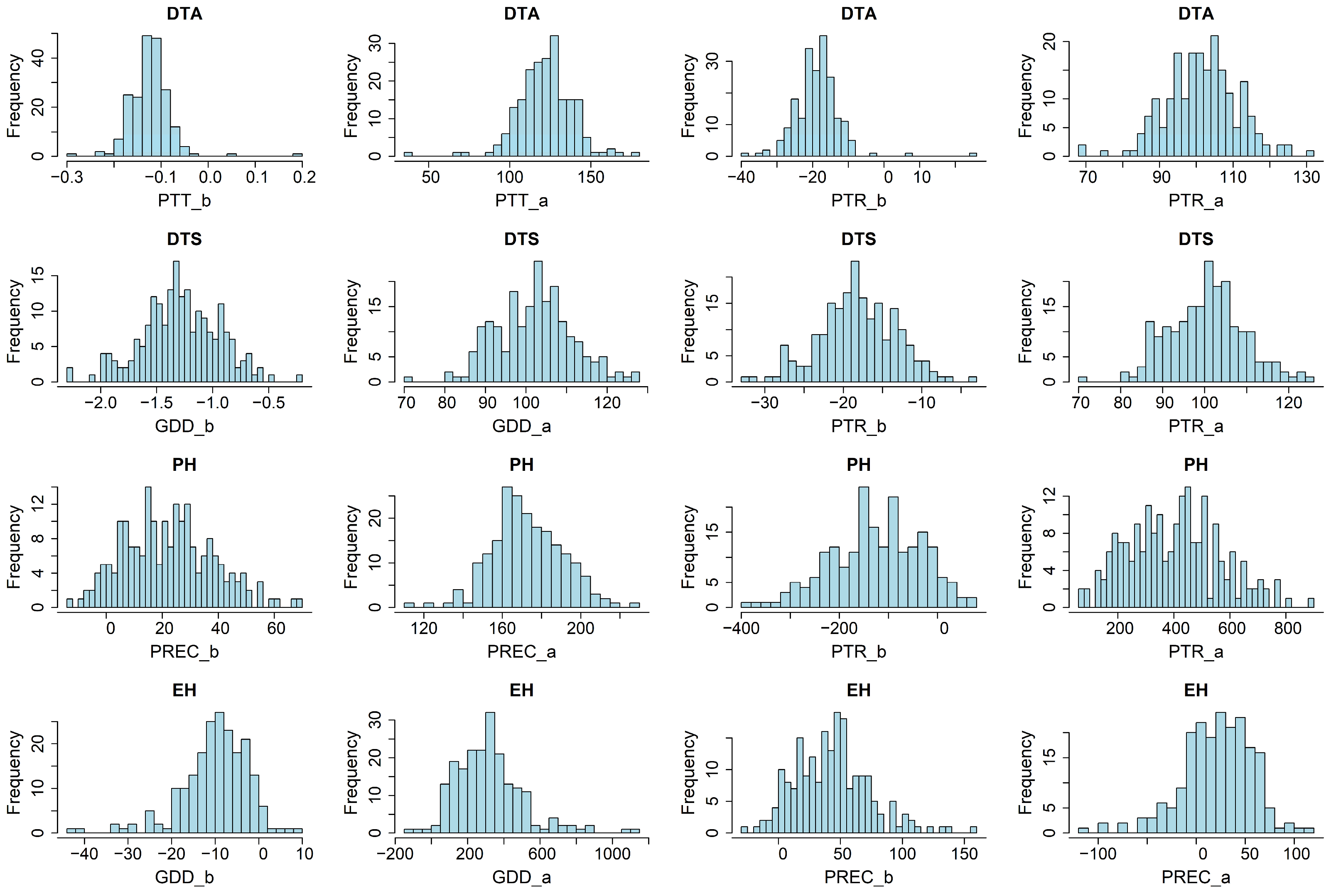
2.2. Phenotype Data Analysis
2.3. Genomic Data Processing
2.4. Meteorological Factors Processing
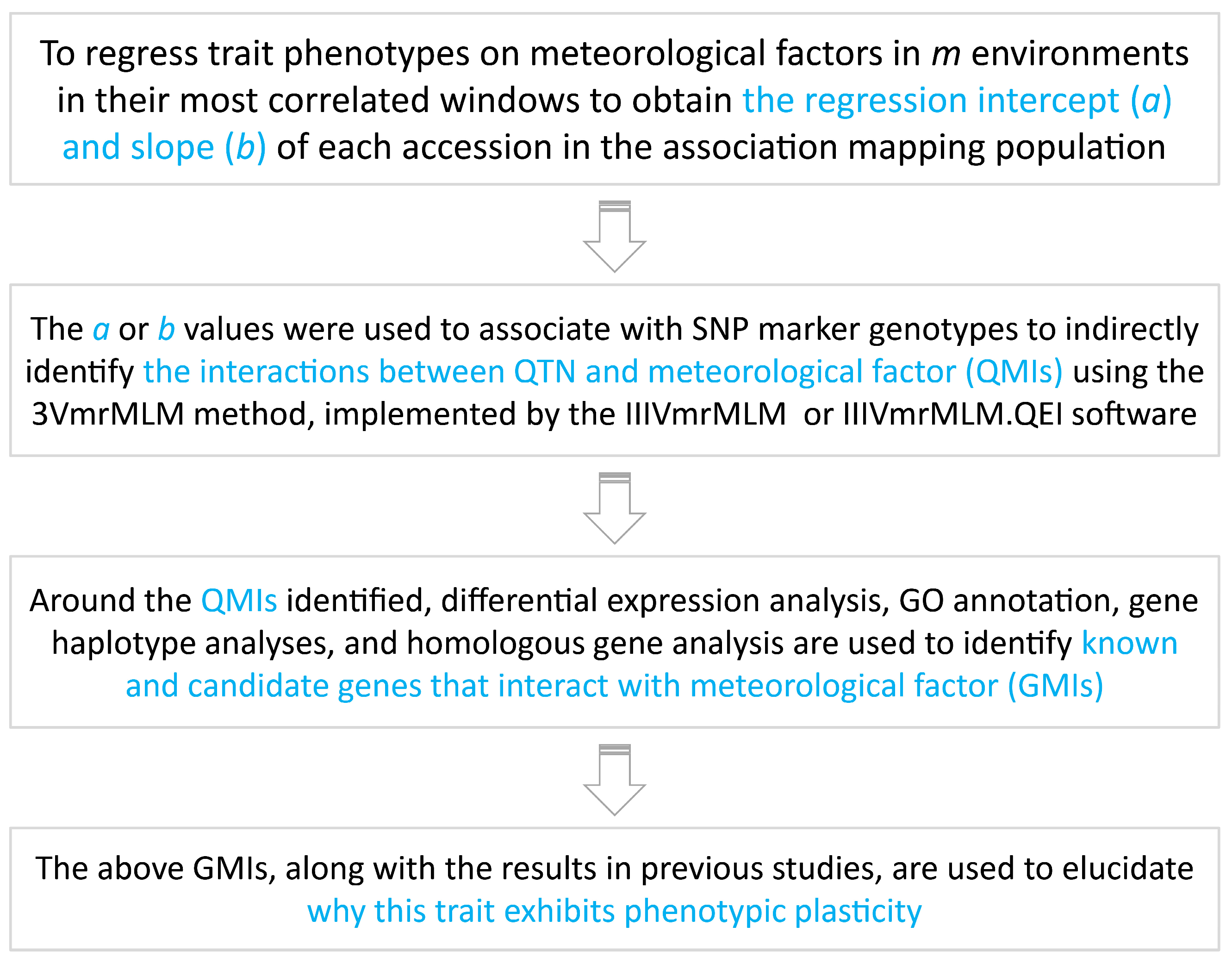
2.5. Critical Window Identification, Phenotypic Plasticity Analysis, and GWAS
2.6. Identifying Known and Candidate Maize Flowering and Height Genes
- Differential expression analysis: Transcriptome data were retrieved from the Gene Expression Omnibus database of NCBI for DTS and supplemented Dataset S4 in Stelpflug et al. [25] for PH, EH, and DTA. In detail, internode and root tissues were used for PH and EH, meiotic tassel and anther tissues for DTA (Dataset S4, Stelpflug et al. [25]), and ear primordium and silk tissues for DTS (GSE50191) [26]. Differentially expressed genes (DEGs) were identified using the R package limma [27], with significance determined by P-adjust < 0.05 and |log2FoldChange| > 1. To incorporate responses to abiotic stress, external datasets of stress-responsive DEGs were integrated, including 7272 DEGs for temperature [28], 11,426 DEGs for photoperiod [29], and 4611 DEGs for water stress [30].
- GO annotation: Candidate DEG protein sequences were annotated via eggNOG-mapper using default parameters (http://eggnog-mapper.embl.de/, accessed on 6 August 2024) [31]. GO annotation results were extracted from eggNOG-mapper using AgBase [32], reserving the biological processes related to phenology, morphology, and abiotic stress.
- Haplotype analysis: For each DEG, SNPs within coding regions and 2 kb promoter regions were used to construct gene haplotypes. Associations between intercept/slope and gene haplotypes were tested by one-way ANOVA in the R function aov(), with the p-value ≤ 0.05. Haplotype distributions were visualized using ggplot2 v3.5.0.
- Homologous gene analysis: Candidate gene protein sequences were blasted against Arabidopsis and Oryza sativa using the TAIR (https://www.arabidopsis.org, accessed on 27 August 2024) and RGAP (https://rice.uga.edu/, accessed on 27 August 2024) databases. A gene was identified as a potential candidate gene if its homolog was implicated in regulatory pathways associated with the target traits and also showed evidence of gene-by-environment interactions.
3. Results
3.1. Phenotypic Plasticity of Flowering Time and Plant Height Across Latitudinal Gradients
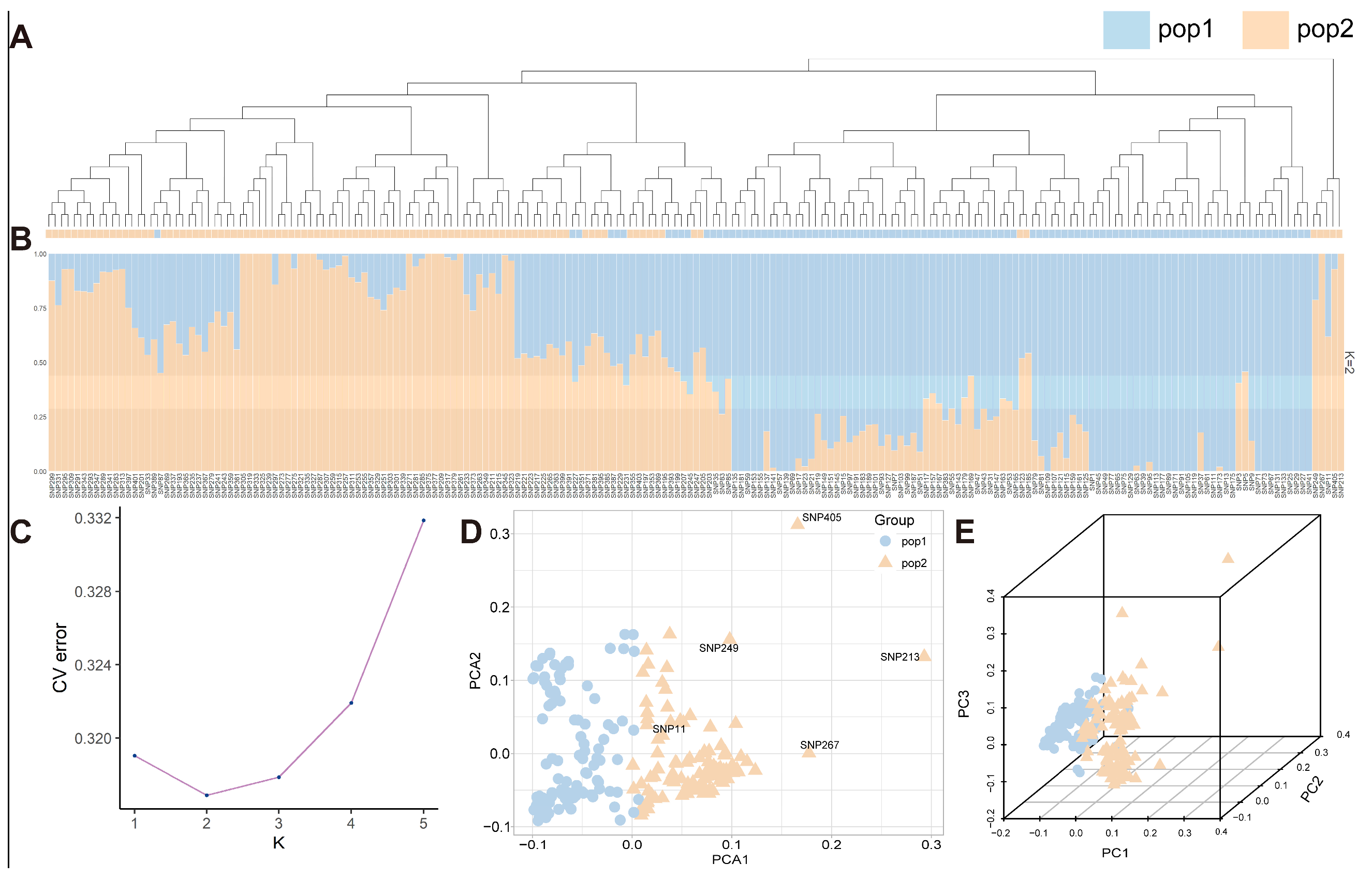
3.2. Determination of Population Structure
3.3. Determination of Meteorological Factors and Their Critical Windows
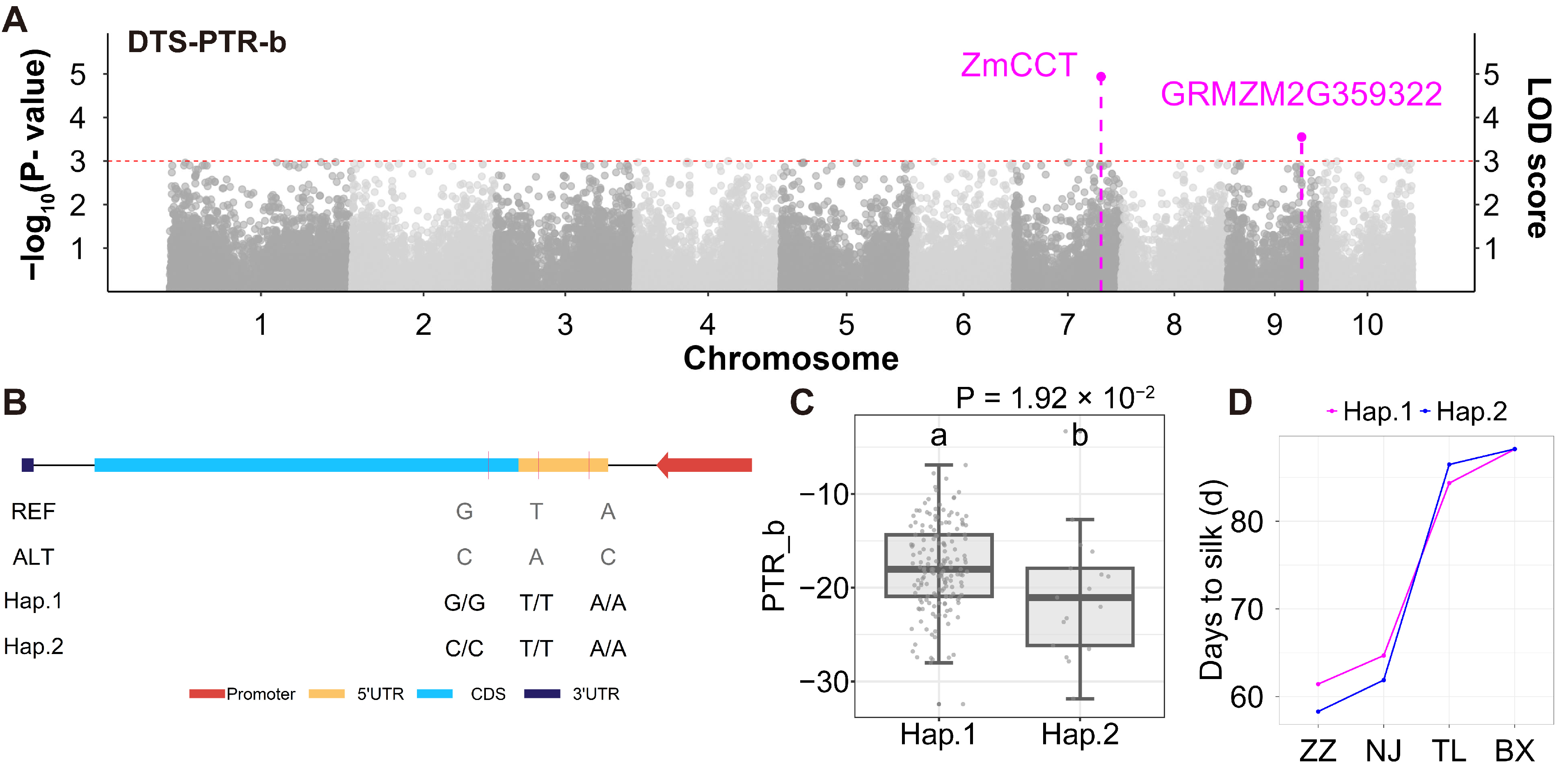
3.4. Indirect Identification of QMIs and Their Candidate Genes for Maize Flowering Time and Plant Height
4. Discussion
4.1. Stage-Dependent Climate Sensitivity
4.2. Genetic Mechanism of Phenotypic Plasticity in Maize Flowering Time
4.3. Genetic Mechanism of Phenotypic Plasticity in Maize Plant Height
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bradshaw, A.D. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar]
- Schlichting, C.D. The evolution of phenotypic plasticity in plants. Annu. Rev. Ecol. Syst. 1986, 17, 667–693. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.E. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 2000, 5, 537–542. [Google Scholar] [CrossRef]
- West-Eberhard, M.J. Developmental plasticity and the origin of species differences. Proc. Natl. Acad. Sci. USA 2005, 102 (Suppl. 1), 6543–6549. [Google Scholar] [CrossRef]
- Scheiner, S.M. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 1993, 24, 35–68. [Google Scholar] [CrossRef]
- Korte, A.; Vilhjálmsson, B.J.; Segura, V.; Platt, A.; Long, Q.; Nordborg, M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012, 44, 1066–1071. [Google Scholar] [CrossRef]
- Qi, Q.; Chu, A.Y.; Kang, J.H.; Jensen, M.K.; Curhan, G.C.; Pasquale, L.R.; Ridker, P.M.; Hunter, D.J.; Willett, W.C.; Rimm, E.B.; et al. Sugar-sweetened beverages and genetic risk of obesity. N. Engl. J. Med. 2012, 367, 1387–1396. [Google Scholar] [CrossRef]
- Huang, T.; Hu, F.B. Gene-environment interactions and obesity: Recent developments and future directions. BMC Med. Genom. 2015, 8 (Suppl. 1), S2. [Google Scholar] [CrossRef]
- Kerin, M.; Marchini, J. Inferring gene-by-environment interactions with a Bayesian whole-genome regression model. Am. J. Hum. Genet. 2020, 107, 698–713. [Google Scholar] [CrossRef]
- Li, X.; Guo, T.; Wang, J.; Bekele, W.A.; Sukumaran, S.; Vanous, A.E.; McNellie, J.P.; Tibbs-Cortes, L.E.; Lopes, M.S.; Lamkey, K.R.; et al. An integrated framework reinstating the environmental dimension for GWAS and genomic selection in crops. Mol. Plant 2021, 14, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Han, X.L.; Li, M.; Chen, Y.; Zhang, Y.M. IIIVmrMLM.QEI: An effective tool for indirect detection of QTN-by-environment interactions in genome-wide association studies. Comput. Struct. Biotechnol. J. 2024, 23, 4357–4368. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.H.; Summerfield, R.J.; Edmeades, G.O.; Roberts, E.H. Photoperiod, temperature, and the interval from sowing to tassel initiation in diverse cultivars of maize. Crop Sci. 1992, 32, 1225–1232. [Google Scholar] [CrossRef]
- Bonhomme, R.; Derieux, M.; Edmeades, G.O. Flowering of diverse maize cultivars in relation to temperature and photoperiod in multilocation field trials. Crop Sci. 1994, 34, 156–164. [Google Scholar] [CrossRef]
- Renfro, B.L.; Ullstrup, A.J. A comparison of maize diseases in temperate and in tropical environments. Proc. Natl. Acad. Sci. USA 1976, 22, 491–498. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, R.; Hou, P.; Li, S.; Zhang, H.; Ming, B.; Long, H.; Liang, S. Phenological responses of maize to changes in environment when grown at different latitudes in China. Field Crops Res. 2013, 144, 192–199. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.W.; Zhang, Z.C.; Xiang, Y.; Liu, M.H.; Zhou, Y.H.; Zuo, J.F.; Zhang, H.Q.; Chen, Y.; Zhang, Y.M. A compressed variance component mixed model for detecting QTNs and QTN-by-environment and QTN-by-QTN interactions in genome-wide association studies. Mol. Plant 2022, 15, 630–650. [Google Scholar] [CrossRef]
- Shu, G.; Wang, A.; Wang, X.; Chen, R.; Gao, F.; Wang, A.; Li, T.; Wang, Y. Identification of QTNs, QTN-by-environment interactions for plant height and ear height in maize multi-environment GWAS. Front. Plant Sci. 2023, 14, 1284403. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Shringarpure, S.S.; Novembre, J.; Lange, K. Admixture 1.3 Software Manual; UCLA Human Genetics Software Distribution: Los Angeles, CA, USA, 2015. [Google Scholar]
- Li, M.; Zhang, Y.W.; Xiang, Y.; Liu, M.H.; Zhang, Y.M. IIIVmrMLM: The R and C++ tools associated with 3VmrMLM, a comprehensive GWAS method for dissecting quantitative traits. Mol. Plant 2022, 15, 1251–1253. [Google Scholar] [CrossRef] [PubMed]
- Masle, J.; Doussinault, G.; Farquhar, G.D.; Sun, B. Foliar stage in wheat correlates better to photothermal time than to thermal time. Plant Cell Environ. 1989, 12, 235–247. [Google Scholar] [CrossRef]
- Zhang, B.; Hautier, Y.; Tan, X.; You, C.; Cadotte, M.W.; Chu, C.; Jiang, L.; Sui, X.; Ren, T.; Han, X.; et al. Species responses to changing precipitation depend on trait plasticity rather than trait means and intraspecific variation. Funct. Ecol. 2020, 34, 2622–2633. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R.; et al. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Li, Y.; Zhang, Y.; Gou, Z.; Qi, X.; Zhang, J. Transcriptomic analysis revealed the common and divergent responses of maize seedling leaves to cold and heat stresses. Genes 2020, 11, 881. [Google Scholar] [CrossRef]
- Fei, J.; Jiang, Q.; Guo, M.; Lu, J.; Wang, P.; Liu, S.; Qu, J.; Ma, Y.; Guan, S. Fine mapping and functional research of key genes for photoperiod sensitivity in maize. Front. Plant Sci. 2022, 13, 890780. [Google Scholar] [CrossRef]
- Kang, J.; Sen, S.; Oliver, M.J.; Sharp, R.E. Comparative transcriptomics reveal metabolic rather than genetic control of divergent antioxidant metabolism in the primary root elongation zone of water-stressed cotton and maize. Antioxidants 2023, 12, 287. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- McCarthy, F.M.; Wang, N.; Magee, G.B.; Nanduri, B.; Lawrence, M.L.; Camon, E.B.; Barrell, D.G.; Hill, D.P.; Dolan, M.E.; Williams, W.P.; et al. AgBase: A functional genomics resource for agriculture. BMC Genom. 2006, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Liang, J.; Abou-Elwafa, S.F.; Cheng, H.; Dou, D.; Ren, Z.; Xie, J.; Chen, Z.; Gao, F.; Ku, L.; et al. ZmCCT regulates photoperiod-dependent flowering and response to stresses in maize. BMC Plant Biol. 2021, 21, 453. [Google Scholar] [CrossRef]
- Li, X.; Guo, X.; Zhao, L.; Zhang, J.; Tang, D.; Zhao, X.; Liu, X. Arabidopsis rad23-4 gene is required for pollen development under UV-B light. Afr. J. Biotechnol. 2012, 11, 10161–10169. [Google Scholar]
- Liu, X.J.; Sun, J.; Huang, Y.; Li, C.; Zheng, P.; Yuan, Y.; Chen, H.; Jan, M.; Zheng, H.; Du, H.; et al. Osj10gBTF3-Mediated import of chloroplast protein is essential for pollen development in rice. Front. Plant Sci. 2021, 12, 713544. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, X.; Zhao, Y.; Li, Y.; Zhang, G.; Peng, Z.; Zhang, J. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 2018, 16, 86–99. [Google Scholar] [CrossRef]
- Lee, S.; Choi, S.C.; An, G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2008, 54, 93–105. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, T.; Pei, F.J.; Chen, Y.; Chang, X.Y.; Mi, J.M.; Zhang, Y.M. Phenotypic plasticity of grain size-related traits in main-crop and ratoon rice. Plant Cell Environ. 2025. [Google Scholar] [CrossRef]
- Ren, H.; Liu, M.; Zhang, J.; Liu, P.; Liu, C. Effects of agronomic traits and climatic factors on yield and yield stability of summer maize (Zea mays L.) in the Huang-Huai-Hai Plain in China. Front. Plant Sci. 2022, 13, 1050064. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of abscisic acid-mediated drought stress responses in plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef]
- Li, B.B.; Chen, X.M.; Deng, T.; Zhao, X.; Li, F.; Zhang, B.C.; Wang, X.; Shen, S.; Zhou, S.L. Timing effect of high temperature exposure on the plasticity of internode and plant architecture in maize. J. Integr. Agric. 2024, 23, 551–565. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, Y.; Wen, X.; Yang, Q.; Liu, L.; Hao, S.; Li, J.; Wu, Z.; Shah, L.; Sohail, A.; et al. The clock component OsLUX regulates rice heading through recruiting OsELF3-1 and OsELF4s to repress Hd1 and Ghd7. J. Adv. Res. 2023, 48, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Tie, S.; Sun, T.; Zhang, L.; Yang, Y.; Qi, J.; Yan, S.; Han, X.; Wang, H.; Shen, C. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 2015, 10, e0118751. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Z.; Li, X.; Chen, Z.; Liu, Y.; Zhang, F.; Li, N. Identification and analysis of the expression of the PIP5K gene family in tomatoes. Int. J. Mol. Sci. 2023, 25, 159. [Google Scholar] [CrossRef]
- Mikami, K.; Katagiri, T.; Iuchi, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J. 1998, 15, 563–568. [Google Scholar] [CrossRef]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Islam, M.A.; Lütken, H.; Haugslien, S.; Blystad, D.R.; Torre, S.; Rolcik, J.; Rasmussen, S.K.; Olsen, J.E.; Clarke, J.L. Overexpression of the AtSHI gene in poinsettia, Euphorbia pulcherrima, results in compact plants. PLoS ONE 2013, 8, e53377. [Google Scholar] [CrossRef][Green Version]
- Deng, W.; Li, R.; Xu, Y.; Mao, R.; Chen, S.; Chen, L.; Chen, L.; Liu, Y.G.; Chen, Y. A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod-and thermo-sensitive dwarfism in rice. J. Exp. Bot. 2020, 71, 1294–1305. [Google Scholar] [CrossRef]
- Han, X.; Wu, X.; Zhang, Y.; Tang, Q.; Zeng, L.; Liu, Y.; Xiang, Y.; Hou, K.; Fang, S.; Lei, W.; et al. Genetic and transcriptome analyses of the effect of genotype-by-environment interactions on Brassica napus seed oil content. Plant Cell 2025, 37, koaf062. [Google Scholar] [CrossRef]



| Trait | Genome-Wide Association Studies | Known Gene | Symbol | Distance to Gene (kb) | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chr. | Posi (bp) | LOD | r2 (%) | p-Value | |||||
| DTS_PTR_b | 7 | 143,109,313 | 4.94 | 1.48 | 1.15 × 10−5 | GRMZM2G179024 | ZmCCT | 4.28 | Su et al. [33] |
| PH_PTR_b | 5 | 206,273,479 | 46.06 | 4.21 | 8.73 × 10−47 | GRMZM2G074267 | ZmPIN1b | 453.67 | Li et al. [34] |
| Trait | GWAS | Gene | Differential Expression Analysis | GO Annotation | Haplotype Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | Position | LOD | r2 (%) | Env | Reference | log2FC | P-adj. | Tissue | ID | Term | |||
| DTA-PTR-b | 9 | 106,405,880 | 13.72 | 4.23 | GRMZM2G035417 | Photo. | Fei et al. [29] | −1.01 | 3.49 × 10−3 | Tassel vs. Anthers | GO:0009266 GO:0032182 | Responding to temperature stimulus; ubiquitin-like protein binding | 2.77 × 10−2 |
| DTA-PTT-a | 7 | 131,784,004 | 5.76 | 2.13 | GRMZM2G069651 | Photo.; Temp. | Fei et al. [29]; Li et al. [28] | −4.54 | 1.75 × 10−5 | Tassel vs. Anthers | GO:0009408 GO:0009507 | Responding to heat; chloroplast | 4.83 × 10−3 |
| DTS-PTR-b | 9 | 122,920,747 | 3.55 | 1.04 | GRMZM2G359322 | Photo. | Fei et al. [29] | 1.77 | 1.63 × 10−3 | Silk vs. Ear primordium | 1.92 × 10−2 | ||
| PH-PREC-a | 1 | 265,522,189 | 6.17 | 1.60 | GRMZM2G062045 | Water | Kang et al. [30] | 1.00 | 2.74 × 10−3 | Internodes vs. Roots | GO:0046488 | PI signaling pathway | 1.79 × 10−4 |
| PH-PTR-b | 4 | 178,674,282 | 12.55 | 4.91 | GRMZM2G370777 | Photo. | Fei et al. [29] | 1.53 | 4.67 × 10−4 | Internodes vs. Roots | GO:0009266; GO:0048367 | Responding to temperature stimulus; shoot system development | 2.35 × 10−2 |
| EH-GDD-a | 8 | 158,116,081 | 6.03 | 3.26 | GRMZM2G035417 | Temp. | Li et al. [28] | 2.03 | 1.70 × 10−3 | First internode vs. Fourth internode | GO:0009725 | Hormonal regulation | 2.43 × 10−2 |
| EH-GDD-b | 8 | 158,116,081 | 8.91 | 5.56 | 1.80 × 10−2 | ||||||||
| EH-GDD-a | 3 | 184,214,740 | 37.36 | 4.60 | GRMZM2G126397 | Temp. | Li et al. [28] | 5.54 | 4.65 × 10−5 | Internodes vs. Roots | GO:0022622; GO:0048367; GO:0009733 | Root development; stem development; responds to auxin | 1.89 × 10−2 |
| EH-GDD-b | 3 | 184,214,740 | 29.75 | 3.63 | 1.80 × 10−2 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Luo, Y.; Shu, G.; Wang, A.; Wang, Y.; Zhang, Y. Phenotypic Plasticity of Maize Flowering Time and Plant Height Using the Interactions Between QTNs and Meteorological Factors. Agronomy 2025, 15, 1078. https://doi.org/10.3390/agronomy15051078
Han X, Luo Y, Shu G, Wang A, Wang Y, Zhang Y. Phenotypic Plasticity of Maize Flowering Time and Plant Height Using the Interactions Between QTNs and Meteorological Factors. Agronomy. 2025; 15(5):1078. https://doi.org/10.3390/agronomy15051078
Chicago/Turabian StyleHan, Xuelian, Yan Luo, Guoping Shu, Aifang Wang, Yibo Wang, and Yuanming Zhang. 2025. "Phenotypic Plasticity of Maize Flowering Time and Plant Height Using the Interactions Between QTNs and Meteorological Factors" Agronomy 15, no. 5: 1078. https://doi.org/10.3390/agronomy15051078
APA StyleHan, X., Luo, Y., Shu, G., Wang, A., Wang, Y., & Zhang, Y. (2025). Phenotypic Plasticity of Maize Flowering Time and Plant Height Using the Interactions Between QTNs and Meteorological Factors. Agronomy, 15(5), 1078. https://doi.org/10.3390/agronomy15051078







